Abstract
Cytauxzoon felis is a virulent, tick-transmitted, protozoan parasite that infects felines. Cytauxzoonosis was previously thought to be uniformly fatal in domestic cats. Treatment combining atovaquone and azithromycin (A&A) has been associated with survival rates of over 60%. Atovaquone, a ubiquinone analogue, targets C. felis cytochrome b (cytb), of which 30 unique genotypes have been identified. The C. felis cytb genotype cytb1 is associated with increased survival rates in cats treated with A&A. The purpose of this study was to design a PCR panel that could distinguish C. felis cytb1 from other cytochrome b genotypes. Primer pairs were designed to span five different nucleotide positions at which single-nucleotide polymorphisms in the C. felis cytb gene had been identified. Through the use of high-resolution melt analysis, this panel was predicted to distinguish cytb1 from other cytb genotypes. Assays were validated using samples from 69 cats with cytauxzoonosis for which the C. felis cytb genotypes had been characterized previously. The PCR panel identified C. felis cytb1 with 100% sensitivity and 98.2% specificity. High-resolution melt analysis can rapidly provide prognostic information for clients considering A&A treatment in cats with cytauxzoonosis.
INTRODUCTION
Cytauxzoonosis is an emerging disease in domestic and wild felids in North and South America, caused by the tick-transmitted apicomplexan parasite Cytauxzoon felis (1–8). Cytauxzoonosis was originally thought to be uniformly fatal in domestic cats (4, 5), but our understanding of the epidemiology of C. felis is evolving. Recent evidence indicates that some cats survive C. felis infections without any evidence of clinical disease and/or history of antiprotozoal therapy (9–13). Whether this change is due to increased recognition of subclinical infections, differences in infectious doses, alternative mechanisms of transmission, or differences in virulence between strains is unclear. For cats presented to veterinary hospitals with acute cytauxzoonosis, however, mortality rates remain high. Even with advances in treatment, mortality rates ranged from 40 to 74% in a prospective randomized clinical trial (14). In that study, atovaquone and azithromycin (A&A) treatment was associated with improved survival rates compared to imidocarb dipropionate treatment (14), which was previously considered the treatment of choice (15, 16). The majority of cats with acute cytauxzoonosis that die do so within 2 to 5 days after presentation (4, 14). Given this rapid clinical course, it is critical to initiate A&A therapy as soon as possible.
Azithromycin is thought to target the mitochondrial ribosomes of the parasite, while atovaquone is presumed to target protozoal cytochrome b (cytb), disrupting electron transport in the parasite mitochondria (17, 18). In related parasites, mutations in the putative atovaquone-binding site of cytb have been associated with responses to A&A treatment (19–21). A recent study identified a C. felis cytb genotype (cytb1) that was associated with increased survival rates in cats treated with A&A (22). Cats that were infected with this genotype (cytb1) and were treated with imidocarb dipropionate, which does not target cytb, did not have a significantly improved survival rate compared to imidocarb dipropionate-treated cats infected with non-cytb1 genotypes (22). Therefore, the improved survival rate of cats infected with C. felis cytb1 appears to be specifically associated with A&A treatment. It is not known whether this association is a direct result of the cytb1 nucleotide or amino acid sequence or whether cytb1 is simply a genetic marker of other mechanisms of susceptibility to A&A treatment (22). Identification of C. felis cytb1 in a clinical sample from a cat with cytauxzoonosis could provide useful prognostic information for a client deciding whether to pursue A&A therapy, which can cost thousands of dollars.
Accurate cytb genotyping may be challenging, due to the location of cytb in the mitochondrial genome. Eukaryotic cells possess multiple copies of mitochondrial genomes, and different mitochondrial sequences can coexist within the same cell or tissue, a condition known as heteroplasmy (23–26). While it is currently not known how many copies of the mitochondrial genome exist in C. felis organisms, up to 150 copies of the mitochondrial genome can exist within one nondividing haploid organism for related apicomplexan parasites (27). Characterization of C. felis cytb genotypes revealed heteroplasmic single-nucleotide polymorphisms (SNPs) in 16 of 30 C. felis cytb genotypes, representing 33% of total samples (23/69 samples) (22). Due to the rapid clinical course, high mortality rates, costs of treatment, and prevalence of heteroplasmy, an assay to detect C. felis cytb1 must be rapid, cost-effective, and sensitive enough to discriminate heteroplasmic SNPs. Quantitative real-time PCR coupled with high-resolution melt (HRM) analysis fulfills these criteria (23, 28–34).
The objective of this study was to develop an assay that can rapidly and accurately identify C. felis cytb1 in clinical samples, using a quantitative real-time PCR panel coupled with HRM analysis. Herein we describe an assay that can detect C. felis cytb1 in feline blood samples with 100% sensitivity and 98.2% specificity. This assay can provide useful prognostic information for owners of cats with cytauxzoonosis.
MATERIALS AND METHODS
Sixty-nine pretreatment DNA samples from cats with cytauxzoonosis were available from a previous study (14). Total DNA was isolated from 200 μl of infected feline whole blood by using a commercial kit (QIAamp DNA Blood minikit; Qiagen Inc., Valencia, CA), according to the kit instructions. All samples were confirmed to be infected through PCR amplification of a portion of the C. felis 18S rRNA gene, as described previously (35). Cytauxzoon felis cytb genotypes had been previously characterized using bidirectional sequencing. Any secondary nucleotide peaks present at a height of ≥30% of the primary nucleotide peak height on DNA sequence chromatograms were detected by a computer program and denoted using IUPAC ambiguity codes (Vector NTI; Invitrogen, Grand Island, NY) (22).
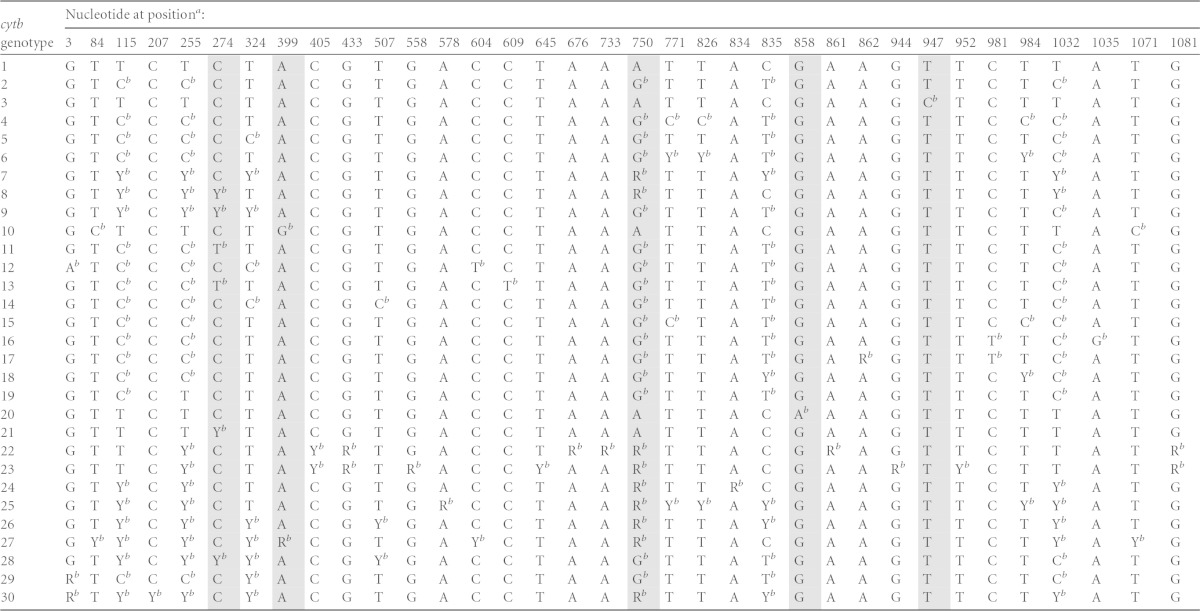
A total of 30 unique C. felis cytb genotypes have been identified; 35 nucleotide positions were determined to have single-nucleotide polymorphisms (SNPs) (22) (Table 1). It was predicted that cytb1 could be distinguished from all other cytb genotypes by assessment of as few as five nucleotide positions (Table 2). Primer pairs were designed to span each of the five nucleotide positions in the C. felis cytb gene (Table 3). Each 25-μl PCR mixture contained 12.5 μl of 2× SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA), 12.5 pmol of each primer, and 1 μl of DNA template. Thermal cycling conditions (CFX96 system; Bio-Rad Laboratories, Inc., Hercules, CA) consisted of an initial denaturation step at 98°C for 3.5 min, 40 amplification cycles of 98°C for 20 s and 60°C for 30 s, and a melt curve step of 62°C to 80°C (increasing in increments of 0.1°C every 10 s). The annealing/extension temperature was optimized to eliminate the production of nonspecific amplicons, and initial PCR products were confirmed via gel electrophoresis. Amplicons that had abnormal amplification curves (e.g., nonsigmoidal) were excluded, and PCRs were repeated prior to high-resolution melt (HRM) analysis (9 repeated PCRs among 1,035 total PCRs). Following PCR, amplicons were analyzed via high-resolution melt (HRM) analysis, with the autocluster detection analysis set at 50% melt curve shape sensitivity and a melting temperature (Tm) difference threshold of 0.10°C (Precision Melt Analysis software; Bio-Rad Laboratories, Inc., Hercules, CA). Assays were performed in triplicate for each sample; only samples that clustered identically with corresponding positive controls in all three replicates were considered true matches to sequence data. Samples that had insufficient DNA volume for triplicate assays were diluted 1:10 in water (4 DNA samples). Positive controls consisted of samples that had been characterized previously as having mitochondrial homoplasmy at the five evaluated C. felis cytb nucleotide positions, and negative controls consisted of C. felis-negative cat DNA and water (no template). Initial amplicon sequences were confirmed via TA cloning (PGEM T Easy; Promega, Madison, WI) and bidirectional sequencing (MCLAB, South San Francisco, CA).
TABLE 1.
Nucleotide positions evaluated for identification of cytb1
SNPs were identified previously at 35 different nucleotide positions in the 1,092-bp C. felis cytb gene. Nucleotide positions selected for assessment by HRM analysis are indicated by shading.
Previously identified SNP.
TABLE 2.
Predicted and actual specificities of PCR panel
| cytb nucleotide(s) analyzed | Predicted cytb1 specificity (%)a | Actual cytb1 specificity (%)a |
|---|---|---|
| 750A | 80.4 (45/56) | 75 (42/56) |
| 750A + 947T | 92.9 (52/56) | 85.7 (48/56) |
| 750A + 947T + 399A | 96.4 (54/56) | 91.1 (51/56) |
| 750A + 947T + 399A + 274C | 98.2 (55/56) | 96.4 (54/56) |
| 750A + 947T + 399A + 274C + 858G | 100 (56/56) | 98.2 (55/56) |
Values in parentheses represent the number of true negatives/(the number of true negatives + the number of false positives).
TABLE 3.
Primer sequences for amplification of five SNP regions analyzed by HRM analysis and full-length C. felis cytb
| Target | Forward primer | Reverse primer | Amplicon size (bp) |
|---|---|---|---|
| Nucleotide 750 | GGAGATGTTGATAATTCAATATTGGC | CATTCAGGAACAATATGCAATGG | 65 |
| Nucleotide 947 | GTCCATTATCATAGAGATTGGACAG | TGACATTCTTCCAATGCATCC | 84 |
| Nucleotide 399 | CAAATGGTCAAATGAGCTATTGG | CCCAATAAAACAAATTTGTAATGACTG | 57 |
| Nucleotide 274 | CTTTATGTTTTTACATGTATTGAAAGGG | CATGACCAAGGTAGATATCTACTAG | 63 |
| Nucleotide 858 | TTAGCAGGACTGATTGCTATG | GCTTCAACCAATGCTACAAG | 59 |
| Full-length cytb | AGGATACAGGGCTATAACCAAC | GTACTCTGGCTATGTCAATTTCTAC | 1,203 |
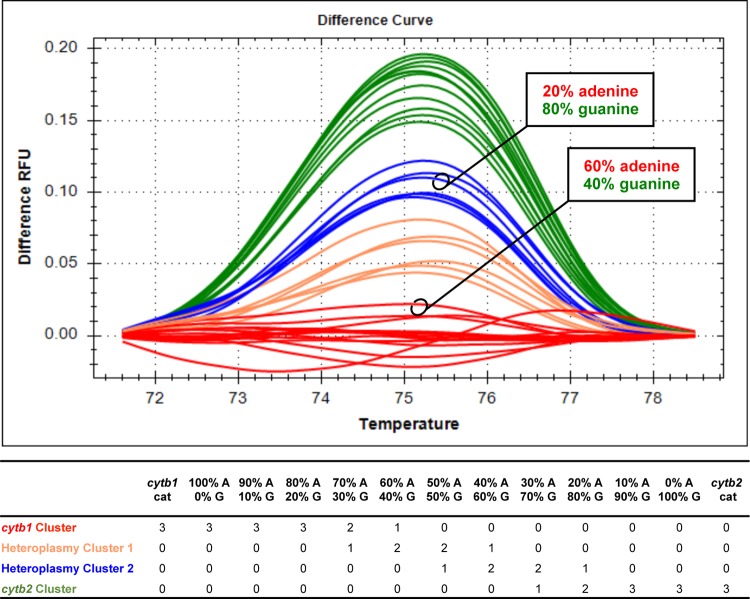
To investigate discrepancies between chromatographic and HRM results for nucleotide 750 that affected the specificity of the PCR panel, HRM analysis was performed using mixtures of cytb clones that simulated mitochondrial heteroplasmy at nucleotide 750. Full-length C. felis cytb1 (adenine at nucleotide position 750) and cytb2 (guanine at nucleotide position 750) were amplified (Table 3) and cloned (PGEM T Easy; Promega, Madison, WI). Plasmids were diluted to clinically relevant concentrations (e.g., cycle threshold results between 15 and 25), mixed in various ratios, and subjected to PCR and HRM analysis of C. felis cytb nucleotide 750, to assess the sensitivity of HRM to differentiate heteroplasmic SNPs at nucleotide position 750 (see Fig. 3).
FIG 3.
Analysis of C. felis cytb clone mixtures confirms the inability of HRM analysis to consistently discriminate heterogeneity in complex mixtures of genotypes. (Top) Difference curves for triplicate analysis of clone mixtures and clinical samples. RFU, relative fluorescence units. (Bottom) Number of replicates per cluster for each sample analyzed.
Predicted and actual specificities of detection of C. felis cytb1 by HRM were calculated for the cumulative assessment of one, two, three, four, or five nucleotide positions (Table 2). Positive predictive values were estimated for the cumulative assessment of each nucleotide position based on the 18.8% prevalence (13/69 samples) of C. felis cytb1 in the sample population and the limits of the 95% confidence interval (CI) (95% CI, 8 to 26.7%) (36) (Table 4).
TABLE 4.
Positive predictive value for identification of C. felis cytb1 is highest when all five nucleotide positions in PCR panel are analyzed
| cytb nucleotide(s) analyzed | Specificity of cytb1 detection (%) | Positive predictive value (%) |
||
|---|---|---|---|---|
| With cytb1 population prevalence of 8% | With cytb1 population prevalence of 18.8% | With cytb1 population prevalence of 26.7% | ||
| 750A | 75 | 25.8 | 48.1 | 59.3 |
| 750A + 947T | 85.7 | 37.8 | 61.8 | 71.8 |
| 750A + 947T + 399A | 91.1 | 49.4 | 72.2 | 80.4 |
| 750A + 947T + 399A + 274C | 96.4 | 70.7 | 86.5 | 91 |
| 750A + 947T + 399A + 274C + 858G | 98.2 | 82.9 | 92.8 | 95.3 |
RESULTS
Overall performance of PCR panel.
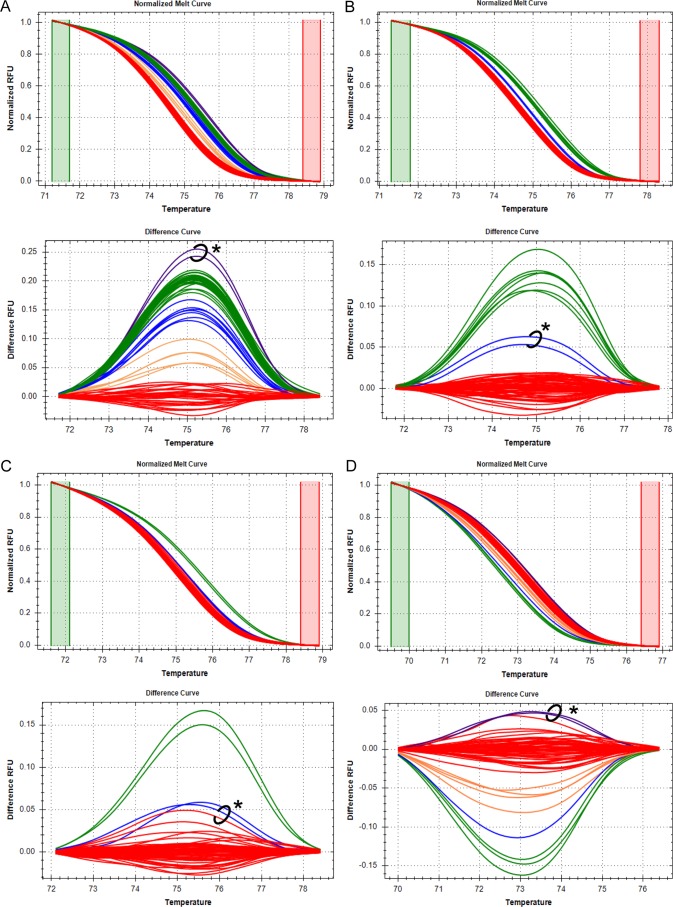
The PCR panel had a sensitivity of 100% (13/13 cytb1 samples correctly identified) and a specificity of 98.2% (55/56 non-cytb1 samples correctly identified) (Table 2). Positive predictive values for identification of C. felis cytb1 by cumulative analysis of each nucleotide position ranged from 25.8% to 95.3% and are summarized in Table 4. Representative high-resolution melt curves and differential fluorescence curves for each nucleotide analyzed are shown in Fig. 1A to E.
FIG 1.
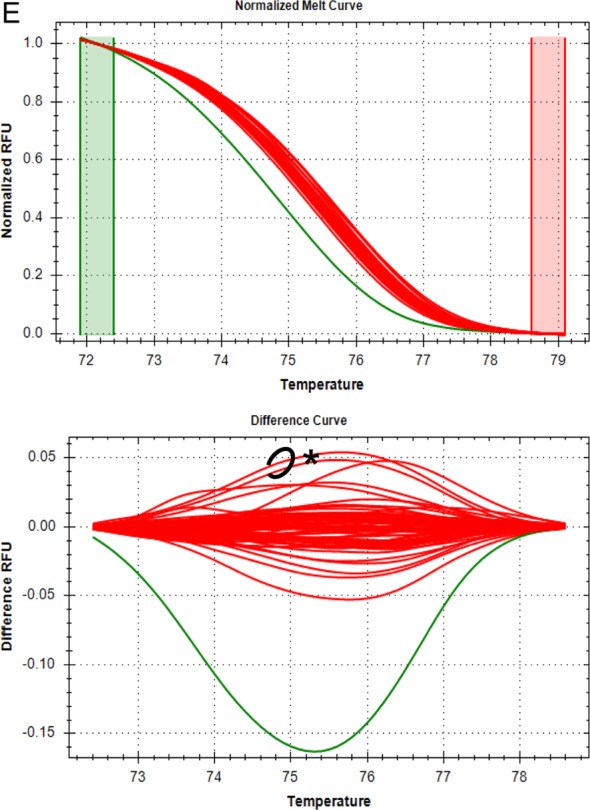
Identification of C. felis cytb1 by HRM analysis of five cytb nucleotides. High-resolution melt curves and differential fluorescence curves of 69 samples are shown for one of three replicate PCRs for each of the five nucleotide positions analyzed. Green and red boxes on melt curves indicate the pre- and postmelt regions, respectively, used for data normalization. RFU, relative fluorescence units. Red clusters of melt curves represent samples with the nucleotide sequence for cytb1 at each position, while clusters of other colors represent samples with SNPs at that location. Two samples (circled on difference curves and denoted with an asterisk) had anomalous clustering patterns that occurred for all nucleotides analyzed in at least one replicate PCR. The clustering patterns of those two samples could not be explained by chromatographic data but did not affect the sensitivity or specificity of the overall assay. (A) Nucleotide position 750. The red cluster represents samples with adenine at nucleotide 750, while the green cluster represents samples with guanine at nucleotide 750. The blue and orange clusters represent samples with apparent heteroplasmy. Based on chromatographic data, the circled samples (purple cluster) had a guanine at nucleotide 750 but clustered outside the expected spectrum of results. (B) Nucleotide position 947. The red cluster represents samples with thymine at nucleotide 947, while the green cluster represents samples with cytosine at nucleotide 947. Based on chromatographic data, the circled samples (blue cluster) had a thymine at nucleotide 947 but clustered between the clusters for thymine and cytosine. (C) Nucleotide position 399. The red cluster represents samples with adenine at nucleotide 399, while the green cluster represents samples with guanine at nucleotide 399. The blue cluster represents samples with apparent heteroplasmy. Based on chromatographic data, the circled samples had an adenine at nucleotide 399 but clustered with a sample confirmed to have heteroplasmy at nucleotide 399 (for replicate PCR data shown, only one sample demonstrated this clustering). (D) Nucleotide position 274. The red cluster represents samples with cytosine at nucleotide 274, while the green cluster represents samples with thymine at nucleotide 274. The blue and orange clusters represent samples with apparent heteroplasmy. Based on chromatographic data, the circled samples (purple cluster) had a cytosine at nucleotide 274 but clustered outside the expected spectrum of results. (E) Nucleotide position 858. The red cluster represents samples with guanine at nucleotide 858, while the green cluster represents samples with an adenine at nucleotide 858. Based on chromatographic data, the circled samples had a guanine at nucleotide 858 but, in 1 of 3 PCR replicates, the samples clustered outside the expected spectrum of results (data from the PCR replicate shown do not have this clustering pattern).
Predicted versus actual performance according to nucleotide position.
Of the five nucleotide positions assessed in this panel, nucleotide 750 was predicted to differentiate the largest cohort of non-cytb1 samples from cytb1 samples. Based on chromatographic data, it was predicted in silico that analysis of nucleotide 750 alone would identify 45/56 non-cytb1 samples (specificity of 80.4%). However, actual HRM analysis of nucleotide 750 alone identified only 42/56 non-cytb1 samples (specificity of 75%). Analysis of nucleotide 750 failed to identify four non-cytb1 samples but additionally identified a single non-cytb1 sample that was predicted to have only a SNP at nucleotide 947 (Table 2).
Based on chromatographic data, it was predicted in silico that analysis of nucleotides 750 and 947 together would identify 52/56 non-cytb1 samples (specificity of 92.9%). When the actual results of HRM analysis of nucleotide 750 were taken into account, it was predicted that analysis of nucleotides 750 and 947 together would identify 48/56 non-cytb1 samples (specificity of 85.7%). The actual results of HRM analysis of nucleotides 750 and 947 were consistent with this prediction (Table 2).
Based on chromatographic data, it was predicted in silico that analysis of nucleotides 750, 947, and 399 together would identify 54/56 non-cytb1 samples (specificity of 96.4%). A total of three samples had SNPs at nucleotide 399, as identified by chromatographic data. One of those samples was predicted to be identified as non-cytb1 by HRM analysis of nucleotide 750; however, HRM analysis of nucleotide 750 failed to identify that sample. Thus, HRM analysis of nucleotide 399 was required to identify the sample as non-cytb1 (Table 2). When this information and the actual results of HRM analysis of the previous two nucleotides were taken into account, it was predicted that analysis of these three nucleotides together would identify 51/56 non-cytb1 samples (specificity of 91.1%). The actual results of HRM analysis were consistent with this prediction (Table 2).
Based on chromatographic data, it was predicted in silico that analysis of nucleotides 750, 947, 399, and 274 together would identify 55/56 non-cytb1 samples (specificity of 98.2%). A total of eight samples had SNPs at nucleotide 274, as identified by chromatographic data. Seven of those samples were predicted to be identified as non-cytb1 by HRM analysis of nucleotide 750; however, HRM analysis of nucleotide 750 failed to identify two of those seven samples. Thus, HRM analysis of nucleotide 274 was required to identify both of those samples as non-cytb1 (Table 2). When this information and the actual results of HRM analysis of the previous three nucleotides were taken into account, it was predicted that analysis of these four nucleotides together would identify 54/56 non-cytb1 samples (specificity of 96.4%). The actual results of HRM analysis were consistent with this prediction (Table 2).
Finally, based on chromatographic data, it was predicted in silico that analysis of nucleotides 750, 947, 399, 274, and 858 together would identify 56/56 non-cytb1 samples (specificity of 100%). When the actual results of HRM analysis of the previous four nucleotides were taken into account, it was predicted that analysis of all five nucleotides together would identify 55/56 non-cytb1 samples (specificity of 98.2%). The actual results of HRM analysis were consistent with this prediction (Table 2).
Agreement between HRM analysis and chromatographic data.
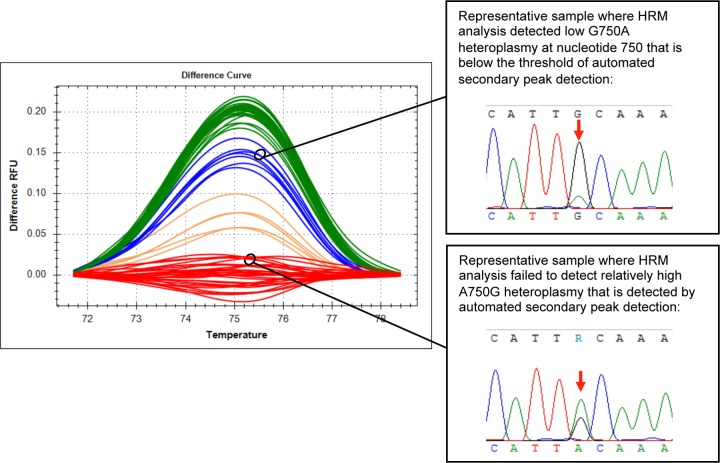
A total of 345 nucleotide positions were analyzed (5 nucleotide positions for 69 samples) (see Table S1 in the supplemental material). The agreement between HRM analysis and automated detection of secondary peaks on chromatograms was fair (kappa = 0.3564). HRM analysis matched automated secondary peak detection at 307/345 nucleotide positions (89%), suggested the presence of additional heteroplasmy at 32/345 nucleotide positions (9.3%), and failed to detect heteroplasmy at 6/345 nucleotide positions (1.7%). Discordant results at nucleotides 947, 399, 274, and 858 did not impair our ability to identify C. felis cytb1. However, failure to detect heteroplasmy at nucleotide position 750 did affect our ability to identify C. felis cytb1; therefore, we further analyzed these discrepancies. HRM analysis of nucleotide 750 failed to detect relatively high A750G heteroplasmy that was identified by automated secondary peak detection (Fig. 2). However, HRM analysis of nucleotide 750 detected low G750A heteroplasmy that was below the limit of automated secondary peak detection but could be identified by careful manual inspection of chromatograms (Fig. 2). These findings were confirmed by HRM analysis of plasmid mixtures simulating mitochondrial heteroplasmy at nucleotide 750 (Fig. 3).
FIG 2.
HRM analysis failed to detect relatively high levels of A750G heteroplasmy but was able to detect low levels of G750A heteroplasmy at C. felis cytb nucleotide 750 in clinical samples. The red cluster is anticipated to have only adenine at nucleotide 750, the green cluster is anticipated to have only guanine at nucleotide 750, and the orange and blue clusters are anticipated to have heteroplasmy at nucleotide 750. RFU, relative fluorescence units.
When analyzed by HRM analysis, two non-cytb1 samples repeatedly had unexplained clustering patterns. For all nucleotide positions analyzed, these samples either were identified as a unique cluster or were placed on the fringe of their expected cluster (Fig. 1A to E). Upon careful manual inspection of chromatograms, however, there was no evidence of SNPs at the targeted nucleotide or at any other nucleotide positions within the amplicon. These results did not affect our ability to detect cytb1.
DISCUSSION
When treated with atovaquone and azithromycin (A&A), cats infected with Cytauxzoon felis cytb1 have had improved survival rates, compared to cats infected with other C. felis cytb genotypes (22). In this study, we developed an assay that can rapidly identify C. felis cytb1 in clinical samples with 100% sensitivity and 98.2% specificity using a quantitative real-time PCR panel and HRM analysis. This test is intended to aid pet owners and clinicians in deciding whether to treat a cat for cytauxzoonosis with A&A, which can cost thousands of dollars. The cost of this assay for clients is likely to be comparable to that of currently offered molecular diagnostic panels ($100 to $200). From the time the sample arrives in the laboratory, the assay can be completed in less than 3 h, which is important for clinical decision-making given the rapid disease progression and high mortality rates associated with acute cytauxzoonosis. While 100% of cats (8/8 cats) infected with C. felis cytb1 survived when treated with A&A, it is important to note that over 50% of cats (20/37 cats) infected with non-cytb1 genotypes also survived when treated with A&A (22). Therefore, while this assay will provide useful prognostic information for treating cats with cytauxzoonosis, other factors also need to be considered when implementing A&A treatment.
First, a variety of factors besides C. felis cytb genotype may contribute to whether a cat will survive infection when treated with A&A. In a previous study that assessed the efficacy of A&A and imidocarb dipropionate for treatment of cats with acute cytauxzoonosis, cats that died tended to have higher levels of parasitemia, higher serum bilirubin levels, and more severe leukopenia compared to cats that survived infection (14). However, treatment groups (A&A versus imidocarb dipropionate) were not considered when these associations were assessed (14). Additionally, the majority of cats that died did so within 24 h after admission to a veterinary clinic (14), which suggests that in some cases the disease might have already progressed beyond the point at which any available treatment would be effective.
Second, given the heterogeneity of the C. felis cytb gene sequence (22), it is likely that uncharacterized C. felis cytb genotypes exist that have SNPs at nucleotide positions that are not assessed by this assay. Some of these uncharacterized genotypes may be A&A resistant but falsely identified as C. felis cytb1 (type I error). Alternatively, some characterized and uncharacterized non-cytb1 genotypes may also be susceptible to A&A treatment. If future studies demonstrate that other C. felis cytb genotypes are also associated with improved survival rates or uncharacterized genotypes are falsely identified as cytb1, then an extended PCR panel analyzing additional nucleotide positions can be developed.
Additionally, the limitations of HRM analysis in detecting heteroplasmy affected this assay. HRM analysis was unable to detect heteroplasmy in one non-cytb1 sample, resulting in one false-positive result and a specificity of 98.2% (Table 2). Although the impact was minimal, this problem also could be solved through development of an extended PCR panel.
Lastly, this study revealed the inability of HRM analysis to discriminate heterogeneity consistently in complex mixtures of genotypes. While these inconsistencies had minimal effects in the current study, the shortcomings of HRM analysis should be considered in the design and interpretation of any HRM assay. We encountered multiple scenarios in which there was disagreement between the chromatographic evaluation and the HRM analysis. First, in some cases HRM analysis was able to detect heteroplasmy that was present in chromatograms but below the limit of automated secondary peak detection (Fig. 2 and 3; see also Table S1 in the supplemental material). This highlights the importance of manual inspection of chromatograms for detection of heteroplasmy. Second, in other cases HRM analysis failed to detect heteroplasmy despite clear evidence of heteroplasmy on chromatograms (Fig. 2 and 3; see also Table S1 in the supplemental material). The reasons behind these discordant results are unclear; however, we hypothesize that the surrounding nucleotide sequences and overall GC content of the amplicon may affect HRM analysis. Third, in some cases HRM analysis detected heteroplasmy for which there was no chromatographic evidence even with manual inspection (Fig. 1; see also Table S1 in the supplemental material). It is unclear whether HRM analysis was detecting actual heteroplasmy beyond the resolution of both automated and manual inspection of chromatograms or whether factors unrelated to amplicon sequences (e.g., individual sample pH or salt concentrations) were skewing the HRM analysis results. Lastly, for some samples the HRM analysis results differed between replicates (see Table S1 in the supplemental material). Thus, if HRM analysis is to be performed on complex mixtures of genotypes, multiple analyses of the same sample may be required for accurate genotyping.
In conclusion, this study describes an assay that can rapidly provide prognostic information to clients considering treatment options for cats with cytauxzoonosis. Through utilization of quantitative PCR and HRM analysis, we were able to develop an assay that identified C. felis cytb1 with 100% sensitivity and 98.2% specificity. Due to the variability of disease progression in individual cases of cytauxzoonosis, the heterogeneity of C. felis cytb sequences, and the inherent limitations of HRM analysis, multiple factors should be considered in the clinical decision-making process.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by the ALSAM Foundation.
No conflicts of interest were present in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00635-15.
REFERENCES
- 1.Glenn BL, Stair EL. 1984. Cytauxzoonosis in domestic cats: report of two cases in Oklahoma, with a review and discussion of the disease. J Am Vet Med Assoc 184:822–825. [PubMed] [Google Scholar]
- 2.Hoover JP, Walker DB, Hedges JD. 1994. Cytauxzoonosis in cats: eight cases (1985–1992). J Am Vet Med Assoc 205:455–460. [PubMed] [Google Scholar]
- 3.Wagner JE. 1976. A fatal cytauxzoonosis-like disease in cats. J Am Vet Med Assoc 168:585–588. [PubMed] [Google Scholar]
- 4.Ferris DH. 1979. A progress report on the status of a new disease of American cats: cytauxzoonosis. Comp Immunol Microbiol Infect Dis 1:269–276. doi: 10.1016/0147-9571(79)90028-6. [DOI] [PubMed] [Google Scholar]
- 5.Birkenheuer AJ, Le JA, Valenzisi AM, Tucker MD, Levy MG, Breitschwerdt EB. 2006. Cytauxzoon felis infection in cats in the mid-Atlantic states: 34 cases (1998–2004). J Am Vet Med Assoc 228:568–571. doi: 10.2460/javma.228.4.568. [DOI] [PubMed] [Google Scholar]
- 6.Jackson CB, Fisher T. 2006. Fatal cytauxzoonosis in a Kentucky cat (Felis domesticus). Vet Parasitol 139:192–195. doi: 10.1016/j.vetpar.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 7.Andre MR, Adania CH, Machado RZ, Allegretti SM, Felippe PA, Silva KF, Nakaghi AC, Dagnone AS. 2009. Molecular detection of Cytauxzoon spp. in asymptomatic Brazilian wild captive felids. J Wildl Dis 45:234–237. doi: 10.7589/0090-3558-45.1.234. [DOI] [PubMed] [Google Scholar]
- 8.MacNeill AL, Barger AM, Skowronski MC, Lanka S, Maddox CW. 19 January 2015. Identification of Cytauxzoon felis infection in domestic cats from southern Illinois. J Feline Med Surg doi: 10.1177/1098612X14567158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haber MD, Tucker MD, Marr HS, Levy JK, Burgess J, Lappin MR, Birkenheuer AJ. 2007. The detection of Cytauxzoon felis in apparently healthy free-roaming cats in the U S A. Vet Parasitol 146:316–320. doi: 10.1016/j.vetpar.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 10.Rizzi TE, Reichard MV, Cohn LA, Birkenheuer AJ, Taylor JD, Meinkoth JH. 2015. Prevalence of Cytauxzoon felis infection in healthy cats from enzootic areas in Arkansas, Missouri, and Oklahoma. Parasit Vectors 8:13. doi: 10.1186/s13071-014-0618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker DB, Cowell RL. 1995. Survival of a domestic cat with naturally acquired cytauxzoonosis. J Am Vet Med Assoc 206:1363–1365. [PubMed] [Google Scholar]
- 12.Meinkoth J, Kocan AA, Whitworth L, Murphy G, Fox JC, Woods JP. 2000. Cats surviving natural infection with Cytauxzoon felis: 18 cases (1997–1998). J Vet Intern Med 14:521–525. doi: 10.1111/j.1939-1676.2000.tb02270.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown HM, Lockhart JM, Latimer KS, Peterson DS. 2010. Identification and genetic characterization of Cytauxzoon felis in asymptomatic domestic cats and bobcats. Vet Parasitol 172:311–316. doi: 10.1016/j.vetpar.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 14.Cohn LA, Birkenheuer AJ, Brunker JD, Ratcliff ER, Craig AW. 2011. Efficacy of atovaquone and azithromycin or imidocarb dipropionate in cats with acute cytauxzoonosis. J Vet Intern Med 25:55–60. doi: 10.1111/j.1939-1676.2010.0646.x. [DOI] [PubMed] [Google Scholar]
- 15.Greene CE, Latimer K, Hopper E, Shoeffler G, Lower K, Cullens F. 1999. Administration of diminazene aceturate or imidocarb dipropionate for treatment of cytauxzoonosis in cats. J Am Vet Med Assoc 215:497–500. [PubMed] [Google Scholar]
- 16.Greene CE. 1998. Infectious diseases of the dog and cat, 2nd ed W. B. Saunders, Philadelphia, PA. [Google Scholar]
- 17.Mather MW, Henry KW, Vaidya AB. 2007. Mitochondrial drug targets in apicomplexan parasites. Curr Drug Targets 8:49–60. doi: 10.2174/138945007779315632. [DOI] [PubMed] [Google Scholar]
- 18.Vaidya AB, Mather MW. 2009. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol 63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- 19.Sakuma M, Setoguchi A, Endo Y. 2009. Possible emergence of drug-resistant variants of Babesia gibsoni in clinical cases treated with atovaquone and azithromycin. J Vet Intern Med 23:493–498. doi: 10.1111/j.1939-1676.2009.0300.x. [DOI] [PubMed] [Google Scholar]
- 20.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. 2000. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother 44:2100–2108. doi: 10.1128/AAC.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siregar JE, Syafruddin D, Matsuoka H, Kita K, Marzuki S. 2008. Mutation underlying resistance of Plasmodium berghei to atovaquone in the quinone binding domain 2 (Qo2) of the cytochrome b gene. Parasitol Int 57:229–232. doi: 10.1016/j.parint.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Schreeg ME, Marr HS, Tarigo J, Cohn LA, Levy MG, Birkenheuer AJ. 2013. Pharmacogenomics of Cytauxzoon felis cytochrome b: implications for atovaquone and azithromycin therapy in domestic cats with cytauxzoonosis. J Clin Microbiol 51:3066–3069. doi: 10.1128/JCM.01407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurelac I, Lang M, Zuntini R, Calabrese C, Simone D, Vicario S, Santamaria M, Attimonelli M, Romeo G, Gasparre G. 2012. Searching for a needle in the haystack: comparing six methods to evaluate heteroplasmy in difficult sequence context. Biotechnol Adv 30:363–371. doi: 10.1016/j.biotechadv.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Sobenin IA, Mitrofanov KY, Zhelankin AV, Sazonova MA, Postnov AY, Revin VV, Bobryshev YV, Orekhov AN. 2014. Quantitative assessment of heteroplasmy of mitochondrial genome: perspectives in diagnostics and methodological pitfalls. Biomed Res Int 2014:292017. doi: 10.1155/2014/292017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White DJ, Wolff JN, Pierson M, Gemmell NJ. 2008. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol 17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- 26.Wong LJ. 2007. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion 7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 27.Wilson RJ, Williamson DH. 1997. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev 61:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong SY, Giffard PM. 2012. Microbiological applications of high-resolution melting analysis. J Clin Microbiol 50:3418–3421. doi: 10.1128/JCM.01709-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vossen RH, Aten E, Roos A, den Dunnen JT. 2009. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery JL, Sanford LN, Wittwer CT. 2010. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn 10:219–240. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- 31.Dobrowolski SF, Hendrickx AT, van den Bosch BJ, Smeets HJ, Gray J, Miller T, Sears M. 2009. Identifying sequence variants in the human mitochondrial genome using high-resolution melt (HRM) profiling. Hum Mutat 30:891–898. doi: 10.1002/humu.21003. [DOI] [PubMed] [Google Scholar]
- 32.Polakova KM, Lopotova T, Klamova H, Moravcova J. 2008. High-resolution melt curve analysis: initial screening for mutations in BCR-ABL kinase domain. Leuk Res 32:1236–1243. doi: 10.1016/j.leukres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 33.van der Stoep N, van Paridon CD, Janssens T, Krenkova P, Stambergova A, Macek M, Matthijs G, Bakker E. 2009. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum Mutat 30:899–909. doi: 10.1002/humu.21004. [DOI] [PubMed] [Google Scholar]
- 34.Vondrackova A, Vesela K, Hansikova H, Docekalova DZ, Rozsypalova E, Zeman J, Tesarova M. 2012. High-resolution melting analysis of 15 genes in 60 patients with cytochrome-c oxidase deficiency. J Hum Genet 57:442–448. doi: 10.1038/jhg.2012.49. [DOI] [PubMed] [Google Scholar]
- 35.Birkenheuer AJ, Marr H, Alleman AR, Levy MG, Breitschwerdt EB. 2006. Development and evaluation of a PCR assay for the detection of Cytauxzoon felis DNA in feline blood samples. Vet Parasitol 137:144–149. doi: 10.1016/j.vetpar.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Rogan WJ, Gladen B. 1978. Estimating prevalence from the results of a screening test. Am J Epidemiol 107:71–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.