Abstract
Background:
Interleukin-16 (IL-16) is an important regulator of T cell activation and was reported to act as a chemoattractant agent. There are evidences that IL16 can control the neuroinflammatory processes in Alzheimer's Disease (AD). This study was performed to investigate the role or association of IL16 polymorphisms, rs11556218 and rs4778889 with the risk of late-onset Alzheimer's disease (LOAD) in Iranian population.
Methods:
Totally, 148 AD patients and 137 nondemented and age-matched subjects were recruited in this study. Genotyping of rs11556218 T/G and rs4778889 T/C polymorphisms was performed by PCR-RFLP method using the NdeI and AhdI restriction enzymes, respectively.
Results:
Statistical analysis of rs11556218 genotypes showed a protective effect against AD in the heterozygote genotype (p=0.001, OR=0.16) as well as rs4778889 (p=0.001, OR=0.23). Frequency of rs11556218 allele T was higher in controls than patients (p=0.001, OR=0.32). However, there was no significant difference in the frequencies of rs4778889 alleles between the AD patients and controls.
Conclusion:
Our results indicate that the rs11556218 and rs4778889 polymorphisms have a protective role in the development of sporadic AD in Iranian population.
Keywords: Alzheimer's disease, Genetic association study, Interleukin-16, Polymorphism
Introduction
Alzheimer's Disease (AD), before all else described by Alois Alzheimer, as a main cause of dementia in the aged population and well-known as the most common type of dementia affects 1 out of 8 persons older than 65 1 , 2 . Mendelian inheritance of 3 genes, PSEN1, PSEN2, and APP, is associated with an early-onset Alzheimer's disease (EOAD). Most cases of Alzheimer's disease are late-onset (LOAD) and are apparently affected by both of genetic and non-genetic factors 2 . This form of AD is strongly associated with ε4 allele of APOE gene 3 . However, existence of some other causative genes responsible for LOAD is obviously expected because about half of LOAD patients do not bear mutations in APOE gene 4 . Inflammation is a response to remove the initial cause of cell injury 5 . Brain inflammation is one of the pathological markers of LOAD 6 . Inflammation occurs in pathologically susceptible areas of the AD brain, in the place in which the expression of acute phase proteins and proinflammatory cytokines is evident. Microglia, neurons, and as trocytes are mainly responsible for the inflammatory reactions 5 . Some studies referred to IL-16 as an immunomoregulatory cytokine that contributes to the regulation of CD4 cell recruitment and activation at regions related to inflammation in correlation with distinct neurodegenerative disorders as well as AD 7 , 8 . Previously, the association between inflammatory pathway genes, including TNFα, CCR2-64I, CCR5 Δ32 and CALHM1 P86L polymorphisms with LOAD was studied 9 , 10 . This study investigated whether the two polymorphisms of IL16 gene, rs11556218 and rs4778889 are associated with the risk of LOAD in an Iranian population.
Materials and Methods
This research was performed in the Genetics Research Center, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran (2013–2014). Totally, 148 LOAD patients and 137 age-matched and unrelated healthy subjects from various old peoples' homes in Tehran, Iran were recruited. The diagnosis of cases was made according to DSM-IV criteria by an expert psychiatrist. In the control group, if they had any important neurologic or psychological disorder, they were excluded from the study. The control subjects were chosen based on extensive evaluation of their medical histories and physical conditions. The volunteers were included if they were older than 65 years and agreed to enter the study. Protocols were confirmed by Ethics Committee of the University of Social Welfare and Rehabilitation Sciences and all participants provided their informed consent.
Nearly, 5 ml of peripheral blood was collected from each subject in an EDTA-coated tube (0.2 ml, 0.5 M). DNA was extracted from the whole blood samples using a standard salting-out method 11 . Genotyping of rs11556218 T/G and rs4778889 T/C polymorphisms was performed using PCR-RFLP technique. PCR amplification and genotyping of the fragment containing the rs11556218 T/G polymorphism was performed by a forward primer 5′-GCTCAGGTTCACAGAGTGTTTCC ATA-3′ and reverse primer 5′-TGTGACAATCACAGC TTGCTTG-3′, followed by digestion with the restriction enzyme NdeI (Fermentas, Germany). The rs4778889 T/C polymorphism was assessed by forward primer5′-CTCCACACTCAAAGCCTTTTGTTCCTATG-3′ and reverse primer 5′-ATCCACGCTGGTTCCTTTCGT-3′, followed by digestion with the restriction enzyme AhdI (Fermentas, Germany).
The PCR reactions were carried out in the final volume of 25 μl containing: 10×PCR buffer (Roche, Germany), 2 mM MgCl2 (Roche, Germany), 0.4 mM of each dNTP (Fermentas, Germany), 5 pmol of each primer, 50 ng template DNA, 1 U Taq DNA polymerase (Roche, Germany) and sterile distilled water up to 25 μl. Amplification conditions started with an initial denaturation step of 5 min at 94 °C, followed by 30 cycles of 30 s denaturation (94 °C), 30 s annealing (60 °C for rs11556218 and 59 °C for rs4778889) and 30 sec extension (72 °C), finished by an ultimate extension for 5 min (72 °C) and finally cooling to 4 °C. All PCR products were subjected to electrophoresis on 2% agarose gel prepared in 1×TAE, stained with ethidium bromide and visualized by exposure to ultraviolet light.
The PCR products of IL16 rs11556218 T/G and IL16 rs4778889 T/C were digested with restriction enzymes NdeI and AhdI at 37 °C overnight, respectively. DNA fragments were subjected to 8% polyacrylamide gel electrophoresis and stained with silver nitrate. Based on figure 1, the G allele of the rs11556218 produced two fragments 147 and 24 bp, whereas the T allele remained undigested (171 bp). In IL16 rs4778889 T/C variant, the C allele was cut into two fragments 146 and 34 bp, while the T allele remained uncut (Figure 2).
Figure 1.
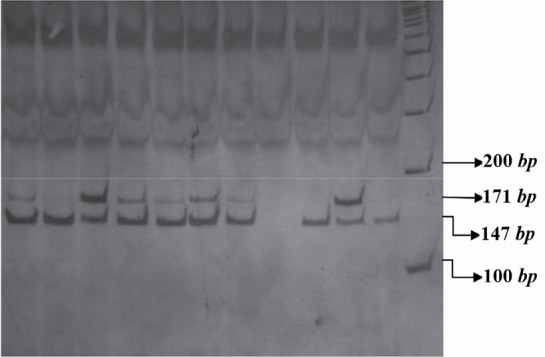
PCR- RFLP profile of IL16 rs11556218 T/G polymorphism digested with NdeI restriction enzyme.
Figure 2.
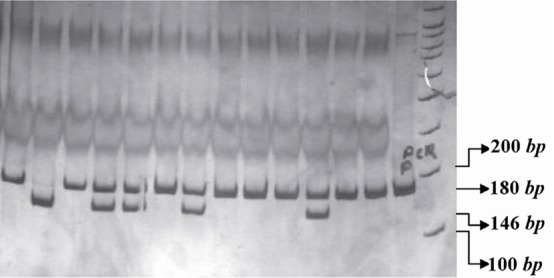
PCR- RFLP profile of IL16 rs4778889 T/C polymorphism digested with AhdI restriction enzyme.
The genotype frequencies in the cases and control subjects were analyzed using the chi square test. Logistic regression analysis was carried out to determine the relationship between LOAD and two polymorphisms (rs11556218 T/G and rs4778889 T/C) using SPSS software (Versian13, Chicago, IL, USA) and Open Epi (Open Source Epidemiologic Statistics for Public Health, Version 2.2.1, Atlanta, GA, USA). A p-value of less than 0.05 was considered statistically significant.
Results
The mean age of the AD group (78.39, SD±7.87) was not significantly different from the control group (77.07, SD±6.97). The distribution of age, sex, job, educational level and genetic background was almost the same in both groups (Table 1).
Table 1.
Comparison of mean age, sex, job, education level and ethnic groups between the AD and control groups using t-test and χ2 test
| Parameter | AD patients (n=146) | Controls (n=148) | p-value |
|---|---|---|---|
| Age | 78.55±7.80 | 77.14±6.95 a | 0.091 |
| Sex (M/F) b | 63/91 | 63/99 | 0.714 |
| Jobs | |||
| Housewife | 82 (55.8%) | 83 (56.2%) | |
| Own business Worker | 34 (23.4%) 13 (9.2%) | 31 (21.0%) 13 (8.6%) | 0.938 |
| Farmer | 5 (3.2%) | 5 (3.1%) | |
| Employee | 12 (8.4%) | 16 (11.1%) | |
| Education level | |||
| Illiterate | 61 (41.6%) | 64 (43.2%) | |
| Primary school | 42 (29.2%) | 44 (29.6%) | 0.427 |
| Secondary school | 24 (16.2%) | 18 (12.3%) | |
| Diploma | 16 (11.1%) | 14 (9.3%) | |
| Academic | 3 (1.9%) | 8 (5.6%) | |
| Ethnic | |||
| Fars | 89 (61.0%) | 94 (63.6%) | |
| Turk Kurd | 37 (25.3%) 6 (3.9%) | 37 (25.3%) 3 (1.8%) | 0.490 |
| Lor | 1 (0.7%) | 4 (2.5%) | |
| Gilak and Mazani | 13 (9.1%) | 10 (6.8%) |
Mean±SD,
Male/Female
The highest and the lowest frequencies of LOAD were observed among the housewives and the farmers, respectively. People with academic education had the lowest incidence and uneducated individuals the highest frequency among the patients and control group, respectively. The subjects consisted of five different Iranian ethnicities in which the Fars was the most common population (Table 1).
The genotype and allele frequencies for the rs11556218 T/G polymorphism were analyzed in LOAD patients and controls. As summarized in table 2, the frequency of GG genotype was not different between the AD patients and the control group (Reference Group). Distribution of TG genotype frequencies showed significant differences between AD patients and control subjects (p=0.001, OR=0.16). Significant difference was observed between allele frequencies in cases and controls (p=0.001, OR=0.32). Analysis of the data stratified by gender revealed that genotypic distribution in women group showed a significant association in the heterozygote genotype frequency in cases and controls as well as in men (Table 3).
Table 2.
The distribution of genotype and allele frequencies for the IL16-rs11556218 T/G and IL16-4778889 T/C polymorphisms
| IL16-rs11556218 T/G | IL16-4778889 T/C | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype allele | Patients (n=137) | Controls (n=147) | p-value | Odds ratio | Genotype allele | Patients (n=146) | Controls (n=148) | p-value | Odds ratio |
| TT | 2 (%1.5) | 0 | 0.934 | Undefined | TT | 111 (%76) | 95 (%64) | Ref* | |
| TG | 35 (%25.5) | 100 (%68) | 0.001 | 0.16 (0.1–0.28) | TC | 7 (%5) | 26 (%17.6) | 0.001 | 0.23 (0.1–0.55) |
| GG | 100 (%73) | 47 (%32) | Ref* | CC | 28 (%19) | 27 (%18.4) | 0.694 | Not applicable | |
| T | 39 (%14) | 100 (%34) | 0.001 | 0.32 (0.21–0.49) | T | 229 (%78.4) | 216 (%73) | 0.123 | 0.74 (0.51, 1.1) |
| G | 235 (%86) | 194 (%66) | C | 63 (%21.6) | 80 (%27) | ||||
Reference Group
Table 3.
The distribution of genotype and allele frequencies for the IL16-rs11556218 T/G polymorphism stratified by sex
| C | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype allele | Patients | Controls | p-value | Odds ratio | Patients | Controls | p-value | Odds ratio |
| TT | 1 (%1.6) | 0 | 0.595* | Undefined | 1 (%1.5) | 0 | 0.99* | Undefined |
| TG | 13 (%21.4) | 33 (%68.75) | 0.001 | OR= 0.13 (0.05–0.3) | 20 (%29.4) | 61 (%71.8) | 0.001 | 0.17 (0.08–0.33) |
| GG | 47 (%77) | 15 (%31.25) | Ref Group | 1 | 47 (%69.1) | 24 (%28.2) | Ref Group | 1 |
| T | 15 (%12.3) | 33 (%65.26) | 0.001 | OR= 0.26 (0.13–0.52) | 22 (%16.2) | 61 (%35.9) | 0.001 | 0.34 (0.2–0.6) |
| G | 107 (%87.7) | 62 (%34.74) | 114 (%83.8) | 109 (%64.1) | ||||
Fisher exact test p-value
There was no significant difference in the CC geno-type of rs4778889 polymorphism; however, the difference in heterozygote genotype was significant (p=0.001, OR=0.23). There was no significant difference in the allele frequencies of rs4778889 polymorphism (p=0.123) (Table 4).
Table 4.
The distribution of genotype and allele frequencies for the IL16-rs4778889 T/C polymorphism stratified by sex
| C | Male | Female | |||||
|---|---|---|---|---|---|---|---|
| Genotype allele | Patients | Controls | p-value | Patients | Controls | p-value | Odds ratio |
| TT | 52 (%80) | 38 (%76) | Ref Group | 52 (%75.4) | 50 (%57.5) | Ref Group | 1 |
| TC | 1 (%1.5) | 3 (%6) | 0.437 | 4 (%5.8) | 22 (%25.3) | 0.001 | 0.17 (0.06–0.54) |
| CC | 12 (%18.5) | 9 (%18) | 0.957 | 13 (%18.8) | 15 (%17.2) | 0.696 | Not applicable |
| T | 105 (%80.8) | 79 (%79) | 0.7397 | 108 (%78.3) | 122 (%68.9) | 0.064 | Not applicable |
| C | 25 (%19.2) | 21 (%21) | 30 (%21.7) | 55 (%31.1) | |||
Analysis of the data stratified by gender revealed that neither female Alzheimer patients nor male Alzheimer patients had significant differences in genotype and allele frequencies compared with the female and male controls.
Discussion
In the present study, IL16 polymorphisms, rs11556218 and rs4778889 were associated with LOAD in an Iranian population. These findings suggest that rs11556218 and rs4778889 can be introduced as useful genetic hallmarks of AD, allowing for identification of related molecular pathways which protect us against LOAD. IL-16 and some other cytokines including IL-1, IL-6, IL-8, IL-11, IL-17, TNFα and TNFβ are well-known as chronic inflammation cytokines. These cytokines operate as the regulators of systemic or tissue-specific inflammation 12 .
Activated astrocytes and microglia are typically found in high quantities in the vicinity of neurons and plaques. This means that inflammation may be involved in LOAD, because glial cells regulate the innate immune response in the Central Nervous System (CNS). Activated astrocytes and microglia yield various proinflammatory signal molecules, including cytokines, growth factors, complement and adhesion molecules 13 , 14 .
The IL16 gene located on chromosome 15 containing 7 exons and 6 introns, is covering approximately 12.8 kb of chromosome 15 15 . The rs11556218 is identified within the intron, whereas rs4778889 is located in coding region. According to Genecards database, IL16 gene is correlated with 84 different disorders containing LOAD. The rs11556218 T/G polymorphism has been shown to be associated with colorectal, gastric and nasopharyngeal cancers 16 , 17 . In 2009, Xue et al showed that individuals carrying the G allele of rs11556218 and the C allele of rs4778889 are at a significantly higher risk of Systemic Lupus Erythematosus (SLE) as compared with those carrying the T allele of rs11556218 and the T allele of rs4778889, respectively. In 2007, Motta et al explained that the association between IL-16 plasma levels and LOAD was dependent on the disease progression. As mentioned before, inflammation has been reported to be involved in the pathogenesis of neurodegenerative disorders. Raised levels of IL-16 in the brain of AD patients confirm that the immune system may play an important role in progression of LOAD 8 , 18 .
Conclusion
Our results confirm the relation between IL16 polymorphisms and development of LOAD in Iranian population. Further studies with bigger sample sizes are required to confirm the present findings. The current study had some limitations. First, the collected sample size was not big enough. Secondly, LOAD is induced by many causative genes and environmental factors, none of which were regarded in this research. Finally, the present study shows that rs11556218 and rs4778889 are associated with LOAD risk in an Iranian population. Further large-scale studies are needed to clarify whether these IL16 polymorphisms interact with non-genetic factors in the development of LOAD.
Acknowledgement
The research was supported by the Genetics Research Center, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, and all our patients and their families and healthy subjects.
References
- 1. Avramopoulos D. Genetics of Alzheimer's disease: recent advances. Genome Med 2009; 1 (3): 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bird TD. Genetic aspects of Alzheimer disease. Genet Med 2008; 10 (4): 231– 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol 2010; 23 (4): 213– 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, et al. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 2000; 66 (1): 196– 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rubio-Perez JM, Morillas-Ruiz JM. A review: inflammatory process in Alzheimer's disease, role of cytokines. ScientificWorld Journal 2012; 2012: 756357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, et al. Inflammation and Alzheimer's disease. Neurobiol Aging 2000; 21 (3): 383– 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cruikshank WW, Kornfeld H, Center DM. Interleukin-16. J Leukoc Biol 2000; 67 (6): 757– 766. [DOI] [PubMed] [Google Scholar]
- 8. Di Rosa M, Dell'Ombra N, Zambito AM, Malaguarnera M, Nicoletti F, Malaguarnera L. Chitotriosidase and inflammatory mediator levels in Alzheimer's disease and cerebrovascular dementia. Eur J Neurosci 2006; 23 (10): 2648– 2656. [DOI] [PubMed] [Google Scholar]
- 9. Manoochehri M, Kamali K, Rahgozar M, Ohadi M, Farrokhi H, Khorshid HR. Lack of Association between Tumor Necrosis Factor-alpha -308 G/A Polymorphism and Risk of Developing Late-Onset Alzheimer's Disease in an Iranian Population. Avicenna J Med Biotechnol 2009; 1 (3): 193– 197. [PMC free article] [PubMed] [Google Scholar]
- 10. Khorram Khorshid HR, Manoochehri M, Nasehi L, Ohadi M, Rahgozar M, Kamali K. Ccr2-64i and Ccr5 Δ32 Polymorphisms in Patients with Late-Onset Alzheimer's disease; A Study from Iran (Ccr2-64i And Ccr5 Δ32 Polymorphisms in Alzheimer's disease). Iran J Basic Med Sci 2012; 15 (4): 937– 944. [PMC free article] [PubMed] [Google Scholar]
- 11. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16 (3): 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azimzadeh P, Romani S, Mohebbi SR, Kazemian S, Vahedi M, Almasi S, et al. Interleukin-16 (IL-16) gene polymorphisms in Iranian patients with colorectal cancer. J Gastrointestin Liver Dis 2011; 20 (4): 371– 376. [PubMed] [Google Scholar]
- 13. Streit WJ. Microglial activation and neuroinflammation in Alzheimer's disease: a critical examination of recent history. Front Aging Neurosci 2010; 2: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffin WS. Inflammation and neurodegenerative diseases. Am J Clin Nutr 2006; 83 (2): 470S– 474S. [DOI] [PubMed] [Google Scholar]
- 15. Tong Z, Li Q, Zhang J, Wei Y, Miao G, Yang X. Association between interleukin 6 and interleukin 16 gene polymorphisms and coronary heart disease risk in a Chinese population. J Int Med Res 2013; 41 (4): 1049– 1056. [DOI] [PubMed] [Google Scholar]
- 16. Gao LB, Rao L, Wang YY, Liang WB, Li C, Xue H, et al. The association of interleukin-16 polymorphisms with IL-16 serum levels and risk of colorectal and gastric cancer. Carcinogenesis 2009; 30 (2): 295– 299. [DOI] [PubMed] [Google Scholar]
- 17. Gao LB, Liang WB, Xue H, Rao L, Pan XM, Lv ML, et al. Genetic polymorphism of interleukin-16 and risk of nasopharyngeal carcinoma. Clin Chim Acta 2009; 409 (1–2): 132– 135. [DOI] [PubMed] [Google Scholar]
- 18. Glass WG, Sarisky RT, Vecchio AM. Not-so-sweet sixteen: the role of IL-16 in infectious and immune-mediated inflammatory diseases. J Interferon Cytokine Res 2006; 26 (8): 511– 520. [DOI] [PubMed] [Google Scholar]


