Abstract
Background:
The human ADIPOQ gene encodes adiponectin protein hormone, which is involved in regulating glucose levels as well as fatty acid breakdown. It is exclusively produced by adipose tissue and abundantly present in the circulation, with concentration of around 0.01% of total serum proteins, with important effect on metabolism.
Methods:
Most deleterious nonsynonymous single nucleotide polymorphisms in the coding region of the ADIPOQ gene were investigated using SNP databases, and detected nonsynonymous variants were analyzed in silico from the standpoint of relevant protein function and stability by using SIFT, PolyPhen-2, PROVEAN and MUpro, I-Mutant2.0 tools, respectively.
Result:
A total of 58 nonsynonymous SNPs consisting of 55 missense variations, 3 nonsense variations were found in the ADIPOQ gene. Next, 14 of the 55 missense variants were predicted to be damaging or deleterious by three different software programs (PolyPhen-2, SIFT, and PROVEAN), and 38 of them were predicted to be less stable (I-Mutant 2.0 and MUpro software). Totally, 10 variants out of 55 missense variants were predicted to be both deleterious and reduce protein stability. Additionally, 3 nonsense variants were predicted to produce a truncated ADIPOQ protein. RMSD and total energy were calculated for 4 nsSNPs out of 10 nsSNPs which were both deleterious and showed a decrease in protein stability.
Conclusion:
rs144526209 has high root-mean-square deviation (RMSD) and lower total energy value compared to the native modeled structure. It was concluded that this nsSNP, potentially functional and polymorphic in the ADIPOQ gene, might be associated with diabetes, obesity, and inflammation.
Keywords: Adiponectin, Diabetes mellitus, Gestational, Nucleotide polymorphism
Introduction
The human ADIPOQ gene is located on chromo-some 3q27.3 and encodes a 244 aminoacid protein hormone with four distinct regions and the first one is a short signal sequence which targets the hormone for secretion outside the cell; next one is a short region that varies between species; the third is a 65-amino acid region with similarity to collagenous proteins; the last is a globular domain, to form these distinct regions and a number of post-translational modifications are required. It is exclusively produced by adipocytes and also froms the placenta in pregnancy and circulates high concentrations in healthy adults and is generally higher in females than males. This sexual differentiation has been attributed to the effect of testosterone on adiponectin secretion.
It is the most abundant circulating hormone secreted by the adipocytes, with putative insulin sensitizing, anti-inflammatory, and antiatherosclerotic properties. In a normal pregnancy, the maternal adiponectin circulating concentration increases in the first half of the pregnancy and then decreases proportionally to weight gain and physiological insulin resistance worsening. Newborn's adiponectin concentrations are higher than maternal circulating levels during pregnancy. Overall, it suggests that adiponectin, in addition to potentially linking excess adiposity to the risk of insulin resistance and type 2 diabetes, has a potential role in pregnancy and fetal growth 1 . Meller et al studied on leptin receptor (LEPR A-D) and adiponectin receptor (ADIPOR 1 & 2) and observed an association between GDM diagnosis and leptin mRNA expression in placental tissues 2 . In screening for GDM by maternal characteristics, the detection rate was 61.6% at a false-positive rate of 20% and the detection increased to 74.1% by the addition of adiponectin and sex hormone binding globulin 3 . A multi-SNP genotype risk score that accounted for 5% of the variance of adiponectin levels exhibited significant association with T2D and markers of insulin resistance, suggesting a shared allelic architecture of adiponectin and other metabolic traits 4 .
As genomic variations among people, Single Nucleotide Polymorphisms (SNPs) exist throughout the genome and can be divided into several groups. Among the different kinds of SNPs, a nonsynonymous SNP in the coding region of a gene is important because it alters the amino acid composition; consequently, such alterations can have an impact on protein structure, function, and subcellular localization. Although pinpointing the effects of the many nonsynonymous SNPs using biochemical analyses is challenging, computational analysis tools predicting their effect on protein activity and stability have been recently developed, such as Polymorphism phenotyping v2 (PolyPhen-2) 5 , Sorting Intolerant From Tolerant (SIFT) 6 , Protein Variation Effect Analyzer (PROVEAN) 7 , I-Mutant 2.0 8 , and MUpro 9 software. The gene was investigated for variants that predispose to type-2 diabetes and insulin sensitivity which leads to Gestational Diabetes Mellitus. Several single nucleotide polymorphisms mutations in the ADIPOQ gene, G84R and G90S mutants, associated with diabetes and hypoadiponectinemia (Vasseur et al, 2002), did not form HMW multimers. R112C and I164T mutants, associated with hypoadiponectinemia, did not assemble into low-molecular-weight trimers, resulting in impaired secretion from the cell 10 associated with type-2 diabetes and obesity. Thus, in the present study, an attempt was made to search for nonsynonymous SNPs in the ADIPOQ gene using genome databases and investigate the impacts of nonsynonymous SNPs on adiponectin protein function and stability using computational tools.
Materials and Methods
Retrieval of nonsynonymous SNPs
Data on nonsynonymous variations of the ADIPOQ gene were collected from the database of SNPs (db SNP) located on the homepage of the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/SNP/) and from the Ensembl genome browser (http://www.ensembl.org/index.html). The reference Transcript ID and the reference protein ID of ADIPOQ are NM_004797 and NP_004788, respectively.
SIFT prediction
The Sorting Intolerant from Tolerant (SIFT) algorithm predicts the effect of coding variants on protein function based on the degree of conservation of amino-acid residues in sequence alignments derived from closely related sequences 6 . It was first introduced in 2001, with a corresponding website that provides users with predictions on their variants. Since its release, SIFT has become one of the standard tools for characterizing missense variation. SIFT is based on the premise that protein evolution is correlated with protein function. Variants that occur at conserved alignment positions are expected to be tolerated less than those that occur at diverse positions. The algorithm uses a modified version of PSIBLAST 11 and Dirichlet mixture regularization 12 to construct a multiple sequence alignment of proteins that can be globally aligned to the query sequence and belong to the same clade. The underlying principle of this program is that it generates alignments with a large number of homologous sequences and assigns scores to each residue, ranging from zero to one. SIFT scores 13 are categorized as potentially intolerant (0.051–0.10), intolerant (0.00–0.05), tolerant (0.201–1.00) or borderline (0.101–0.20). The higher the tolerance index of a particular amino acid substitution, the lesser is its likely impact (Table 1).
Table 1.
PolyPhen-2, SIFT, and PROVEAN results for the 55 missense variants of the ADIPOQ gene
| Nucleotide | Protein | dbSNP ID | SIFT prediction (score) | PolyPhen-2 prediction (score) | PROVEAN prediction (Score) |
|---|---|---|---|---|---|
| c.13G>A | p.Gly5Arg | rs201248773 | Tolerated (0.32) | Benign (0.001) | Neutral (−.106) |
| c.26T>A | p.Leu9Gln | rs114155159 | Damaging (0.04) | Probably damaging (0.995) | Neutral (−1.052) |
| c.65C>T | p.Thr22Ile | rs201223375 | Tolerated (0.31) | Benign (0.138) | Neutral (−0.306) |
| c.76G>A | p.Gly26Arg | rs200006814 | Tolerated (0.52) | Possibly damaging (0.616) | Neutral (0.369) |
| c.101G>T | p.Gly34Val | rs201392172 | Tolerated (0.07) | Benign (0.259) | Neutral (−2.168) |
| c.113G>A | p.Gly38Asp | rs144448520 | Tolerated (0.61) | Benign (0.434) | Neutral (0.475) |
| c.122C>T | p.Ala41Val | rs200936740 | Tolerated (0.42) | Probably damaging (0.975) | Neutral (−1.501) |
| c.133G>C | p.Gly45Arg | rs200573126 | Damaging (0.00) | Probably damaging (1.00) | Deleterious (−7.247) |
| c.140C>T | p.Pro47Leu | rs372597136 | Damaging (0.03) | Probably damaging (1.00) | Deleterious (−5.655) |
| c.143G>A | p.Gly48Asp | rs182223755 | Damaging (0.00) | Probably damaging (1.00) | Deleterious (−6.347) |
| c.161G>T | p.Gly54Val | rs13061862 | Damaging (0.00) | Probably damaging (1.00) | Deleterious (−8.068) |
| c.163C>T | p.Arg55Cys | rs138227502 | Damaging (0.05) | Probably damaging (1.00) | Deleterious (−4.459) |
| c.164G>A | p.Arg55His | rs143606172 | Tolerated (0.13) | Probably damaging (1.00) | Deleterious (−2.989) |
| c.191A>G | p.Glu64Gly | rs147185738 | Tolerated (0.33) | Possibly damaging (0.470) | Deleterious (−4.166) |
| c.221T>C | p.Ile74Thr | rs138835949 | Tolerated (0.57) | Benign (0.000) | Neutral (−0.884) |
| c.223G>T | p.Gly75Cys | rs199670988 | Damaging (0.00) | Probably damaging (0.998) | Deleterious (−7.542) |
| c.245A>G | p.Glu82Gly | rs200935936 | Tolerated (0.27) | Benign (0.005) | Deleterious (−3.613) |
| c.250G>A | p.Gly84Arg | rs199646033 | Damaging (0.00) | Probably damaging (1.00) | Deleterious (−7.287) |
| c.253G>T | p.Val85Leu | rs376862518 | Tolerated (0.67) | Benign (0.027) | Neutral (−0.469) |
| c.256C>G | p.Pro86Ala | rs371274243 | Tolerated (0.41) | Benign (0.003) | Neutral (−0.544) |
| c.268G>A | p.Gly90Ser | rs62625753 | Damaging (0.00) | Probably damaging (1.00) | Deleterious (−5.678) |
| c.271C>T | p.Pro91Ser | rs200130041 | Tolerated (0.27) | Probably damaging (0.977) | Deleterious (−4.046) |
| c.272C>G | p.Pro91Arg | rs200470297 | Tolerated (0.17) | Probably damaging (0.997) | Deleterious (−4.911) |
| c.290T>C | p.Ile97Thr | rs370574236 | Tolerated (0.61) | Benign (0.000) | Neutral (0.888) |
| c.323C>T | p.Ala108Val | rs72563731 | Tolerated (0.26) | Possibly damaging (0.670) | Neutral (−2.073) |
| c.326A>G | p.Tyr109Cys | rs201989364 | Tolerated (0.15) | Probably damaging (0.989) | Neutral (−2.314) |
| c.331T>C | p.Tyr111His | rs17366743 | Tolerated (0.54) | Benign (0.006) | Neutral (−1.502) |
| c.334C>T | p.Arg112Cys | Rs121917815 | Damaging (0.00) | Probably damaging (1.000) | Deleterious (−6.382) |
| c.335G>T | p.Arg112Pro | rs79645624 | Damaging (0.01) | Probably damaging (1.000) | Deleterious (−4.631) |
| c.335G>C | p.Arg112Leu | rs79645624 | Damaging (0.02) | Probably damaging (0.997) | Deleterious (−5.354) |
| c.353G>A | p.Gly118Glu | rs202043211 | Damaging (0.02) | Probably damaging (1.000) | Deleterious (−6.544) |
| c.355T>C | p.Leu119Met | rs146386537 | Tolerated (0.07) | Probably damaging (1.000) | Neutral (−1.919) |
| c.359A>C | p.Glu120Ala | rs200433818 | Tolerated (0.26) | Benign (0.001) | Neutral (0.429) |
| c.367G>A | p.Val123Ile | rs367717792 | Tolerated (0.37) | Benign (0.145) | Neutral (−0.188) |
| c.371C>A | p.Thr124Asn | rs199656636 | Tolerated (0.06) | Benign (0.008) | Neutral (−0.905) |
| c.374T>A | p.Ile125Asn | rs370120250 | Tolerated (0.45) | Benign (0.332) | Neutral (−1.831) |
| c.392G>A | p.Arg131His | rs78685763 | Tolerated (0.21) | Probably damaging (1.000) | Deleterious (−3.945) |
| c.391C>T | p.Arg131Cys | rs202200116 | Tolerated (0.06) | Probably damaging (1.000) | Deleterious (−6.301) |
| c.425A>T | p.His142Leu | rs199547839 | Damaging (0.00) | Probably damaging (1.000) | Deleterious (−8.400) |
| c.436T>A | p.Ser146Thr | rs375589933 | Tolerated (0.7) | Benign (0.001) | Neutral (0.893) |
| c.463C>T | p.Pro155Ser | rs200546423 | Tolerated (0.07) | Probably damaging (1.000) | Deleterious (−5.144) |
| c.482C>T | p.Ala161Val | rs113716447 | Tolerated (0.14) | Benign (0.440) | Neutral (−2.088) |
| c.491T>C | p.Ile164Thr | rs185847354 | Damaging (0.01) | Possibly damaging (0.942) | Deleterious (−3.852) |
| c.541G>T | p.Ala181Ser | rs372548575 | Tolerated (0.73) | Benign (0.371) | Neutral (−0.296) |
| c.568C>G | p.Gln190Glu | rs200035452 | Tolerated (1.00) | Benign (0.030) | Neutral (−1.637) |
| c.593C>T | p.Ser198Phe | rs375480082 | Damaging (0.00) | Probably damaging (1.000) | Deleterious (−5.888) |
| c.595G>A | p.Gly199Ser | rs144526209 | Damaging (0.02) | Probably damaging (1.000) | Deleterious (−3.664) |
| c.623G>A | p.Gly208Asp | rs200107352 | Tolerated (0.08) | Possibly damaging (0.815) | Deleterious (−6.460) |
| c.626A>G | p.Asp209Gly | rs199733477 | Damaging (0.00) | Probably damaging (1.000) | Deleterious (−6.730) |
| c.658G>C | p.Glu220Gln | rs183590709 | Tolerated (0.27) | Possibly damaging (0.728) | Neutral (−0.796) |
| c.661C>A | p.Arg221Cys | rs138773406 | Tolerated (0.18) | Possibly damaging (0.855) | Neutral (0.443) |
| c.661C>T | p.Arg221Ser | rs138773406 | Tolerated (0.40) | Benign (0.002) | Neutral (0.150) |
| c.665A>G | p.Asn222Ser | rs374868336 | Tolerated (0.29) | Benign (0.066) | Deleterious (−3.868) |
| c.722A>C | p.His241Pro | rs141205818 | Tolerated (1.00) | Benign (0.001) | Neutral (3.024) |
| 726C>G | p.Asp242Glu | rs200424832 | Tolerated (0.25) | Possibly damaging (0.909) | Neutral (−1.650) |
Reference transcript ID, NM_004797.
Reference protein ID, NP_001171271.
PROVEAN prediction
PROVEAN (Protein Variation Effect Analyzer) predicts the functional impact for all classes of protein sequence variations not only single aminoacid substitutions but also insertions, deletions, and multiple substitutions on the alignment-based score 7 . The score measures the change in sequence similarity of a query sequence to a protein sequence homolog between without and with an amino acid variation of the query sequence. If the PROVEAN score ≤−2.5, the protein variant is predicted to have a “deleterious” effect, while if the PROVEAN score is >−2.5, the variant is predicted to have a “neutral” effect (Table 1). Both types of softwares are available on the homepage of the J. Craig Venter Institute; the SIFT tool is at http://sift.jcvi.org, and the PROVEAN tool is at http://provean.jcvi.org.
PolyPhen-2 prediction
PolyPhen 14 is a computational tool for identification of potentially functional nsSNPs. Predictions are based on a combination of phylogenetic, structural, and sequence annotation information characterizing a substitution and its position in the protein. For a given aminoacid variation, PolyPhen performs several steps: (a) extraction of sequence-based features of the substitution site from the UniProt database, (b) calculation of profile scores for two aminoacid variants, and (c) calculation of structural parameters and contacts of a substituted residue. PolyPhen scores were classified as “benign”, “possibly damaging” or “probably damaging” 13 (Table 1). Input options for the PolyPhen server are protein sequence or accession number together with sequence position with two aminoacid variants.
Mutant2.0
I-Mutant2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html) is a support vector machine-based tool for the prediction of protein stability changes upon nonsynonymous variations 8 . The tool evaluates the stability change upon nonsynonymous SNP starting from the protein structure or from the protein sequence. The DDG value (difference in free energy of mutation) is calculated from the unfolding Gibbs free energy value of the variant protein minus the unfolding Gibbs free energy value of the wild type (kcal/mol), and scores <0 are predicted by the algorithm to indicate decreased stability, whereas scores >0 are considered to indicate increased stability (Table 2).
Table 2.
I-Mutant and MUpro results for the 55 missense variants of the ADIPOQ gene
| Protein | I-Mutant 2.0 prediction (DDG) | Mupro prediction (Score) |
|---|---|---|
| p.Gly5Arg | Decrease (−0.48) | Decrease (−0.12568759) |
| p.Leu9Gln | Decrease (−0.81) | Decrease (−0.69667764) |
| p.Thr22Ile | Increase (0.08) | Decrease (−0.71481575) |
| p.Gly26Arg | Decrease (−1.75) | Increase (0.90615093) |
| p.Gly34Val | Decrease (−0.58) | Decrease(−0.1274136) |
| p.Gly38Asp | Decrease (−1.06) | Increase (0.48011262) |
| p.Ala41Val | Decrease (−0.29) | Increase (0.3896755) |
| p.Gly45Arg | Decrease (−0.24) | Decrease (−0.38721241) |
| p.Pro47Leu | Decrease (−0.55) | Increase (1) |
| p.Gly48Asp | Decrease (−0.34) | Decrease (−0.025777872) |
| p.Gly54Val | Decrease (−0.81) | Increase (0.38553968) |
| p.Arg55Cys | Decrease (−0.29) | Decrease (−1) |
| p.Arg55His | Increase (0.2) | Decrease (−1) |
| p.Glu64Gly | Decrease (−0.54) | Decrease (−1) |
| p.Ile74Thr | Decrease (−0.01) | Decrease (−1) |
| p.Gly75Cys | Decrease (−1.81) | Decrease (−1) |
| p.Glu82Gly | Decrease (−0.49) | Decrease (−1) |
| p.Gly84Arg | Decrease (−1.38) | Decrease (−0.32531487) |
| p.Val85Leu | Decrease (−1.03) | Decrease (−0.32141618) |
| p.Pro86Ala | Decrease (−0.84) | Decrease (−0.23579969) |
| p.Gly90Ser | Decrease (−0.32) | Decrease (−0.99667156) |
| p.Pro91Ser | Decrease (−0.81) | Decrease (−1) |
| p.Pro91Arg | Decrease (−0.55) | Decrease (−0.8319191) |
| p.Ile97Thr | Decrease (−0.96) | Decrease (−1) |
| p.Ala108Val | Increase (0.01) | Decrease (−1) |
| p.Tyr109Cys | Increase (0.01) | Decrease (−0.080615393) |
| p.Tyr111His | Decrease (−1.29) | Decrease (−0.48825376) |
| p.Arg112Leu | Increase (0.01) | Increase (0.71689609) |
| p.Arg112Pro | Decrease (−0.89) | Decrease (−0.16765113) |
| p.Arg112Cys | Decrease (−1.07) | Decrease (−0.30172448) |
| p.Gly118Glu | Decrease (−0.13) | Increase (0.23834727) |
| p.Leu119Met | Decrease (−1.09) | Decrease (−1) |
| p.Glu120Ala | Decrease (−1.47) | Increase (0.22586738) |
| p.Val123Ile | Decrease (−1.33) | Decrease (−0.38486441) |
| p.Thr124Asn | Decrease (−0.89) | Decrease (−0.38899978) |
| p.Ile125Asn | Decrease (−1.3) | Decrease (−0.70044733) |
| p.Arg131His | Decrease (−1.74) | Decrease (−1) |
| p.Arg131Cys | Decrease (−0.98) | Decrease (−0.47318314) |
| p.His142Leu | Increase (0.02) | Increase (0.18807228) |
| p.Ser146Thr | Decrease (−0.26) | Increase (0.084784406) |
| p.Pro155Ser | Decrease (−2.15) | Decrease (−1) |
| p.Ala161Val | Decrease (−0.54) | Increase (0.55854501) |
| p.Ile164Thr | Decrease (−3.27) | Decrease (−1) |
| p.Ala181Ser | Decrease (−1.78) | Decrease (−1) |
| p.Gln190Glu | Decrease (−0.18) | Increase (0.38789463) |
| p.Ser198Phe | Increase (0.42) | Increase (0.55978538) |
| p.Gly199Ser | Decrease (−1.37) | Decrease (−0.46462892) |
| p.Gly208Asp | Decrease (−0.42) | Decrease (−0.70348214) |
| p.Asp209Gly | Decrease (−1.55) | Decrease (−0.45580758) |
| p.Glu220Gln | Decrease (−0.74) | Decrease (−0.51532966) |
| p.Arg221Cys | Decrease (−0.93) | Decrease (−0.026809162) |
| p.Arg221Ser | Decrease (−3.09) | Decrease (−0.45650595) |
| p.Asn222Ser | Decrease (−1.57) | Decrease (−0.073805266) |
| p.His241Pro | Decrease (−0.86) | Decrease (−0.4625189) |
| p.Asp242Glu | Increase (0.46) | Increase (0.74495989) |
Reference protein ID, NP_001171271.
MUpro
MUpro (http://www.ics.uci.edu/~baldig/mutation.html) is also a support vector machine-based tool for the prediction of protein stability changes upon non-synonymous SNPs 9 . The value of the energy change is predicted, and a confidence score between −1 and 1 for measuring the confidence of the prediction is calculated. A score <0 means the variant decreases the protein stability; conversely, a score >0 means the variant increases the protein stability (Table 2).
Modeling of mutant structures and calculation of their RMSD values
To evaluate the structural stability between the native and mutant, protein structure analysis was performed based on the availability of X-ray crystallo-graphic structure of a protein in any database. In case of ADIPOQ, 3D crystallographic structure was not available in PDB. Therefore, a structure of human adiponectin globular domain was created by homology modeling using a 30 kDa adipocyte complement-related protein precursor -ACRP30 (PDB 1C28), the most suitable template identified by blast searches, as the template showed 91.97% of sequence identity 15 . Mutant models were prepared by FASTA format sequence submitted in SWISS-MODEL expasy ( http://swissmodel.expasy.org/ ). Energy minimization was done for both mutant and native models through DESMOND server with 2000 iterations. Further free energy and RMSD values were calculated by swiss PDB viewer and SuperPose online server, respectively.
Results
SNP analysis
By examining ADIPOQ gene using the dbSNP and HGVD databases, a total of 58 nonsynonymous SNPs were found. These SNPs consist of 55 missense variations and 3 nonsense variations.
Prediction of deleterious nsSNPs
In PolyPhen-2 analysis, 26 (47.8%) of the 55 variants were predicted to be probably damaging, and the others were predicted to be benign or possibly damaging, whereas in SIFT, 18 variants (32.7%) were predicted to be damaging, and others were predicted to be tolerated. By PROVEAN analysis, 27 variants (49.1%) were predicted to be deleterious, but the others were neutral (Figure 1). Among the above, 16 (29%) common ADIPOQ gene variants, namely, c.133G>C (p. Gly45Arg), c.140C>T (p.Pro47Leu), c.143G>A (p. Gly48Asp), c.161G>T (p.Gly54Val), c.163C>T (p.Arg 55Cys), c.223G>T (p.Gly75Cys), c.250G>A (p.Gly 84Arg), c.268G>A (p.Gly90Ser), c.334C>T (p.Arg 112Cys) c.335G>C (p.Arg112Leu), c.335G>T (p.Arg 112Pro), c.353G>A (p.Gly118Glu), c.425A>T (p.His 142Leu), c.593C>T (p.Ser198Phe), c.595G>A (p.Gly 199Ser), andc.626A>G (p.Asp209Gly) were found.
Figure 1.
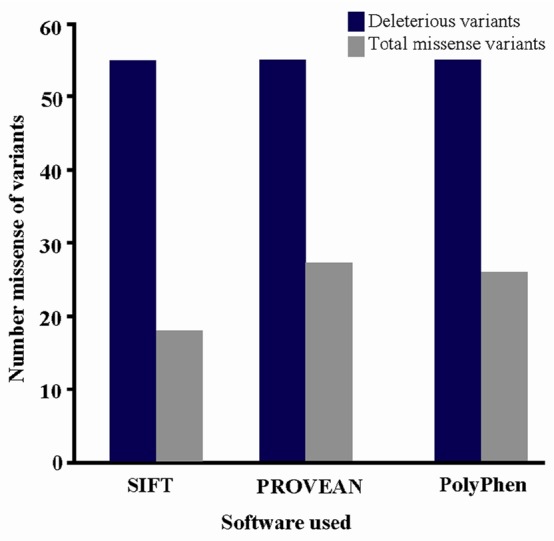
Graphical representation of deleterious variations.
Identification of functional nsSNP
Changes in the protein stability of missense variants were examined using I-Mutant 2.0 and MUpro software (Figure 2). In I-Mutant 2.0 prediction, 47 (85.4%) of 55 variants and in case of MUpro analysis, 41 (74.5%) variants were predicted to decrease protein stability. A total of 37 variants (67.2%) out of the 55 missense variants, including 10 out of 16 common damaging or deleterious variants namely c.133G>C (p. Gly45Arg), c.143G>A (p.Gly48Asp), c.163C>T (p. Arg55Cys), c.223G>T (p.Gly75Cys), c.250G>A (p. Gly84Arg), c.268G>A (p.Gly90Ser), c.334C>T (p. Arg112Cys) c.335G>C (p.Arg112Leu), c.595G>A (p. Gly199Ser), and c.626A>G (p.Asp209Gly) as determined using PolyPhen-2, SIFT, and PROVEAN software applications, were predicted to be less stable using both the I-Mutant 2.0 and the MUpro software.
Figure 2.
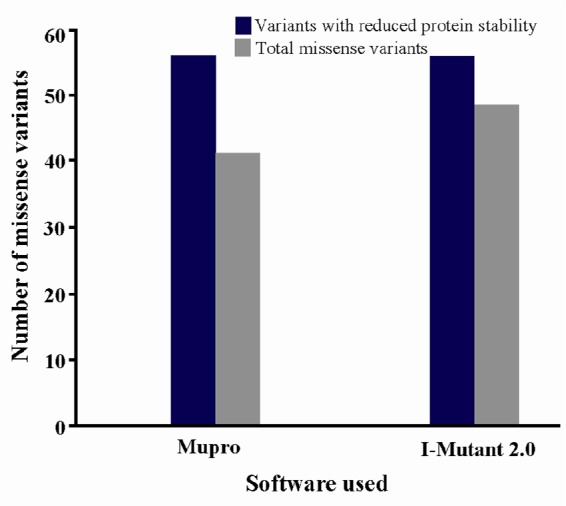
Graphical representation of protein stability analysis.
Three nonsense variations in the ADIPOQ gene were predicted to produce a truncated ADIPOQ protein. The c.274C>T (p.Arg92Ter, c.635G>A (p. Trp212Ter), and c.658G>T (p.Glu220Ter) variants predicted to truncate the protein production are given in table 3.
Table 3.
Summary of nonsense variations of ADIPOQ gene
| dbSNP ID | Nucleotide | Protein |
|---|---|---|
| rs139024247 | c.274C>T | p Arg92Ter |
| rs202013088 | c.635G>A | p. Trp212Ter |
| rs183590709 | c.658G>T | p.Glu220Ter |
Reference protein ID, NP_001171271.
Modeling of mutant proteins
The mutations which were both deleterious with less protein stability in the ADIPOQ gene were executed by swiss PDB viewer independently to get modeled structures. Then, energy minimization was achieved by DESMOND server for native and mutant structures. The total energy and RMSD values for the native and mutated structures are given in table 4. The higher the RMSD value is, the more the deviation between the two structures is, which in turn changes their functional activity. The total energies and RMSD values were higher for one mutant structure compared to the homology modeled structure (Table 4); these nsSNPs could affect the structure of the proteins.
Table 4.
RMSD and total energy of modeled structure and its mutant forms
| Total energy (Kcal/mol) | RMSD (A°) | |
|---|---|---|
| Native model (1c28.A) | −581.9624761 | - |
| Mutant model (G45R) | - | - |
| Mutant model (G48D) | - | - |
| Mutant model (R55C) | - | - |
| Mutant model (G75C) | - | - |
| Mutant model (G84R) | - | - |
| Mutant model (G90S) | - | - |
| Mutant model (R112C) | −1161.836281 | 0.93 |
| Mutant model (R112P) | −1136.55282 | 0.97 |
| Mutant model (G199S) | −1309.268642 | 2.80 |
| Mutant model (D209G) | −1210.319551 | 0.94 |
Discussion
Our analysis revealed 58 nonsynonymous variants out of 55 missense and other three were nonsense variants. 10 variants namely c.133G>C (p.Gly45Arg), c.143G>A (p.Gly48Asp), c.163C>T (p.Arg55Cys), c. 223G>T (p.Gly75Cys), c.250G>A (p.Gly84Arg), c.268 G>A (p.Gly90Ser), c.334C>T (p.Arg112Cys), c.335 G>T (p.Arg112Pro), c.595G>A (p.Gly199Ser), andc. 626A>G (p.Asp209Gly) out of 55 missense variants showed deleterious scores by SIFT, PROVEAN, PolyPhen (Table 1) and decreasing the protein stability upon their aminoacid changes by I Mutant 2.0 and MUpro (Table 2). Mutant models were built by swiss model by using template 1c28.A to 4 nsSNPs out of 10 nsSNPs which is common to both deleterious and less protein stability due to the template predicted by complement component C1q domain region of the ADIPOQ protein only. Further energy minimization was done by Desmond server and total energy was calculated by swiss PDB viewer and RMSD values were calculated by SuperPose online server. The RMSD value of mutant (G199S) model was high compared to the native model. In case of total energy, mutant models show lower energy than the native models as given in table 4. Three nonsense variations in the ADIPOQ gene were predicted to produce a truncated protein. The c.274C>T (p.Arg92Ter) variant in collagen region, c.635G>A (p.Trp212Ter), and c.658G>T (p.Glu220 Ter) variants in complement component C1q domain were predicted to truncate the protein production; these results suggested that p.Arg92Ter nonsense variant truncates the whole region of the complement C1q domain and the remaining two variants such as p. Trp212Ter and p.Glu220Ter terminate the partial complement C1q domain of the ADIPOQ protein synthesis.
Adiponectin, an endogenous insulin-sensitizing hormone and the most abundant adipokine produced especially by the human adipose tissue, is linked to metabolic syndrome, type-2 diabetes, insulin resistance, obesity, and inflammation as well as several types of cancers. Adiponectin has anti-inflammatory and antilipogenic effects, while Tumor Necrosis Factor alpha (TNF-alpha) reduces insulin sensitivity and has proinflammatory effects 16 . In general, a lower level of adiponectin concentration in blood circulation correlates with an increased body mass index (BMI) and insulin resistance. A higher BMI leads to a higher risk for obesity. Greater insulin resistance increases risk for type 2 diabetes mellitus (T2DM). Two particular variants such as rs17300539 and rs266729 in the promotor region of the ADIPOQ cause cells to make less adiponectin. Decreased adiponectin means less glucose utilization and less efficient fat burning and therefore a greater risk of developing obesity and T2DM 17 .
Genetic factors such as single nucleotide polymorphisms in the adiponectin gene and environmental factors such as a high-fat diet and inactivity are associated with low adiponectin concentrations and may contribute to the development of insulin resistance, type 2 diabetes, and atherosclerosis. Adiponectin automatically self-associates into larger structures with high molecular weight. Initially, three adiponectin molecules bind together to form a homotrimer and they continue to self-associate and form hexamers or dodecamers. High-molecular-weight adiponectin was further found to be associated with a lower risk of diabetes with similar magnitude of association as total adiponectin 18 . However, coronary artery disease has been found to be positively associated with high-molecular-weight adiponectin, but not with low-molecular-weight adiponectin 19 . Evaluation of adiponectin levels with the ratio of High Molecular Weight (HMW)/Low Molecular Weight (LMW) and (MMW) and consideration of different ethnic genetic backgrounds are of importance in the translational research of adiponectin. Two novel nonsynonymous ADIPOQ variations i.e. P32L, and R55C were achieved using an extreme phenotype sequencing approach. Individuals with these novel variations had low adiponectin and exhibited reduced HMW structures compared to individuals without these variations. Although each variation is present in the heterozygous state, dominant negative effects may exist 20 . The high-molecular-weight isoform adiponectin is believed to be the biologically active form that activates downstream events in both skeletal muscle and the liver 21 . Several rare ADIPOQ gene mutations affecting the multimerization and consequently the biological function of the protein have been characterized. For example, the Arg112Cys and Ile164Thr mutants do not assemble into trimers, leading to the clinic symptom hypoadiponectinemia. The Gly84Arg and Gly 90Ser mutants are able to assemble into trimers and hexamers but are unable to form the high-molecular-weight multimers, leading directly to diabetes 21 . R55H, G84R, and G90S variations did not disturb adiponectin trimeric and hexameric formations but obstructed their multimerization. These variants are not close to interdisulfide bond forming site (Cys36) and they were still capable of forming hexamers. However, they might cause conformational change and conceal the remaining free thiol from interacting with other hexamers 15 .
Therefore, plasma/serum adiponectin levels and genomic DNA polymorphisms in the ADIPOQ gene can be used as the biomarkers for early diagnosis and clinical prediction of diabetes, obesity, diabetic complications and other metabolic disorders.
Conclusion
In the present study, a total of 58 nonsynonymous SNPs in ADIPOQ gene involved in diabetes, obesity and inflammation were analyzed. Out of the 58 nsSNPs, 55 were found to be missense variations and 3 were nonsense variations. Further in silico analysis using different softwares (PolyPhen 2, SIFT and PROVEAN) predicted that 16 of the 55 missense variants were damaging or deleterious. Also, in silico analysis (I-Mutant 2.0 and MUpro) was carried out and 37 variants were identified that were predicted to be less stable. In addition, 3 nonsense variants were predicted to lead to the production of a truncated ADIPOQ protein. Further total energy and RMSD values were calculated for 4 nsSNPs out of 10 nsSNPs which were both deleterious and showed a decrease in protein stability. Mutant model G199S (rs144526209) showed high RMSD with low total energy which can be considered as the most deleterious variant of ADIPOQ gene.
Acknowledgement
The authors are thankful for management of K L University for providing the facilities to undertake this study.
References
- 1. Brochu-Gaudreau K, Rehfeldt C, Blouin R, Bordignon V, Murphy BD, Palin MF. Adiponectin action from head to toe. Endocrine 2010; 37 (1): 11– 32. [DOI] [PubMed] [Google Scholar]
- 2. Meller M, Qiu C, Vadachkoria S, Abetew DF, Luthy DA, Williams MA. Changes in placental adipocytokine gene expression associated with gestational diabetes mellitus. Physiol Res 2006; 55 (5): 501– 512. [DOI] [PubMed] [Google Scholar]
- 3. Nanda S, Savvidou M, Syngelaki A, Akolekar R, Nicolaides KH. Prediction of gestational diabetes mellitus by maternal factors and biomarkers at 11 to 13 weeks. Prenat Diagn 2011; 31 (2): 135– 141. [DOI] [PubMed] [Google Scholar]
- 4. Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet 2012; 8 (3): e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7 (4): 248– 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4 (7): 1073– 1081. [DOI] [PubMed] [Google Scholar]
- 7. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012; 7 (10): e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gromiha MM, An J, Kono H, Oobatake M, Uedaira H, Prabakaran P, et al. ProTherm, version 2.0: thermodynamic database for proteins and mutants. Nucleic Acids Res 2000; 28 (1): 283– 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005; 33 (Web Server issue): W72– 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasseur F, Helbecque N, Dina C, Lobbens S, Delannoy V, Gaget S, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet 2002; 11 (21): 2607– 2614. [DOI] [PubMed] [Google Scholar]
- 11. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25 (17): 3389– 3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sjölander K, Karplus K, Brown M, Hughey R, Krogh A, Mian IS, et al. Dirichlet mixtures: a method for improved detection of weak but significant protein sequence homology. Comput Appl Biosci 1996; 12 (4): 327– 345. [DOI] [PubMed] [Google Scholar]
- 13. Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 2004; 83 (6): 970– 979. [DOI] [PubMed] [Google Scholar]
- 14. Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30 (17): 3894– 3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jungtrakoon P, Plengvidhya N, Tangjittipokin W, Chimnaronk S, Salaemae W, Chongjaroen N, et al. Novel adiponectin variants identified in type 2 diabetic patients reveal multimerization and secretion defects. PLoS One 2011; 6 (10): e26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hui JM, Hodge A, Farrell GC, Kench JG, Kriketos A, George J. Beyond insulin resistance in NASH: TNF-alpha or adiponectin? Hepatology 2004; 40( 1): 46– 54. [DOI] [PubMed] [Google Scholar]
- 17. Izadi M, Goodarzi MT, Samari Khalaj HR, Khorshidi D, Doali H. Correlated with insulin resistance in obese men with type 2 diabetes. Int J Endocriol Metab 2011; 9 (1): 253– 257. [Google Scholar]
- 18. Zhu N, Pankow JS, Ballantyne CM, Couper D, Hoogeveen RC, Pereira M, et al. High-molecular-weight adiponectin and the risk of type 2 diabetes in the ARIC study. J Clin Endocrinol Metab 2010; 95 (11): 5097– 5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rizza S, Gigli F, Galli A, Micchelini B, Lauro D, Lauro R, et al. Adiponectin isoforms in elderly patients with or without coronary artery disease. J Am Geriatr Soc 2010; 58 (4): 702– 706. [DOI] [PubMed] [Google Scholar]
- 20. Kottyan LC, Woo JG, Keddache M, Banach W, Crimmins NA, Dolan LM, et al. Novel variations in the adiponectin gene (ADIPOQ) may affect distribution of oligomeric complexes. Springerplus 2012; 1 (1): 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadowaki T, Yamauchi T. Adiponectin and adiponectin receptors. Endocr Rev 2005; 26 (3): 439– 451. [DOI] [PubMed] [Google Scholar]


