Abstract
This study represents a complete comparative analysis of the most widely used African swine fever (ASF) diagnostic techniques in the European Union (EU) using field and experimental samples from animals infected with genotype II ASF virus (ASFV) isolates circulating in Europe. To detect ASFV, three different PCRs were evaluated in parallel using 785 field and experimental samples. The results showed almost perfect agreement between the Universal ProbeLibrary (UPL-PCR) and the real-time (κ = 0.94 [95% confidence interval {CI}, 0.91 to 0.97]) and conventional (κ = 0.88 [95% CI, 0.83 to 0.92]) World Organisation for Animal Health (OIE)-prescribed PCRs. The UPL-PCR had greater diagnostic sensitivity for detecting survivors and allows earlier detection of the disease. Compared to the commercial antigen enzyme-linked immunosorbent assay (ELISA), good-to-moderate agreement (κ = 0.67 [95% CI, 0.58 to 0.76]) was obtained, with a sensitivity of 77.2% in the commercial test. For ASF antibody detection, five serological methods were tested, including three commercial ELISAs, the OIE-ELISA, and the confirmatory immunoperoxidase test (IPT). Greater sensitivity was obtained with the IPT than with the ELISAs, since the IPT was able to detect ASF antibodies at an earlier point in the serological response, when few antibodies are present. The analysis of the exudate tissues from dead wild boars showed that IPT might be a useful serological tool for determining whether or not animals had been exposed to virus infection, regardless of whether antibodies were present. In conclusion, the UPL-PCR in combination with the IPT was the most trustworthy method for detecting ASF during the epidemic outbreaks affecting EU countries in 2014. The use of the most appropriate diagnostic tools is critical when implementing effective control programs.
INTRODUCTION
African swine fever (ASF) is a complex and lethal viral disease affecting swine and has a significant socioeconomic impact on both the developed and developing world. It has a major negative effect on national, regional, and international trade and constrains pig production in affected areas. The devastating acute form of the disease is characterized, among other features, by functional and congestive-hemorrhagic disorders of the digestive and respiratory systems and causes around 100% mortality in infected pigs (1). Both European wild boars (Sus scrofa) and feral pigs are susceptible and exhibit clinical signs and mortality rates similar to those of domestic pigs. In contrast, African wild pigs (Phacochoerus and Potamochoerus spp.) are resistant to the disease (2–10).
The causative agent of the disease, the ASF virus (ASFV), is a large double-stranded DNA virus and the only member of the Asfarviridae family, genus Asfivirus (11, 12). The virus genome is 170 to 192 kb long (13–17). ASF is endemic in sub-Saharan Africa, where it was first described in 1921 (18). Several outbreaks have occurred since then in Europe and South and Central America. In most non-African countries, the disease has been successfully eradicated, the only exception being Sardinia (Italy), where the disease is still endemic (19, 20). In April 2007, the disease spread from East Africa to the Republic of Georgia (21), and outbreaks occurred in Armenia, Azerbaijan, and the Russian Federation (22). The ongoing spread of ASFV into adjacent eastern European countries, such as Ukraine (23, 24) and Belarus (25), and the situation in Russia affecting both wild boars and domestic pigs placed neighboring areas in the European Union (EU) at risk for the spread of ASFV. The first cases of ASF in wild boars in Lithuania and Poland were reported in early 2014 in areas bordering Belarus (26–30). According to the World Organisation for Animal Health (OIE), during 2014, nearly 260 ASF cases or outbreaks in wild boars and domestic pigs were detected in EU countries (Latvia, Lithuania, Estonia, and Poland). This situation, combined with the uncertainty present in Belarus, has created a permanent risk of reintroducing ASF into the EU via wild boars or the illegal trade of contaminated pork products and waste (31).
No vaccine is available to prevent ASF infection. The control and eradication measures applicable are based on classical disease control methods, including surveillance, epidemiological investigation, tracing of pigs, and stamping out in infected holdings. These measures must be combined with strict quarantine and biosecurity measures in domestic pig holdings and animal movement control. Due to the characteristics of the disease, passive surveillance based on investigation of clinical signs and high fatality rate of pigs plays a pivotal role in the early diagnosis of ASF. In addition, given a certain proportion of animals may also survive the infection, active surveillance also provides very valuable data on the evolution of the disease and guidance on the assessment of the effectiveness of the control measures. However, to be successful, surveillance must have adequate laboratory support for a rapid diagnosis, which in combination, will allow the early detection of the disease and therefore its spread (32).
ASF diagnosis requires the identification of animals that are or were previously infected with ASFV (1, 19). Thus, an appropriate diagnosis involves the detection and identification of ASFV-specific antigens or DNA and antibodies (33–35). The OIE-recommended tests for virus detection include virus isolation, fluorescent antibody tests (FAT), and both real-time and conventional PCR assays (33–37). These PCRs are the most widely used at the national reference laboratory (NRL) level within the EU. New real-time PCRs developed in recent years have been shown to provide greater sensitivity for detecting animals that have survived infection (38, 39). Other assays, such as antigen detection enzyme-linked immunosorbent assay (ELISA), which allows for large-scale testing of samples, are also available at the NRL level but have been reported as having lower analytical sensitivity than that of PCR tests (34).
For the detection of ASF antibodies, the OIE-prescribed assays involve the use of an ELISA for antibody screening, backed up by immunoblotting (IB) or indirect immunofluorescence (IIF) as confirmatory tests (33, 40). The indirect immunoperoxidase test (IPT), validated by the European Union Reference Laboratory (EURL) for ASF, has effective analytical and diagnostic sensitivity and can be used as an alternative confirmatory test for the diagnosis of ASF using porcine sera. In addition, it can be applied with ease to a large number of samples and does not require expensive fluorescence microscope equipment (41). Currently, three commercial ELISA kits are available for the detection of ASF antibodies (Ingenasa, IDvet, and Svanovir), of which the Ingezim PPA Compac, K3 from Ingenasa is the most widely used at the EU level (C. Gallardo, personal communication).
The techniques currently in use for ASF diagnosis give a confident diagnosis of the disease in any epidemiological situation (34, 35, 40, 42). However, ASF diagnosis is complex due to the wide range of clinical forms and the similarity of its symptoms to those of other viral infections, such as classical swine fever (CSF) (1). The current epidemic situation of ASF in the EU has created a need to review the sensitivity and specificity of current diagnostic tests and their ability to diagnose ASF in both domestic and wild Suidae in affected areas. To this end, the EURL has performed, in collaboration with the NRLs of the four affected EU countries, a comparative study using all the ASF diagnostic tests that are currently being used across the EU to analyze experimental and field samples collected from both domestic and wild pigs. This paper reviews the performance characteristics, including sensitivity and specificity, of current ASF diagnostic tests in order to guide effective actions for rapid identification and further control of ASF in affected countries.
MATERIALS AND METHODS
Viruses, cell cultures, and virus propagation.
For the experimental in vivo studies, three ASFV isolates belonging to p72 genotype II obtained from the outbreaks in Armenia in 2007 (Arm07), Ukraine in 2012 (Ukr12/Zapo), and Lithuania in 2014 (LT14/1490) were used. The ASFV isolates were propagated (2 to 6 passages) in porcine blood monocytes (PBM) recovered from naive domestic pigs (43, 44). Titrations of ASFV stocks were performed using a hemadsorption assay to monitor the endpoint dilution of ASFV isolates into PBM. Titers were estimated using Reed and Muench's method (45) and expressed as 50% hemadsorbing doses per milliliter (HAD50/ml) per sample.
The stable monkey (MS) kidney cell line (ECACC, 91070510) was used for conventional soluble cytoplasmic antigen production after infection with the ASFV MS-adapted E70 isolate (E70 MS 48), as described in the OIE Terrestrial Manual 2012 (33), and in the preparation of the ASFV-coated 96-well plates used as the antigen in the IPT (41).
Samples included in the study. (i) Porcine samples from field ASFV-infected areas within the EU.
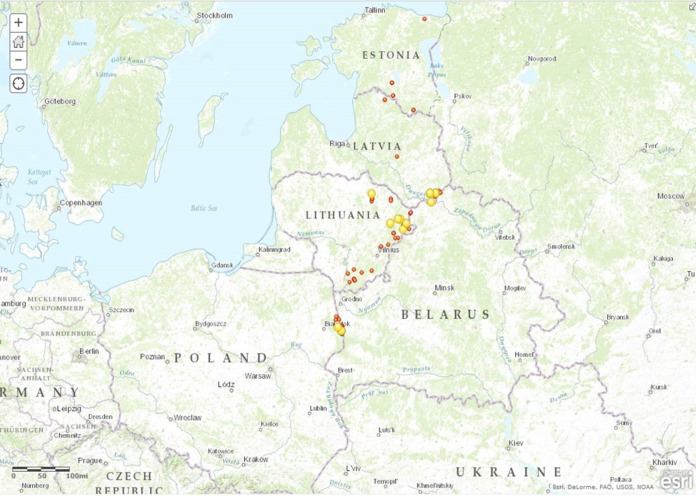
A panel of 314 field samples collected during the 2014 outbreaks in EU countries (Latvia, Lithuania, Poland, and Estonia) were used in this study (Fig. 1). These samples were taken from a total of 125 infected animals (91 wild boars and 34 domestic pigs) previously identified by the NRLs. Specifically, in the case of wild boars, 182 tissue samples, mainly from the spleen (32.2%) and bone marrow (21.3%), and 22 blood, 5 serum, and 2 fluid samples from the peritoneal cavity were analyzed (Table 1). For domestic pigs, the tests included 70 tissue, 17 blood, and 16 serum samples (Table 1). The tissues tested comprised 28 spleen (40%), 16 kidney (22.9%), 11 lung (15.7%), 10 lymph node (14.3%), 4 tonsil (5.7%) and 1 diaphragm muscle (1.4%) sample.
FIG 1.
Map of the sites in EU countries where samples were collected from wild boar (red) and domestic pigs (yellow).
TABLE 1.
Description of the 314 tested field samples obtained from epidemic outbreaks in EU countries in 2014
| Country by sample typea | No. of animals tested | Total no. of samples | No. of samples analyzed by type |
|||
|---|---|---|---|---|---|---|
| Tissue | Blood | Serum | Fluid | |||
| Wild boar samples | ||||||
| Poland | 27 | 114 | 97 | 15 | 0 | 2 |
| Lithuania | 45 | 48 | 41 | 5 | 2 | 0 |
| Latvia | 8 | 26 | 23 | 1 | 2 | 0 |
| Estonia | 11 | 23 | 21 | 1 | 1 | 0 |
| Total WB | 91 | 211 | 182 | 22 | 5 | 2 |
| Domestic pig samples | ||||||
| Poland | 6 | 51 | 40 | 6 | 5 | 0 |
| Lithuania | 21 | 32 | 13 | 11 | 8 | 0 |
| Latvia | 7 | 20 | 17 | 0 | 3 | 0 |
| Estonia | 0 | 0 | 0 | 0 | 0 | 0 |
| Total DP | 34 | 103 | 70 | 17 | 16 | 0 |
| Total samples | 122 | 314 | 252 | 39 | 21 | 2 |
WB, wild boar; DP, domestic pig.
(ii) Porcine samples from ASFV experimental studies.
One hundred fifty paired serum and blood-EDTA samples were collected at regular intervals until the end of the study from three independent experimental infections with virulent ASFV isolates belonging to P72 genotype II. In addition, 15 different types of tissues and organs were obtained from each necropsied animal (liver, spleen, tonsil, heart, lung, kidney, and submandibular, retropharyngeal, inguinal, popliteal, mesenteric, mediastinal, gastrohepatic, splenic, and renal lymph nodes). This gave a total of 450 tissue samples that were used in the comparative studies for both antibody and ASFV detection. The number and type of experimental samples are summarized in Table 2. Animal experiments were conducted at the biosafety level 3 (BSL3) animal facilities at the Instituto Nacional de Tecnologia Agraria y Alimentaria-Centro de Investigación en Sanidad Animal (INIA-CISA), in accordance with EC Directive 86/609/EEC (46) regarding the accommodation and care of animals used for experimental and other scientific purposes, as described in the recommendation. A short description of the experimental design is shown below.
TABLE 2.
Description of the tested experimental samples collected from animals infected with ASFV genotype II viruses
Experimental infection with ASFV Arm07 isolate.
Four Landrace x Large White pigs were inoculated intramuscularly with 10 HAD50/ml of the ASFV Armenia 2007 (Arm07) isolate. Two untreated pigs were maintained in contact and housed in the same box as the inoculated animals. Inoculated and in-contact animals developed acute forms of clinical disease and were slaughtered or died as a result of the infection between 7 to 9 days postinoculation (dpi) (inoculated group) or 16 days postexposure (dpe) (in-contact group) (39).
Experimental infection with the ASFV Ukr12/Zapo isolate.
Four domestic pigs were inoculated by the intramuscular route with 10 HAD50/ml of the Ukraine ASFV Ukr12/Zapo isolate. The inoculated pigs were placed in contact with two naive pigs being housed in the same box. All the pigs developed a peracute to acute form of the disease and died, or were slaughtered in an ethical manner, between 4 and 10 dpi (infected pigs) and 11 to 12 dpe (in-contact pigs) (C. Gallardo, personal communication, 2013).
Experimental infection with ASFV LT14/1490 isolate.
Ten naive pigs were placed in contact with eight pigs experimentally inoculated by the intramuscular route with 10 HAD50/ml of the Lithuanian LT14/1490 strain. The Lithuanian ASFV strain induced an acute disease, which resulted in 94.5% mortality. Seven of the eight inoculated animals died or were euthanized due to the severity of symptoms between 7 and 9 dpi. One inoculated pig showed a delayed course of the disease, resembling the same as that seen in the in-contact animals, which died or were slaughtered from 14 to 22 dpe. One in-contact pig remained asymptomatic throughout the experiment and was slaughtered at day 61 (47).
ASFV detection tests. (i) Samples tested.
The number and type of samples tested in parallel by the selected ASFV detection tests are summarized in Table 3. Briefly, 295 field-collected samples and 600 samples obtained from the experimental infections were tested in parallel by the PCR assays. The field samples consisted of 252 tissue, 39 blood-EDTA, two serum, and two fluid samples, while 150 blood and 450 tissue samples were analyzed from the experimental infections.
TABLE 3.
Number of field and experimental samples tested by the ASFV and antibody detection tests
| Sample type | No. of samples by type and test |
|||||||
|---|---|---|---|---|---|---|---|---|
| ASFV detection tests |
ASF antibody detection tests |
|||||||
| ASFV genome (PCRs) |
ASFV antigen detection (Ag-ELISA)a |
ELISAs |
IPT |
|||||
| Field samples | Experimental samples | Field samples | Experimental samples | Field samples | Experimental samples | Field samples | Experimental samples | |
| Tissue | 252 | 450 | 67 | 30 | 0 | 300b | 140 | 300b |
| Blood-EDTA | 39 | 150 | 25 | 150 | 0 | 0 | 26 | 0 |
| Serum | 2 | 0 | 0 | 0 | 21 | 150 | 21 | 150 |
| Fluid | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Total | 295 | 600 | 92 | 180 | 21 | 450 | 188 | 450 |
Ag, antigen.
Corresponds to spleen, lung, and liver samples obtained from 90 ASFV genotype II-infected domestic pigs and 210 samples obtained from 70 non-ASFV-infected animals.
In addition, 272 samples were tested to detect ASF antigen by ELISA and included 92 field samples from 79 animals (67 spleen and 25 blood samples) and 180 experimental samples from 30 infected domestic pigs (30 spleen and 150 blood samples). The type of samples tested in the antigen ELISA included spleen and blood, as is recommended by the manufacturers.
(ii) ASFV genome detection by PCR.
The field and experimental samples were tested to detect the ASFV genome using the OIE conventional PCR (33, 36), the OIE real-time PCR (33, 37), and the Universal ProbeLibrary (UPL) real-time PCR (38). Briefly, 10% (wt/vol) clarified homogenized tissue suspensions were prepared in phosphate-buffered saline using field and experimental tissue samples. The DNA was extracted from each tissue homogenate and from blood, serum, and fluid samples using the High Pure PCR template preparation kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany). For amplification of the ASFV genomic DNA, the PCRs were carried out using undiluted and 1:10 diluted extracted DNA for each sample.
(iii) ASFV antigen detection by ELISA.
A total of 272 samples were tested to detect ASF antigen using the commercially available antigen detection ELISA, the double-antibody sandwich (DAS) ELISA manufactured by Ingenasa (Ingenasa-Ingezim PPA DAS K2; Ingenasa, Madrid, Spain). The samples were analyzed undiluted and in a 1:10 dilution, according to the manufacturer's instructions, and the results obtained were compared to those obtained using the PCR assays.
(iv) ASF virus isolation and titration.
Virus isolation was assessed in PCR-positive experimental and field samples using a hemadsorption assay into PBM, as described in the OIE Terrestrial Manual (33). The plates were examined for hemadsorption over a period of 6 days. The samples were blind passaged three times. Titers were estimated by endpoint dilution, as described in “Viruses, Cell Cultures, and Virus Propagation” above.
ASF antibody detection tests. (i) Samples tested.
The number and type of samples tested in parallel by the selected ASF antibody detection tests are shown in Table 3. In summary, 150 experimental and 21 field serum samples were tested using the ELISAs and IPT. The performance of each technique for detecting ASF antibodies in tissue exudate was initially assessed by the analysis of 90 experimental exudates obtained from the spleens, livers, and lungs of the ASFV genotype II-infected domestic pigs. To determine the specificity of the assays, a panel of 210 negative tissue exudates obtained from 70 non-ASF-infected animals were included in the study. In addition, 140 field tissue exudates, 26 blood, and one fluid sample taken during the outbreaks within the EU were tested by IPT.
(ii) ELISAs.
The four different ELISAs included in the comparative study were the OIE indirect ELISA based on the ASFV semipurified antigen (33) and the three following tests: the (i) blocking Ingenasa-ELISA based on the use of monoclonal antibody against the P72 ASFV protein (Ingenasa-Ingezim PPA Compac K3; Ingenasa, Madrid, Spain), (ii) IDvet-ELISA, a multiantigen indirect ELISA kit for the detection of antibodies against P32, P62, and P72 ASFV proteins (ID Screen African swine fever indirect assay; Grabels, France), and (iii) indirect Svanova-ELISA based on recombinant P30 ASFV proteins as antigen (Svanovir ASFV-Ab; Boehringer Ingelheim Svanova, Uppsala, Sweden). Sera and exudate tissues were tested by each ELISA, according the manufacturer's instructions for serum samples.
(iii) IPT.
For the preparation of IPT antigen plates, 80% confluent monolayers of MS cells were grown in Eagle fresh medium without serum in 96-well tissue culture-grade microtiter plates. The plates were infected with a multiplicity of infection (MOI) of 20 with ASFV MS adapted-E70 isolate (E70 MS 48) and incubated in a humidified atmosphere containing 5% CO2 at 37°C for 24 h. After incubation, the inocula were removed by vacuum suction, and the ASFV-infected cell sheets were fixed with a cold solution containing 70% methanol and 30% acetone for 10 min. Finally, the plates were washed with phosphate-buffered saline (PBS) for 20 min, sealed with tape, and stored at −20°C until further use. Experimental and field samples were tested by IPT using the standardized operating procedure, as described by the EURL (48).
Data analysis.
The concordance between each test was the overall percentage agreement between the results of the two assays calculated using two-by-two contingency tables. Kappa coefficient (κ) statistics were used to evaluate the significance of the level of concordance between results beyond that expected by chance, with κ values of 0.81 to 1.00 representing almost perfect agreement, values of 0.61 to 0.80 representing substantial agreement, values of 0.41 to 0.60 representing good agreement, values of 0.21 to 0.40 representing moderate agreement, values of 0.01 to 0.20 representing slight agreement, and values of 0.00 representing no agreement (49). From the overall analysis of the results, the final sensitivity and specificity were calculated using the results of the IPT and the UPL-PCR as a reference for antibody and virus detection, respectively. All samples that gave a doubtful result in the ELISAs (those with results in the cutoff interval) were considered positive.
RESULTS
Comparison of PCRs and antigen ELISA in the detection of ASFV. (i) Experimental samples from animals infected with genotype II ASFV.
(a) Analysis of blood samples.
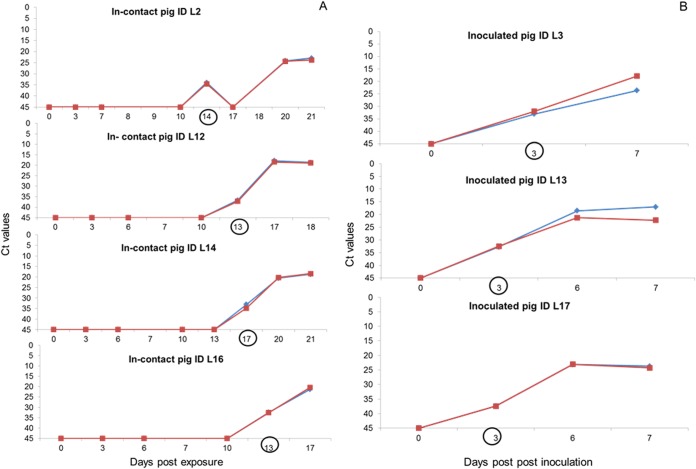
Out of the 150 blood samples collected at different times from the 30 experimentally infected pigs, the number of positives was 62 (41.3%) using the UPL-PCR, 59 (39.9%) using the OIE real-time PCR, and 52 (34.7%) using the OIE conventional PCR. When the samples were analyzed with the antigen ELISA, 48 tested positive (32%) according to the threshold recommended by the ELISA kit protocol (Table 4). Good correlation was observed between the UPL-PCR and the OIE real-time PCR, with just three of the 62 UPL positive samples (with high cycle threshold [CT] values of >36) testing negative in the OIE real-time PCR. Three false-negative blood samples were collected at 17, 34, and 38 dpe from in-contact animals exposed to the virulent Lithuanian 2014 ASFV isolate (LT14/1490), which remained asymptomatic throughout the experimental infection (47). The false-negative results detected with both conventional PCR and/or antigen ELISA were correlated with blood samples with real-time PCR cycle threshold (CT) values of >30, mainly collected at the initial stages of infection (Fig. 2).
TABLE 4.
Comparison of UPL real-time PCR, OIE real-time PCR, OIE conventional PCR, and antigen ELISA results for the detection of ASFV in blood and tissues collected from pigs experimentally infected with genotype II ASFV isolates
| ASFV strain by sample type | No. of pigs examined | No. of samples examined | UPL-PCR |
OIE real-time PCR |
OIE conventional PCR |
Ag-ELISA (Ingenasa) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of positive samples | % | No. of positive samples | % | No. of positive samples | % | No. of positive samples/total no. of animals | % | |||
| Blood samples | ||||||||||
| Ukr12/Zapo | 6 | 19 | 10 | 52.6 | 10 | 52.6 | 10 | 52.6 | 9 | 47.3 |
| Arm07 | 6 | 20 | 10 | 50 | 10 | 50 | 10 | 50 | 10 | 50 |
| LT14/1490 | 18 | 111 | 42 | 37.83 | 39 | 35.13 | 32 | 29.8 | 29 | 26.12 |
| Total | 30 | 150 | 62 | 41.3 | 59 | 39.3 | 52 | 34.7 | 48 | 32.0 |
| Tissue samples | ||||||||||
| Ukr12/Zapo | 6 | 90 | 90 | 100 | 90 | 100 | 90 | 100 | 6/6a | 100 |
| Arm07 | 6 | 90 | 90 | 100 | 90 | 100 | 90 | 100 | 6/6a | 100 |
| LT14/1490 | 18 | 270 | 260 | 96.29 | 255 | 94.4 | 255 | 94.4 | 16/18a | 88.8 |
| Total | 30 | 450 | 440 | 97.8 | 435 | 96.6 | 435 | 96.6 | 28 | 93.3 |
In Ag-ELISA, spleen samples for each pig were included in the study.
FIG 2.
Comparative viremia results determined by the OIE real-time PCR (red) and UPL real-time PCR (blue) in blood samples collected from exposed (A) and inoculated (B) pigs using the ASFV genotype II Lithuania 2014 isolate. The black circles indicate the day postexposure/postinoculation at which a false-negative result was obtained with the OIE conventional PCR and antigen ELISA.
(b) Analysis of tissue samples.
A second panel of 450 tissue samples collected during necropsy was analyzed in parallel using the three PCR assays. The ASFV genome was detected by the UPL-PCR in 97.8% of the tested tissues, while in the OIE-prescribed PCR assays, the positive percentage decreased to 96.6%. The 15 negative samples corresponded to organs collected from two pigs exposed to the Lithuania 2014 strain (47). Of the 30 spleen samples tested using the antigen ELISA, 28 were positive and two were negative, which enabled us to detect 93.3% of the infected animals (Table 4).
(ii) Field samples collected during the EU epidemic outbreaks.
Using the UPL-PCR, the 295 tested field samples were positive. Compared with the real-time and conventional OIE-PCR-prescribed assays, the percentages of positive results decreased to 98.98% and 96.3%, respectively (Table 5). The OIE real-time PCR failed to identify two blood samples and one bone marrow sample that were UPL positive, with CT values of >35. Using the OIE conventional PCR test, the number of negative samples increased to 10, all with CT values being >30 when tested with the real-time PCRs.
TABLE 5.
Comparison of the three PCR tests used to detect the ASFV genome in field-collected samples from wild boars and domestic pigs during the epidemic outbreaks in EU countries
| Sample type | No. of samples tested | UPL-PCR |
OIE real-time PCR |
OIE conventional PCR |
|||
|---|---|---|---|---|---|---|---|
| No. of positive samples | % | No. of positive samples | % | No. of positive samples | % | ||
| Tissue | 252 | 252 | 99.6 | 250 | 99.2 | 246 | 97.6 |
| Blood-EDTA | 39 | 39 | 100 | 38 | 97.4 | 36 | 92.3 |
| Serum | 2 | 2 | 100 | 1 | 50 | 0 | 0 |
| Fluid | 2 | 2 | 100 | 2 | 100 | 2 | 100 |
| Total | 295 | 295 | 99.7 | 291 | 98.64 | 284 | 96.3 |
The performance using the antigen ELISA in detecting field-infected animals was assessed by the analysis of 92 field samples (67 spleen and 25 blood samples), all previously tested as positive using the UPL-PCR. The percentage of positive results with the antigen ELISA was 71.74%, corresponding to the detection of 66 samples (52 spleen and 14 blood samples) obtained from 65 of the 79 investigated pigs (data not shown).
(iii) Analysis of the results.
Taking the UPL-PCR test as a reference method able to detect 100% of the infected or exposed animals, 12 false negatives were detected out of 797 ASF positive samples with the OIE real-time PCR (98.5% sensitivity, [95% confidence interval {CI}, 97.4 to 99.1%]) and 26 with the OIE conventional PCR (96.7% sensitivity, [95% CI, 95.3 to 97.8%]). The overall analysis of experimental and field samples showed almost perfect agreement between the UPL-PCR and the real-time (κ = 0.94 [95% CI, 0.91 to 0.97]) and conventional (κ = 0.88 [95% CI, 0.83 to 0.92) OIE-prescribed PCRs. Good-to-moderate agreement (κ = 0.67 [95% CI, 0.58 to 0.76]) between the antigen ELISA and the UPL test gave a sensitivity of 77.2% (95% CI, 70.6 to 82.6%). Comparative sensitivity values are shown in Table 6.
TABLE 6.
Comparative sensitivity results obtained using the OIE-prescribed PCRs and the commercial antigen ELISA (Ingenasa) used for analyzing field and experimental samples tested previously as positives with UPL-PCR as a reference test
| Sample type | OIE real-time PCR |
OIE conventional PCR |
Ag-ELISA Ingenasa |
|||
|---|---|---|---|---|---|---|
| No. of positive samples/total no. | Ss (% [95% CI])a | No. of positive samples/total no. | Ss (% [95% CI]) | No. of positive samples/total no. | Ss (% [95% CI]) | |
| Experimental | 494/502 | 98.4 | 487/502 | 97.0 | 76/92 | 82.6 |
| Field | 291/295 | 98.6 | 284/295 | 96.3 | 66/92 | 71.7 |
| Total | 785/797 | 98.5 (97.4–99.1) | 771/797 | 96.7 (95.3–97.8) | 142/184 | 77.2 (70.6–82.6) |
Ss, sensitivity.
Virus isolation versus UPL real-time PCR results.
Virus isolation was attempted in the 502 UPL-PCR-positive blood and tissue samples obtained from the domestic pigs experimentally infected with genotype II. ASFV was successfully isolated after three passages in PBM cells in 96.81% of the tested samples showing the characteristic hemadsorption ASFV pattern. No viable virus was detected in samples with UPL CT values of 36 ± 3.5, mainly collected at the initial stage of infection from pigs infected with the Lithuanian ASFV isolate. In contrast, when 185 field-derived samples representative of each positive domestic pig and wild boar were subjected to virus isolation, the virus was successfully isolated in 77 cases (41.62%). Samples that exhibited unsuccessful results were mainly derived from wild boars and resulted in 27 ASFV viruses isolated (30.7%) from a total of 91 animals tested. In domestic pigs, viruses were recovered after three passages in 29 out of 34 domestic pigs (86%), although no relationship was established between the negative samples and the CT values reported by the UPL-PCR. It is important to note that all ASFV field viruses were hemadsorbing.
Comparison of the ELISAs and IPT assay for antibody detection using pig sera. (i) Experimental sera from animals infected with genotype II ASFV.
Due to the acute nature of the disease induced by the ASFV genotype II isolates, most of the animals died or were slaughtered prior to the development of measurable antibodies. The analysis of sera collected from inoculated and in-contact animals revealed a detectable antibody response by IPT between 16 and 21 dpi/dpe in 23.3%, which corresponds to 7 out of the 30 domestic pigs. The level of detection was reduced to three (10%) with the Ingenasa-ELISA and two (6.6%), with the OIE-, IDvet-, and Svanova-ELISAs (Table 7).
TABLE 7.
Comparative IPT and ELISA results obtained in serum samples from seroconverted animals experimentally infected with genotype II ASFV isolates
| ASFV isolate | Animal identification | dpi/dpea | Result forb: |
||||
|---|---|---|---|---|---|---|---|
| IPT | OIE-ELISA | Ingenasa-ELISA | IDvet-ELISA | Svanova-ELISA | |||
| Arm07 | Contact pig 5 | 16 | Pos | Neg | Pos | Neg | Neg |
| LT14/1490 | Inoculated pig 6 | 18 | Pos | Neg | Neg | Neg | Neg |
| LT14/1490 | Contact pig 2 | 21 | Pos | Neg | Neg | Neg | Neg |
| LT14/1490 | Contact pig 10 | 17 | Pos | Neg | Neg | Neg | Neg |
| LT14/1490 | Contact pig 11 | 17 | Pos | Neg | Neg | Neg | Neg |
| LT14/1490 | Contact pig 11 | 18 | Pos | Pos | Pos | Pos | Pos |
| LT14/1490 | Contact pig 12 | 18 | Pos | Neg | Neg | Neg | Neg |
| LT14/1490 | Contact pig 15 | 17 | Pos | Pos | Pos | Pos | Pos |
dpi, days postinfection; dpe, days postexposure.
Pos, positive; Neg, negative.
(ii) Field serum samples from pigs in affected areas in the EU.
Twenty-one samples collected in 2014 from domestic pigs (16) and wild boars (5) from affected EU countries were analyzed in parallel using the four ELISAs and the IPT. As with the results obtained using the experimental serum samples, due to the acute nature of the disease, the number of positive sera detected by IPT was 10 (47.62%), whereas the number decreased to six (37.5%) with the Ingenasa-ELISA, four (25%) with the IDvet- and Svanova-ELISAs, and two (9.52%) with the OIE-prescribed ELISA. ASFV antibody titers were determined in sera using IPT by endpoint dilution, as described by EURL (2014) (48). The results showed that the ELISAs were unable to detect the infected pigs with antibody titers (reported as 2-fold dilutions by IPT) of <1:640 (9.32 log2) in the Ingenasa-ELISA, <1:5,210 (12.32 log2) for the IDvet and Svanova tests, and <1:20,480 (14.32 log2) in the case of the OIE-ELISA (data not shown).
(iii) Analysis of the results.
From the results obtained in both experimental and field samples collected from animals with known infectious status, the IPT was selected as the reference method. Of the 18 ASF-positive samples, there were 14 false negatives from the OIE-ELISA, 12 from the IDvet- and Svanova-ELISAs, and 9 from the Ingenasa-ELISA. Due to the low number of positive samples, the sensitivity values ranged from 22.22% using the OIE-ELISA to 50% with the Ingenasa test. Kappa values from 0.34 (95% CI, 0.01 to 0.67) to 0.64 (95% CI, 0.42 to 0.87) showed moderate-to-substantial agreement between the IPT and the ELISAs. The comparative sensitivity values are shown in Table 8.
TABLE 8.
Comparative sensitivity results obtained using the commercial ELISAs to analyze field and experimental sera tested previously as 18 positives, with IPT as a reference test
| Sample type | OIE-ELISA |
Ingenasa-ELISA |
IDvet-ELISA |
Svanova-ELISA |
||||
|---|---|---|---|---|---|---|---|---|
| No. of positive samples/total no. | Ss (%) | No. of positive samples/total no. | Ss (%) | No. of positive samples/total no. | Ss (%) | No. of positive samples/total no. | Ss (%) | |
| Experimental | 2/8 | 25 | 3/8 | 37.5 | 2/8 | 25 | 2/8 | 25 |
| Field | 2/10 | 20 | 6/10 | 60 | 4/10 | 40 | 4/10 | 40 |
| Total | 4/18 | 22.22 | 9/18 | 50 | 6/18 | 33.3 | 6/18 | 33.3 |
Performance of the ELISAs and IPT assays in antibody detection in exudate tissue samples. (i) Experimental infections.
Ninety exudate tissues obtained from animals infected with genotype II ASFV isolates were subjected to the ELISA and the IPT assay for antibody detection. Specific anti-ASFV antibodies were detected with Ingenasa-ELISA in 25 out of 90 exudates from 18 out of 30 experimentally infected animals. The IPT detected 10 of the infected pigs with 20 positive exudates, whereas the OIE-, Svanova-, and IDvet-ELISAs detected a decreasing number of positive animals, with 5 (6 samples), 3 (6 samples), and 2 (3 samples) animals, respectively.
To confirm the specificity (Sp) of the results obtained in the analysis of experimental samples, a total of 210 exudates from negative spleen, lung, and liver tissues from 70 healthy domestic pigs were analyzed in parallel with the ELISAs and IPT. No specific antibody response was detected with the IPT or IDvet-ELISA, thereby showing 100% specificity. The number of false-positive samples was five with the OIE-ELISA (Sp, 97.6%), 18 with the Svanova-ELISA (Sp, 91.4%), and 33 using the Ingenasa-ELISA (Sp, 84.3%). The false-positive samples were associated with hemolyzed exudates, mainly obtained from the spleen (72.7% of false positives) and liver (24.2% of false positives).
Statistically, the Ingenasa-ELISA showed greater sensitivity than that of IPT and the other ELISAs used to detect ASF antibodies in positive exudate samples. However, its specificity (84.3%) was the lowest in detecting negative exudate samples. Combining the results obtained from all tested exudates of the infected and noninfected animals, the positive predictive value (PPV) of this technique was found to be 17.2% and its negative predictive value (NPV) to be 95.9%. Therefore, the accuracy of Ingenasa-ELISA compared to IPT was found to be 80.7%.
(ii) Field infections.
Taking into consideration the results obtained from experimental samples, the IPT was selected for the analysis of exudate tissue samples collected in the field during the epidemic EU outbreaks. A total of 140 field tissue exudates, 26 blood, and 1 fluid sample from 32 domestic pigs and 51 wild boars were tested. A specific antibody response was detected in 87 out of the 167 samples from 30 wild boars (58.8%) and 11 domestic pigs (34.37%). A 2-fold serial dilution of the samples containing specific antibodies determined high ASF antibody IPT titers ranging from 1:1,280 to 1:40,960 (10.32 to 15.32 log2) in nine of the 30 positive wild boars (30%), whereas only three domestic pigs had titers >10.32 log2.
DISCUSSION
ASF is a very complex disease for which no vaccine is available. Prevention and control of ASF are based on two main principles, early detection (based on epidemiological, clinical, and laboratory findings) and strict sanitary measures. Therefore, the use of the most appropriate diagnostic tools that are updated to be applicable to all scenarios is critical for the implementation of effective control programs. Previous studies have examined the performance of serological (40, 42) and virological (34) ASF diagnostic tests with a relatively large number of samples, which were mainly obtained from the African continent. The recent emergence of ASF in 2014 in some eastern EU countries has highlighted the need to assess ASF diagnostic tests to check their ability to detect the disease via viral DNA, antigens, and antibodies in clinical specimens in infected areas. This study provides a complete analysis that compares the most typical ASF diagnostic assays used at the EU level with a set of samples collected from field and experimentally infected animals with genotype II ASFV isolates currently circulating in eastern and central Europe (21, 28, 50).
Three PCR assays for the detection of the ASFV genome (33, 36–38) were tested in parallel using 785 field and experimental samples obtained from genotype II-infected pigs. In the data obtained, 3.3% more samples were shown to be ASFV positive with the UPL-PCR method (38) than with the OIE conventional PCR (33, 36). The DNA was easily detected by both PCRs when high levels of virus were present in blood and tissues in the clinical phase of infection. However, the OIE conventional PCR failed to give positive results for samples with CT values of >30, especially those collected during early stages of the disease. This lower sensitivity might be due to the presence of one nucleotide mismatch close to the 3′ end in the forward primer, identified in the target sequence of recent ASFV genotype II isolates, which include Georgia 2007 (GenBank accession no. FR682468), Krasnodar 2012 (GenBank accession no. KJ195685), Lithuania 2014, and Poland 2014 (data not shown).
There was a slight difference (P = 0.015) between the total number of UPL-PCR-positive samples and the total number of OIE real-time PCR (33, 37)-positive results. The OIE-prescribed assay failed to detect the asymptomatic pig experimentally exposed to Lithuanian ASFV (49) and three field-derived samples from hunted wild boars in Lithuania and Latvia. Although no virus was recovered from these samples, the presence of antibodies indicates ASFV exposure in these wild boars and confirms the specificity of the results obtained. Overall, the comparative PCR results presented in this study agree with previous work in which the UPL-PCR was able to detect more positive samples than the OIE-PCR tests, thereby confirming its superior diagnostic sensitivity in the detection of survivors and its ability to rapidly detect the disease even when the typical clinical signs are as yet not evident (38).
Published data on the sensitivity and specificity of the currently available commercial antigen ELISA kit (ELISA Ingezim K2; Ingenasa, Madrid, Spain) are limited to the results obtained with a few African samples (34). This antigen ELISA is cheaper to set up than PCR methods and allows for large-scale testing of samples to be conducted quickly without any special laboratory equipment. In this study, we compared the performance of this assay with the three PCR methods described above and analyzed a panel of 277 samples from experimentally infected pigs and field samples from wild boars and domestic pigs in infected areas in the EU. The sensitivity of the antigen ELISA was poor (77.2%) compared to that of the UPL-PCR, most of all in the case of field-derived samples, even when there was a high virus load. This corresponds with previous studies that have shown that field samples in poor conditions can decrease the effective sensitivity of the test (34, 51). It is also important to note that the formation of the antigen-antibody complexes in the tissues of seropositive animals can block the interaction between the ASFV antigen and ASFV conjugate and therefore affect its sensitivity. In the majority of cases, the severe nature of the epidemic disease affecting EU countries leads to mortality with high levels of viral presence in all tissues (9, 10, 47, 52, 53). It is therefore highly likely that the virus will be detected using the antigen ELISA in a high proportion of samples taken from dead pigs during an acute outbreak of disease. However, from the results obtained in this study, the use of the antigen ELISA is recommended as a herd test and should be accompanied by other virological tests.
The attempt to isolate infectious virus from each UPL-PCR-positive animal gave irregular results in experimental and field samples. Although the virus was easily isolated in experimental samples when CT values remained at <36 ± 3.5, variable results were obtained when field-derived samples were subjected to virus isolation, even despite the higher values of viral DNA detected. This low effectiveness in virus isolation in field samples might be related to the poor state of samples, which will affect the viability of the virus, especially if we bear in mind the high percentage of material received from hunted or dead wild boars. To compare acute-phase-sample assays, virus isolation is not likely to be the most fruitful approach, as it is more expensive than other techniques and requires both specialized facilities and training. However, virus isolation and identification by hemadsorption tests (HAD) are recommended as a reference test when ASF has been confirmed by other methods, particularly in the event of a primary outbreak or a case of ASF (54). In addition, virus isolation is essential if the objective is to obtain virus stocks for future molecular and biological characterization studies.
Serological assays are the most commonly used diagnostic tests due to their simplicity, relatively low cost, and need for little specialized apparatus or few facilities. For ASF diagnosis, this is particularly relevant given that no vaccine is available against ASFV, which means that the presence of anti-ASFV antibodies always indicates infection. Furthermore, anti-ASFV antibodies appear soon after infection and persist for up to several months or even years (35). The present study compares the performance of the four available screening ELISAs with the confirmatory IPT in detecting ASF antibodies in field and experimental serum samples.
The examination of sequential serum samples from the 30 domestic pigs experimentally infected with genotype II ASFV indicated greater sensitivity for the IPT than for the ELISAs. The IPT was able to detect ASF antibodies at an earlier stage of serological response than the ELISA. The same result was obtained from the analysis of field-derived samples despite the limited number of tested sera. The sensitivity of the ELISAs, based on the number of positive samples detected by each assay, was significantly lower than that of the IPT as a reference test and ranged from 22.22% (OIE-ELISA) to 50% seropositive pigs detected (Ingenasa-ELISA). Kappa analysis showed that the IDvet- and Svanova-ELISAs were not statistically different from each other and were both superior to the OIE-ELISA. The sensitivities of the assays determined by this study were lower than or equivalent to those in other published reports, especially to those using the OIE-recommended ELISA (42). Although this study was limited by the small positive sample size, this low sensitivity may be due to the fact that samples were collected from acutely infected animals prior to the development of the antibodies that are measurable by the ELISAs. Notably, sera showing ASF IPT antibody titers of >14.32 log2 were found to be positive in all the ELISAs.
Our tests fell into two distinct methodological categories, ELISA and IPT, each with its advantages and disadvantages. The ELISA format permits the rapid testing and interpretation of a large number of samples and can be easily automated. However, the relatively lower sensitivity observed for the ELISA compared to that of IPT suggests that seroprevalence rates may be underestimated when the ELISA is employed for surveillance in areas where virulent strains inducing acute infections are circulating, as is the case in certain EU countries. The data presented here reaffirm IPT as a useful serological tool for the early diagnosis of ASF, even when few antibodies are present. Nevertheless, it should be kept in mind that IPT is a labor-intensive method that requires individual microscopic examination of all samples and that interpretation can vary according to the examiner. Therefore, the development of new screening ELISAs capable of detecting low ASF antibody titers should be contemplated.
The spread of ASF into the wild boar population of the EU has highlighted the need for targeted surveillance and early warning actions (32, 55). Samples usually obtained from hunted/captured animals, animals found dead, and animal debris are tested to determine the presence of the disease. Based on the above-mentioned results, the presence of the ASFV genome is easily determined by PCR even when bone material is tested. However, the search for antibodies from hunted or dead animals is essential for obtaining a complete picture of the epidemiology in question at the time of these epidemic outbreaks and for determining the date of infection. In this study, the performance of the IPT and the four ELISAs was evaluated to detect specific ASFV antibodies in tissue exudates, thereby exploring their potential as alternative diagnostic samples. These tissue exudate samples were obtained from (i) naive, (ii) experimentally inoculated, and (iii) field-infected domestic and wild pigs. An initial evaluation based on the analysis of experimental samples revealed the Ingenasa-ELISA to be the most sensitive kit, as it detected antibodies in 60% of the infected pigs. Using the IPT, the percentage decreased to 30% and to <20% when using the OIE-, IDvet-, and Svanova-ELISAs. However, our data demonstrate that when the validation experiment was extended to include additional negative samples, only the specificity index of IPT and IDvet-ELISA remained high (100%), since the Ingenasa-ELISA generated a significantly reduced specificity (84.3%). Several studies have previously reported that hemolyzed sera may influence ELISA performance. Also, the manufacturers do not recommend the use of this type of sample in the ELISA. Therefore, these divergent results were most likely due to the use of hemolyzed samples obtained from tissues (51). Our results also underline the fact that positive ELISA results should always be confirmed by alternative methods, such as IPT, FAT, or IB tests, as is recommended by the OIE (33).
To determine the date of infection, that is, when infected animals were exposed to ASFV (regardless of whether antibodies were present), samples from wild boars and domestic pigs were tested using IPT. From the overall analysis of the results (including those obtained from sera), the presence of antibodies was confirmed in 60.37% of the wild boars and in 46.8% of the domestic pigs. Moreover, the IPT antibody titers stressed that wild boars generally had higher levels of antibody titers than those of domestic pigs. An interesting finding is that animals with the highest antibody titers were previously diagnosed at the limit of detection with the UPL-PCR test but resulted negative when tested with the OIE-prescribed virological assays. These data suggest that despite the fact that the ASFV isolates affecting EU countries correspond to a virulent strain that leads to high mortality in affected wild and domestic pigs, some animals can survive for over a month and are able to recover from the infection or even remain subclinically infected (47, 56). The role of such animals in the transmission and maintenance of the disease needs to be further investigated.
In conclusion, although a number of good validated ASF diagnostic techniques are available, the data presented in this paper show that the UPL-PCR in combination with the IPT are the most trustworthy methods for the early detection of the ASFV genome and antibodies in affected EU countries. These techniques are accurate and specific for a reliable diagnosis of ASF, though the OIE real-time PCR test can be also used with great confidence, since it shows almost perfect agreement with the UPL-PCR. Virus isolation produces highly variable results when field samples are tested but is still the test of choice to confirm an outbreak in a free country and to obtain virus for subsequent detailed analysis. Antibody detection techniques are needed in order to get complete information that will assist control and eradication programs. Despite its shortcomings, the ELISA is the most commonly used type of test to screen for ASF antibodies, although further confirmation of positive and inconclusive ELISA results is always required. In this context, the IPT is the best test given its superior sensitivity and its performance to test blood, serum, and/or exudate tissue samples, which is particularly relevant for wild boar surveillance and control programs. In light of the current epidemic situation in EU countries, we recommend that IPT be used from wild boars and to test for ASF antibodies when sick or dead domestic pigs appear. The success of surveillance activities is dependent on the availability of the most appropriated diagnostic tests that can provide reliable information for feasible control and eradication programs.
ACKNOWLEDGMENTS
We thank our colleagues from the National Reference Laboratories for ASF in Poland, Lithuania, Latvia, and Estonia, and from the European Union reference laboratory for ASF for their intellectual and practical contributions.
This study was supported by the European Union Reference laboratory for ASF (grant UE- LR PPA/03).
REFERENCES
- 1.Sánchez-Vizcaíno JM, Mur L, Gomez-Villamandos JC, Carrasco L. 2015. An updated on the epidemiology and pathology of African swine fever. J Comp Pathol 152:9–21. doi: 10.1016/j.jcpa.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Plowright W, Parker J, Peirce MA. 1969. African swine fever virus in ticks (Ornithodoros moubata, Murray) collected from animal burrows in Tanzania. Nature 221:1071–1073. doi: 10.1038/2211071a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson PJ. 1986. Epidemiology of African swine fever. Rev Sci Tech Off Int Epiz 5:487–493. [DOI] [PubMed] [Google Scholar]
- 4.Penrith ML, Thomson GR, Bastos AD, Phiri OC, Lubisi BA, Du Plessis EC, Macome F, Pinto F, Botha B, Esterhuysen J. 2004. An investigation into natural resistance to African swine fever in domestic pigs from an endemic area in southern Africa. Rev Sci Tech 23:965–977. [DOI] [PubMed] [Google Scholar]
- 5.Penrith ML, Vosloo W. 2009. Review of African swine fever: transmission, spread and control. J S Afr Vet Assoc 80:58–62. [DOI] [PubMed] [Google Scholar]
- 6.Penrith ML, Vosloo W, Jori F, Bastos AD. 2013. African swine fever virus eradication in Africa. Virus Res 173:228–246. doi: 10.1016/j.virusres.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Jori F, Vial L, Penrith ML, Pérez-Sánchez R, Etter E, Albina E, Michaud V, Roger F. 2013. Review of the sylvatic cycle of African swine fever in sub-Saharan Africa and the Indian Ocean. Virus Res 173:212–227. doi: 10.1016/j.virusres.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Costard S, Mur L, Lubroth J, Sanchez-Vizcaino JM, Pfeiffer DU. 2013. Epidemiology of African swine fever virus. Virus Res 173:191–197. doi: 10.1016/j.virusres.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 9.Blome S, Gabriel C, Dietze K, Breithaupt A, Beer M. 2012. High virulence of African swine fever virus Caucasus isolate in European wild boars of all ages. Emerg Infect Dis 18:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blome S, Gabriel C, Beer M. 2013. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res 173:122–130. doi: 10.1016/j.virusres.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Dixon LK, Escribano JM, Martins C, Rock DL, Salas ML, Wilkinson PJ. 2005. Asfarviridae, p 135–143. In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (ed), Virus Taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press, London, United Kingdom. [Google Scholar]
- 12.Arias M, Sánchez-Vizcaíno JM. 2012. African swine fever, p 396–404. In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW (ed), Diseases of swine, 10th ed John Wiley and Sons, West Sussex, United Kingdom. [Google Scholar]
- 13.de Villiers EP, Gallardo C, Arias M, da Silva M, Upton C, Martin R, Bishop RP. 2010. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 400:128–136. doi: 10.1016/j.virol.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Chapman DA, Tcherepanov V, Upton C, Dixon LK. 2008. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J Gen Virol 89:397–408. doi: 10.1099/vir.0.83343-0. [DOI] [PubMed] [Google Scholar]
- 15.Chapman DA, Darby AC, Da Silva M, Upton C, Radford AD, Dixon LK. 2011. Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus. Emerg Infect Dis 17:599–605. doi: 10.3201/eid1704.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portugal R, Coelho J, Hoper D, Little NS, Smithson C, Upton C, Martins C, Leitao A, Keil GM. 2015. Related strains of African swine fever virus with different virulence: genome comparison and analysis. J Gen Virol 96:408–419. doi: 10.1099/vir.0.070508-0. [DOI] [PubMed] [Google Scholar]
- 17.Bishop RP, Fleischauer C, de Villiers EP, Okoth EA, Arias M, Gallardo C, Upton C. 2015. Comparative analysis of the complete genome sequences of Kenyan African swine fever virus isolates within p72 genotypes IX and X. Virus Genes 50:303–309. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery RE. 1921. On a form of swine fever occurring in British East Africa (Kenya Colony). J Comp Pathol Therap 34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- 19.Sánchez-Vizcaíno JM, Mur L, Martínez-López B. 2012. African swine fever: an epidemiological update. Transbound Emerg Dis 59(Suppl 1):S27–S35. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Vizcaíno JM, Mur L, Martínez-López B. 2013. African swine fever (ASF): five years around Europe. Vet Microbiol 165:45–50. doi: 10.1016/j.vetmic.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, Dwarka R, Onashvili T, Albina E, Dixon LK. 2008. African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis 14:1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Food and Agriculture Organization of the United Nations. 2013. African swine fever in the Russian Federation: risk factors for Europe and beyond. EMPRES Watch 28:1–13. http://www.fao.org/docrep/018/aq240e/aq240e.pdf. [Google Scholar]
- 23.World Organisation for Animal Health. 2012. Immediate notification report. Ref OIE: 12168. World Organisation for Animal Health, Paris, France: http://web.oie.int/wahis/reports/en_imm_0000012168_20120731_134719.pdf. [Google Scholar]
- 24.World Organisation for Animal Health. 2014a. African swine fever, Ukraine. World Organisation for Animal Health, Paris, France: http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=14625. [Google Scholar]
- 25.World Organisation for Animal Health. 2013. Immediate notification report. Ref OIE: 13663. World Organisation for Animal Health, Paris, France: http://www.oie.int/wahis_2/temp/reports/en_imm_0000013663_20130624_102939.pdf. [Google Scholar]
- 26.World Organisation for Animal Health. 2014. Immediate notification report. Ref OIE: 14690. World Organisation for Animal Health, Paris, France: http://www.oie.int/wahis_2/temp/reports/en_imm_0000014690_20140127_143257.pdf. [Google Scholar]
- 27.World Organisation for Animal Health. 2014c. Immediate notification report. Ref OIE: 14793. World Organisation for Animal Health, Paris, France. http://www.oie.int/wahis_2/temp/reports/en_imm_0000014793_20140218_131143.pdf. [Google Scholar]
- 28.Gallardo C, Fernández-Pinero J, Pelayo V, Gazaev I, Markowska-Daniel I, Pridotkas G, Nieto R, Fernández-Pacheco P, Bokhan S, Nevolko O, Drozhzhe Z, Perez C, Soler A, Kolvasov D, Arias M. 2014. Genetic variation among African swine fever genotype II viruses, eastern and central Europe. Emerg Infect Dis 20:1544–1547. doi: 10.3201/eid2009.140554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejsak Z, Truszczyński M, Kozak E, Markowska-Daniel I. 2014a. Epidemiological analysis of two first cases of African swine fever in wild boars in Poland. Medycyna Weter 70:369–372. [Google Scholar]
- 30.Pejsak Z, Truszczyński M, Niemczuk K, Kozak E, Markowska-Daniel I. 2014. Epidemiology of African swine fever in Poland since the detection of the first case. Pol J Vet Sci 17:665–672. [DOI] [PubMed] [Google Scholar]
- 31.European Food Safety Authority (EFSA). 2014. Scientific review on African swine fever. EFSA J 12:3628. [Google Scholar]
- 32.European Commission, Health and Consumers Directorate-General (7138/2013). Guidelines on surveillance and control of African swine fever in feral pigs and preventive measures for pig holdings. European Commission, Health and Consumers Directorate-General, Brussels, Belgium: http://ec.europa.eu/food/animal/diseases/controlmeasures/docs/sanco_7138_2013_asf_wb_en.pdf. [Google Scholar]
- 33.World Organisation for Animal Health. 2012. African swine fever. In OIE terrestrial manual 2012. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 34.Oura CA, Edwards L, Batten CA. 2013. Virological diagnosis of African swine fever–comparative study of available tests. Virus Res 173:150–158. doi: 10.1016/j.virusres.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez-Vizcaíno JM, Mur L. 2013. African swine fever diagnosis update. Dev Biol (Basel) 135:159–165. [DOI] [PubMed] [Google Scholar]
- 36.Agüero M, Fernández J, Romero L, Sánchez Mascaraque C, Arias M, Sánchez-Vizcaíno JM. 2003. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J Clin Microbiol 41:4431–4434. doi: 10.1128/JCM.41.9.4431-4434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, Bastos AD, Drew TW. 2003. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods 107:53–61. doi: 10.1016/S0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 38.Fernández-Pinero J, Gallardo C, Elizalde M, Robles A, Gomez C, Bishop R, Heath L, Couacy-Hymann E, Fasina FO, Pelayo V, Soler A, Arias M. 2013. Molecular diagnosis of African swine fever by a new real-time PCR using universal probe library. Transbound Emerg Dis 60:48–58. doi: 10.1111/j.1865-1682.2012.01317.x. [DOI] [PubMed] [Google Scholar]
- 39.Tignon M, Gallardo C, Iscaro C, Hutet E, Van der Stede Y, Kolbasov D, De Mia GM, Le Potier MF, Bishop RP, Arias M, Koenen F. 2011. Development and inter-laboratory validation study of an improved new real-time PCR assay with internal control for detection and laboratory diagnosis of African swine fever virus. J Virol Methods 178:161–170. doi: 10.1016/j.jviromet.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Cubillos C, Gomez-Sebastian S, Moreno N, Nuñez MC, Mulumba-Mfumu LK, Quembo CJ, Heath L, Etter EM, Jori F, Escribano JM, Blanco E. 2013. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res 173:159–167. doi: 10.1016/j.virusres.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Gallardo C, Nieto R, Martín E, Pelayo V, Arias M. 2012. Validation of indirect immunoperoxidase technique (IPT) as alternative confirmatory test for African swine fever antibody detection. Proc IX Int Cong Vet Virol (ESVV), 4 to 7 September 2012, Madrid, Spain. [Google Scholar]
- 42.Gallardo C, Soler A, Nieto R, Carrascosa AL, De Mia GM, Bishop RP, Martins C, Fasina FO, Couacy-Hymman E, Heath L, Pelayo V, Martin E, Simon A, Martin R, Okurut AR, Lekolol I, Okoth E, Arias M. 2013. Comparative evaluation of novel African swine fever virus (ASF) antibody detection techniques derived from specific ASF viral genotypes with the OIE internationally prescribed serological tests. Vet Microbiol 162:32–43. doi: 10.1016/j.vetmic.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Carrascosa AL, Bustos MJ, de Leon P. 2011. Methods for growing and titrating African swine fever virus: field and laboratory samples. Curr Protoc Cell Biol Chapter 26:Unit 26.14. [DOI] [PubMed] [Google Scholar]
- 44.de Leon P, Bustos MJ, Carrascosa AL. 2013. Laboratory methods to study African swine fever virus. Virus Res 173:168–179. doi: 10.1016/j.virusres.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 46.European Commission. 1986. Council directive of 24 November 1986 on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes. EC Directive 86/609/EEC European Commission, Brussels, Belgium: http://ec.europa.eu/food/fs/aw/aw_legislation/scientific/86-609-eec_en.pdf. [Google Scholar]
- 47.Gallardo C, Soler A, Nieto R, Cano C, Pelayo V, Sánchez MA, Pridotkas G, Fernandez-Pinero J, Briones V, Arias M. 22 March 2015. Experimental infection of domestic pigs with African swine fever virus Lithuania 2014 genotype II field isolate. Transbound Emerg Dis; doi: 10.1111/tbed.12346. [DOI] [PubMed] [Google Scholar]
- 48.European Union Reference Laboratory (EURL) for ASF. Standard operating procedure for the detection of antibodies against African swine fever by indirect immunoperoxidase technique. European Union Reference Laboratory (EURL) for ASF, Centro de Investigación en Sanidad Animal (CISA-INIA), Madrid, Spain: http://asf-referencelab.info/asf/images/files/PROTOCOLOS-EN/SOP-ASF-IPT-1(1).pdf. [Google Scholar]
- 49.Everitt BS. 1989. Statistical methods for medical investigations. Oxford University Press, New York, NY. [Google Scholar]
- 50.Malogolovkin A, Yelsukova A, Gallardo C, Tsybanov S, Kolbasov D. 2012. Molecular characterization of African swine fever virus isolates originating from outbreaks in the Russian Federation between 2007 and 2011. Vet Microbiol 158:415–419. doi: 10.1016/j.vetmic.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Gallardo C, Nieto R, Pelayo V, Fernández-Pacheco P, Soler A, Simón A, Pérez C, Martín E, Markowska-Daniel I, Pridotkas G, Tignon M, Fernández-Pinero J, Arias M. 2014. Assessment of ASF diagnostic tests in domestic pig and wild boar sample analysis. Some considerations on diagnostic interpretation. In Workshop on laboratory diagnosis of African and Classical swine fever (ASF and CSF), 2 to 3 June 2014, Madrid, Spain. [Google Scholar]
- 52.Gabriel C, Blome S, Malogolovkin A, Parilov S, Kolbasov D, Teifke JP, Beer M. 2011. Characterization of African swine fever virus Caucasus isolate in European wild boars. Emerg Infect Dis 17:2342–2345. doi: 10.3201/eid1712.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guinat C, Reis AL, Netherton CL, Goatley L, Pfeiffer DU, Dixon L. 2014. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Vet Res 45:93. doi: 10.1186/s13567-014-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.European Council. 2002. Council Directive 2002/60/EC of 27 June 2002 laying down specific provisions for the control of African swine fever and amending Directive 92/119/EEC as regards Teschen disease and African swine fever (text with EEA relevance). European Council, Brussels, Belgium: http://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32002L0060&from=EN. [Google Scholar]
- 55.De la Torre A, Bosch J, Iglesias I, Muñoz MJ, Mur L, Martinez-Lopez B, Martinez M, Sanchez-Vizcaino JM. 2013. Assessing the risk of African swine fever introduction into the European Union by wild boar. Transbound Emerg Dis 62:272–279. [DOI] [PubMed] [Google Scholar]
- 56.Mur L, Igolkin A, Varentsova A, Pershin A, Remyga S, Shevchenko I, Zhukov I, Sanchez-Vizcaino JM. 30 November 2014. Detection of African swine fever antibodies in experimental and field samples from the Russian Federation: implications for control. Transbound Emerg Dis; doi: 10.1111/tbed.12304. [DOI] [PubMed] [Google Scholar]