Abstract
Clostridium difficile infection (CDI) became a public health problem for the global spreading of the so-called hypervirulent PCR ribotypes (RTs) 027 and 078, associated with increases in the transmission and severity of the disease. However, especially in Europe, several RTs are prevalent, and the concept of hypervirulence is currently debated. We investigated the toxin and resistance profiles and the genetic relatedness of 312 C. difficile strains isolated in a large Italian teaching hospital during a 5-year period. We evaluated the role of CDI-related antibiotic consumption and infection control practices on the RT predominance in association with their molecular features and transmission capacity. Excluding secondary cases due to nosocomial transmission, RT018 was the predominant genotype (42.4%) followed by RT078 (13.6%), while RT027 accounted for 0.8% of the strains. RT078 was most frequently isolated from patients in intensive care units. Its prevalence significantly increased over time, but its transmission capacity was very low. In contrast, RT018 was highly transmissible and accounted for 95.7% of the secondary cases. Patients with the RT018 genotype were significantly older than those with RT078 and other RTs, indicating an association between epidemic RT and age. We provide here the first epidemiological evidence to consider RT018 as a successful epidemic genotype that deserves more attention in clinical practice.
INTRODUCTION
Clostridium difficile infection (CDI) is one of the most common hospital-acquired infections and the main known cause of antibiotic-associated diarrhea in hospitals in industrialized countries (1). Previous antibiotic treatment, hospitalization, old age, and underlying diseases represent major risk factors for CDI. The disease spectrum ranges from mild diarrhea to severe pseudomembranous enterocolitis, sepsis, and death (2, 3).
In the past decade, many countries in North America and Europe have registered an increase in CDI rates and related severity and mortality (4, 5). These epidemiological changes coincided with the emergence and epidemic spread of C. difficile strains belonging to the PCR ribotype (RT) 027/North American pulsed-field gel electrophoresis type 1 (NAP1) (6). Indeed, RT027 became the predominant strain in many geographical regions, as documented in different epidemiological studies, and, since 2005, several countries have conducted epidemiological surveillance to monitor its spread (7, 8). It has been reported to occur in at least 38 U.S. states and in 16 European countries, being also responsible for outbreaks (7, 9). Moreover, an increase in community-acquired CDI has been reported; in Europe, this phenomenon correlates with the diffusion of RT078/NAP7 or NAP8 which, like RT027, has been associated with higher disease severity and attributable mortality (1, 10).
RT027 and RT078 are often referred to as hypervirulent genotypes of C. difficile, but this definition is not without controversy. Though this term denotes an increase in virulence, it has often been misused to identify epidemic strains that cause outbreaks (7, 11). The success of an epidemic strain relies not only on its ability to cause disease (virulence) but also on its transmissibility. Indeed, depending on the country and the time period, different RTs, such as RT001, RT017, and RT014/020, have been reported as prevalent and associated with severe infections and large outbreaks in hospital settings (12–15).
In Italy, we reported in 2010 for the first time the presence of RT027 and RT078; later, these RTs were detected as minor genotypes (16–19). RT018 has been reported as the predominant genotype circulating in Italian hospitals (20). Longitudinal surveillance data on C. difficile epidemiology are lacking, as systematic isolation and molecular typing of C. difficile strains are not routinely performed. Between 2009 and 2013, we characterized for infection control purposes 312 C. difficile strains, representing 66% of all the CDI cases identified at the San Raffaele hospital. The aims of this study were to analyze the epidemiology of CDI in our hospital during a 5-year period and to investigate the influence of antibiotic consumption and infection control practice on the predominance of different RTs in association with their molecular characteristics and transmission capacity.
MATERIALS AND METHODS
Definitions.
We included in this study the first strain isolated from the first stool sample collected from symptomatic patients; subsequent strains or samples from the same patient, defined as duplicates, were excluded. CDI was defined as the acute onset of diarrhea with a positive toxin A and B assay (Vidas C. difficile toxin A&B; bioMérieux) and no other documented cause of diarrhea (21, 22). Health care-associated CDI (HA-CDI) was defined as the development of CDI >48 h after hospital admission or within 4 weeks after discharge. Community-associated CDI (CA-CDI) was defined as the development of CDI within 48 h of hospital admission in patients with no documented prior hospitalization or long-term care facility stay in the preceding 3 months. Indeterminate CDI was defined as the development of CDI between 4 to 12 weeks after hospital discharge (23). For the epidemiological investigation, the patient case histories were analyzed. An outbreak was defined as ≥2 related CDI cases presenting the same RT and a link confirmed by classical epidemiological investigation. The incidence of CDI was defined as the number of cases per 1,000 admissions or 10,000 patient-days. These data were compared with the CDI incidence registered in other hospitals of the region with similar characteristics. The transmission index for each RT was calculated as the total number of secondary cases divided by the total number of index cases (24).
Bacterial strains, study setting, and data collection.
Between January 2009 and December 2013 at the San Raffaele hospital (OSR), a total of 312 nonduplicate toxigenic C. difficile strains were isolated from unformed Vidas toxin A&B-positive stool samples. Clostridium difficile selective agar (CDSA) plates (Becton Dickinson) were used for strain recovery. The OSR is a large (1,400-bed) private university hospital with close to 45,000 mean admissions per year and is located in the northeastern area of Milan, in the Lombardy region of Italy. The annual consumption of clindamycin, cephalosporins including extended-spectrum cephalosporins, β-lactamase-resistant β-lactams, macrolides, and fluoroquinolones was expressed in defined daily doses (DDDs) per 100 patients per day to measure the percentage of patients exposed every day to antimicrobials. Patient genders, ages, dates of admission, wards, and dates of CDI diagnosis were collected as epidemiological data.
PCR ribotyping.
The clonal relatedness of the C. difficile strains was analyzed using PCR ribotyping (16) and by comparing the identified RT to the following reference strains: strains N1 (RT001, NCTC11204) and R20291 (RT027, NCTC13366) from the National Collection of Type Cultures (Health Protection Agency, United Kingdom); strain 630 (RT012, BAA-1382) from LGC Standards (United Kingdom); strains 1470 (RT017), R7605/78 (RT078), and BII5 (RT126), kindly provided by M. Rupnik (Maribor University, Slovenia); strains RT002, RT014/020, and RT018, kindly provided by F. Agnoletti (Istituto Zooprofilattico Sperimentale delle Venezie, Italy); and strains RT056, RT106, and RT137, kindly provided by V. Pasquale (Parthenope University of Naples, Italy). The genomic DNA fingerprinting patterns produced by PCR ribotyping were analyzed with InfoQuest FP v5.1 (Bio-Rad Laboratories).
Detection of tcdA, tcdB, and cdtA and cdtB genes and tcdC gene analysis.
Genes encoding toxin A (tcdA), toxin B (tcdB), and binary toxin (cdtA and cdtB) were detected by multiplex PCR (21, 25). Deletions and mutations in the tcdC gene were detected in C. difficile strains belonging to RT078 and RT027 by PCR amplification and sequencing with primers C1 (5′-TTAATTAATTTTCTCTACAGCTATCC-3′) and C2 (5′-TCTAATAAAAGGGAGATTGTATTATG-3′) of a 718-bp fragment encompassing the entire tcdC gene (21). The sequences were analyzed and compared to the wild-type tcdC sequence from strain VPI 10463 (GenBank accession number Y10689).
Detection of ermB gene and fluoroquinolone resistance.
Since ermB is known to be predominant in macrolide-lincosamide-streptogramin B-resistant C. difficile, the presence of this gene was investigated by PCR using the primers and protocol by Sutcliffe et al. (26, 27). The C245T mutation in the quinolone resistance-determining region (QRDR) of the gyrA gene, which is associated with a high level of resistance (MIC, ≥32 μg/ml) to fluoroquinolones, was detected by PCR (28).
Statistical analysis.
A linear trend test was performed over time using the Cochran-Armitage trend test to evaluate significant changes in CDI incidence and isolation of the major C. difficile RT identified during the study period. Spearman's correlation (rho) was used to assess the relationship between the antibiotic consumption and the incidence of C. difficile and the major RT detected. Categorical variables were analyzed using the chi-square test or Fisher's exact test, when appropriate. The nonparametric two-tailed Student's t test was used to compare the median age of patients infected with a major RT genotype. P values of <0.05 were considered statistically significant.
RESULTS
Incidence of CDI, antibiotic consumption and infection control measures.
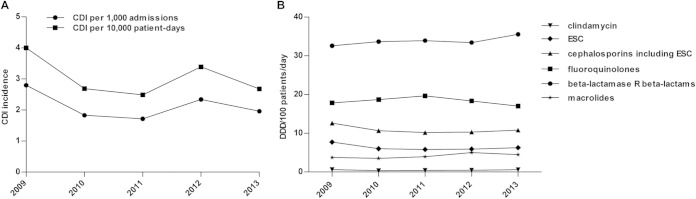
Of 8,649 stool samples tested during the study period, 642 (7.4%) were positive for C. difficile. Of these, 473 were from individual patients with CDI, while 169 were duplicate samples. Overall, there was a 22% reduction of samples tested through the study period, from 1,964 samples in 2009 to 1,532 samples in 2013. In total, 312 nonduplicate C. difficile strains were available, representing 66% of all CDI cases identified at the OSR between 2009 and 2013. Figure 1 shows the yearly incidence of CDI (A) and the consumption data of antibiotics that have been considered risk factors for the occurrence and increasing rate of CDI (B). A mean incidence of 2.13 CDI episodes per 1,000 admissions or 3.05 per 10,000 patient-days was determined in our hospital. The CDI rate decreased between 2009 and 2011 and then due to the occurrence of several epidemic events, peaked in 2012, and fell to a preepidemic level in 2013. Overall, through the whole study period, there were no significant changes in either the CDI incidence or antibiotic use (Fig. 1). We did not find any association between CDI rate and antibiotic consumption. According to data provided for the Lombardy region, the CDI incidence in OSR and in other hospitals with similar characteristics was comparable (data not shown).
FIG 1.
CDI rate and antibiotic consumption from 2009 to 2013. (A) Distribution of CDI case incidence per 1,000 admissions or 10,000 patient-days. (B) Data on antibiotic consumption presented as defined daily doses (DDDs) per 100 patients per day for clindamycin, extended-spectrum cephalosporins (ESCs) (cefotaxime, ceftazidime, and ceftriaxone), cephalosporins, including ESC (cefazolin, cefoxitin, and cefotetan), fluoroquinolones (ciprofloxacin and levofloxacin), beta-lactamase-resistant (R) beta-lactams (ampicillin-sulbactam, amoxicillin-clavulanic acid, ticarcillin-clavulanic acid, and piperacillin-tazobactam), and macrolides (erythromycin, clarithromycin, and azithromycin).
C. difficile epidemiological and interpatient transmission data.
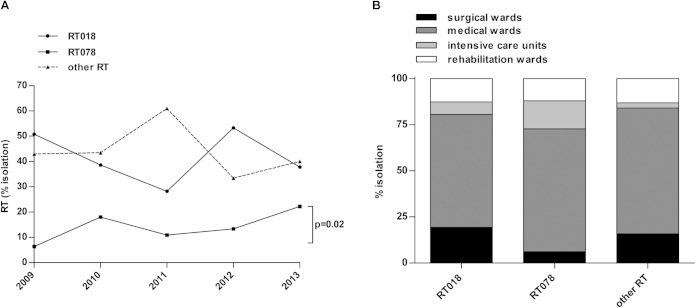
PCR ribotyping and epidemiological investigation distinguished 243 C. difficile strains isolated from index cases and 69 strains originating from secondary cases due to nosocomial transmission. Table 1 shows the distribution of the 243 C. difficile strains from index cases among the different RTs. We identified two major RTs accounting for 56% of the C. difficile strains: RT018 (42.4%; 103 of 243 strains) and RT078 (13.6%; 33 of 243 strains). The remaining 44% of the strains belonged to 8 identified RTs (12.3%; 30 of 243 strains) and 46 unidentified RTs (other RTs, 31.7%; 77 of 243 strains); only 2 strains belonged to RT027 (0.8%) (Table 1). Figure 2A shows the isolation trend of C. difficile strains (index cases) belonging to RT018, RT078, and all other RTs (non018 and non078) between 2009 and 2013; while RT018 displayed a fluctuating trend, RT078 presented a significantly increasing trend (P = 0.02).
TABLE 1.
PCR ribotypes accounting for C. difficile strains isolated from index case patientsa
| PCR ribotype | No. (%) of strains |
|---|---|
| 018 | 103 (42.4) |
| 078 | 33 (13.6) |
| 014/020 | 14 (5.8) |
| 002 | 4 (1.7) |
| 001 | 3 (1.2) |
| 056 | 3 (1.2) |
| 012 | 2 (0.8) |
| 027 | 2 (0.8) |
| 137 | 1 (0.4) |
| 106 | 1 (0.4) |
| Other | 77 (31.7) |
n = 243.
FIG 2.
Isolation and distribution of predominant PCR ribotypes. Percentage of C. difficile strains belonging to RT018, RT078, and all other (non-018 and non-078) RTs isolated from index case patients (n = 243) between 2009 and 2013 (A) and distribution in surgical, medical, and rehabilitation wards and intensive care units (B).
The majority of CDI index cases (65%; 158 of 243 cases) were detected in medical wards, followed by surgical (16%; 39 of 243 cases) and rehabilitation (12.8%; 31 of 243 cases) wards and intensive care units (6.2%; 15 of 243 cases). In particular, as shown in Fig. 2B, the distribution of C. difficile strains belonging to RT018, RT078, and all other RTs was similar in medical and rehabilitation wards. In contrast, in intensive care units, RT078 was detected more frequently than RT018 (15.2% versus 6.8%) and all other RTs (15.2% versus 2.8%; P = 0.02).
Ten of 243 C. difficile strains (4.1%) originated from patients with CA-CDI. Of these, 3 were RT018 (3 of 103; 2.9%), 1 was RT078 (1 of 33; 3%), and 6 belonged to other RTs (6 of 107; 5.6%).
Nosocomial transmission events were caused by three RTs, RT018, RT078, and RT014/020, which accounted for 95.7% (66 of 69), 2.9% (2 of 69), and 1.4% (1 of 69) of the secondary cases, respectively. The transmission index of RT018 (0.640) was significantly higher than that of RT078 (0.0606; P < 0.0001) or RT014/020 (0.0093; P = 0.0328).
Patients infected with RT018 were significantly older than those infected with RT078 or all other RTs. The median ages of patients with RT018, RT078, and all other RTs were 78 years, 71 years, and 69 years, respectively (RT018 versus RT078, P = 0.0074; RT018 versus all other RTs, P < 0.0001). Only 12.4% of patients with RT018 were younger than 65 years compared to 28.6% of patients with RT078 (P = 0.0255) and 38% of those with all other RTs (P < 0.0001).
Molecular characteristics of C. difficile strains.
Table 2 shows the molecular characteristics of C. difficile strains isolated from index cases. All strains tested positive for toxins A and B. The prevalence of strains with binary toxin was 18.5%; binary toxin was present in all RT078 strains and in 11.2% of other RT strains. The gyrA mutation C245T, conferring a high level of resistance to fluoroquinolones, was present in 56.8% of the strains. In particular, the presence of this mutation was associated with the two major RTs, 100% of the RT018 and 66.6% of the RT078 C. difficile strains, while only 12.1% of the strains belonging to all other RTs carried this mutation (RT018 and RT078 versus other RTs; P < 0.001). The presence of the ermB gene was observed in 23% of the strains. It was mainly detected in RT078 (42.4%) and in other RT strains (29%); only 10.7% of the RT018 strains were ermB positive (RT078 and other RTs versus RT018; P < 0.001). We did not observe significant changes in the frequency of detection of the gyrA mutation C245T or the gene ermB during the study period.
TABLE 2.
Molecular characteristics of C. difficile strains isolated from index cases
| PCR ribotype | Total no. of strains | No. (%) of strains with molecular characteristic: |
|||
|---|---|---|---|---|---|
| tcdA and tcdB | cdtA and cdtB | C245T in gyrA | ermB | ||
| 018 | 103 | 103 (100) | 0 (0) | 103 (100) | 11 (10.7) |
| 078 | 33 | 33 (100) | 33 (100) | 22 (66.6) | 14 (42.4) |
| Othera | 107 | 107 (100) | 12 (11.2) | 13 (12.1) | 31 (29) |
| Total | 243 | 243 (100) | 45 (18.5) | 138 (56.8) | 56 (23) |
Including RTs 014/020, 001, 002, 056, 012, 027, 137, and 106 and unidentified RTs.
All RT078 strains presented the 39-bp deletion and the nonsense mutation C184T in the tcdC gene. The two RT027 strains carried the binary toxin genes, the 18-bp deletion, the nucleotide deletion at position 117 in the tcdC gene, and the gyrA mutation C245T, and they were ermB negative.
DISCUSSION
Currently, longitudinal data on the burden of CDI in Italy are largely missing and uncertain. A very recent study, conducted in five hospitals in Rome over a 6-year period (2006 to 2011), reported an increase in the CDI incidence from 0.84 to 2.3 per 10,000 patient-days (29). In our study, we observed higher CDI rates, with an average value of 3.05 episodes per 10,000 patient-days, perfectly overlapping with incidence data of other similar hospitals of the Lombardy region. In addition, in our hospital between 2009 and 2013, we noticed a reduction in the CDI incidence that did not reach statistical significance due to the occurrence of an increasing peak in 2012. This peak was associated with the detection of a major number of nosocomial transmission events compared to the other years, partly due to changes in the routine workflow of the microbiology laboratory, which shortened the time to culture the positive toxin A&B stool samples, thus increasing the rate of positive C. difficile culture and improving the detection of outbreaks and epidemiological monitoring. In 2006, the infection control committee of the OSR finalized the first 5-year plan (2007 to 2011) for the management and prevention of health care-associated infections. This plan allowed implementing several important strategies: designation of infection control nurses and doctors in each ward; revision of the guidelines on hand hygiene, environment cleaning, and antiseptic and disinfectant use; introduction of guidelines on antibiotic prophylaxis; and education and training activities of health care workers. Moreover, molecular typing of the major multidrug-resistant organisms (MDROs), including C. difficile, was introduced to rapidly identify and limit epidemic clusters. The implementation of the guidelines on isolation and contact precautions was strengthened to control the spreading of MDROs and reduce the risk of transmission. Although none of these measures specifically targeted C. difficile, they could have contributed, at least in part, to the lowering of the number of CDI episodes between 2009 and 2011.
The use of broad-spectrum β-lactams, cephalosporins, fluoroquinolones, clindamycin, and macrolides has been associated with increasing CDI incidence (30–32). Through the study period, there were no significant changes in the consumption of these antibiotics, and, although infectious disease consultants were regularly involved in antibiotic therapy prescribing for infected patients in all wards, no antimicrobial stewardship programs were implemented at that time. Overall, we did not find any correlation between either the CDI trend or RT prevalence and the consumption of the different classes of antibiotics over time. The high number of interpatient transmission episodes and, consequently, the increase in the CDI incidence observed in 2012 did not seem to be linked to any specific antibiotic class consumption trend.
Although RT027 has been associated with the current CDI epidemic, there has been evidence recently that, in some European countries, the prevalence of this RT is starting to wane (18, 33). Our group reported the presence of RT027 in Italy in 2010; since then, it has been only sporadically detected (16, 17, 19). Here, we confirmed that the prevalence of RT027 is low, accounting for only 0.8% of all the C. difficile strains analyzed. As demonstrated by several studies, the molecular features of C. difficile are related to country and may change over time; thus, it is likely that other epidemic strains of C. difficile could emerge (9, 34–37). In agreement with the few Italian reports available, in our setting the predominant genotype during the whole study period was RT018, which accounted for 42.4% of the strains, followed by RT078, which was responsible for 13.6% of the CDI index cases (18, 20).
As previously reported, the fact that the prevalence of the gyrA mutation C245T was significantly higher in RT018 and RT078 strains than in other RT strains suggested that the use of fluoroquinolones could be a driving force for the selection and spread of successful C. difficile genotypes (2, 20). The ermB gene was observed with a significantly higher frequency in RT078. We also noticed that RT078 was most frequently detected in intensive care units compared to both RT018 and other RTs. Unfortunately, we were not able to collect patient clinical data to evaluate the severity of the C. difficile-associated disease. In Europe, RT078 has been reported to be the predominant RT identified in CA-CDI cases (11, 18), but we did not find in our population study any association between RT078 and a source of CDI in the community. This could be partially explained because the majority of our CDI cases were health care associated; however, the proportion of RT078 and RT018 strains responsible for CA-CDI was similar. Overall, the prevalence of RT078 significantly increased over time, but the interpatient transmissibility was very low, as highlighted by the transmission index (0.0606). In contrast, RT018 accounted for 95.7% of the secondary cases and was highly transmissible, presenting a transmission index 10-fold higher than that of RT078 (0.640). We also found that patients infected with an RT018 strain were significantly older than those infected with RT078 or other RT strains, confirming that old age represents an important risk factor for CDI and suggesting an association between the epidemic success of an RT and the elderly (3).
The hypervirulence of RT027 and RT078 has recently become a matter of debate. Indeed, there has been growing evidence that RT027 was not a significant predictor of severe CDI and poor clinical outcome in nonepidemic settings or in settings with a low RT027 prevalence (38–40). In addition, there has often been a misuse of the term hypervirulent as a synonym of epidemic, because most of the works supporting the hypothesis that RT027 strains are more virulent and related to more severe disease have been conducted in outbreak settings (41, 42). For pathogenic bacteria, lower virulence could favor host survival and, consequently, transmission to new susceptible hosts, and the success of an epidemic strain relies on its transmission capacity. In this study, we provided for the first time epidemiological evidence for inclusion of RT018 among successful epidemic genotypes. Our findings were further corroborated by recent works showing that RT018 was not only predominant and responsible for outbreaks in Asia, particularly in Korea and Japan, but also associated with CDI relapse (43–45).
This study has some limitations. Our data originate from a single, but very representative, health care setting. Also, only 70% of the isolates from the CDI cases diagnosed during the study time were available for the study.
Despite these limitations, the presented results indicate that C. difficile genotypes other than RT027 (e.g., RT018) deserve more attention in clinical practice for their epidemic potential and underline the importance of local surveillance program to identify and monitor areas of endemicity and epidemic C. difficile strains. Additionally, care should be taken when defining RT027 or RT078 as hypervirulent and using this term as a synonym for epidemic. More studies are needed to understand the epidemiological dynamics of C. difficile genotypes worldwide and to identify the features correlated to the epidemic behavior of a C. difficile strain. Until then, all genotypes should be monitored.
ACKNOWLEDGMENTS
We thank M. Rupnik (Maribor University, Slovenia), F. Agnoletti (Istituto Zooprofilattico Sperimentale delle Venezie, Italy), and V. Pasquale (Parthenope University of Naples, Italy) for providing representative C. difficile strains.
REFERENCES
- 1.Dubberke ER, Olsen MA. 2012. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55 (Suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors, and management. Nat Rev Gastroenterol Hepatol 8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 4.Kuijper EJ, Coignard B, Tüll P, ESCMID study group for Clostridium difficile, EU Member States, European Centre for Disease Prevention and Control. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12(Suppl 6):S2–S18. [DOI] [PubMed] [Google Scholar]
- 5.Walker AS, Eyre DW, Wyllie DH, Dingle KE, Griffiths D, Shine B, Oakley S, O'Connor L, Finney J, Vaughan A, Crook DW, Wilcox MH, Peto TE. 2013. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 56:1589–1600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Connor JR, Johnson S, Gerding DN. 2009. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136:1913–1924. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 7.Hunt JJ, Ballard JD. 2013. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol Mol Biol Rev 77:567–581. doi: 10.1128/MMBR.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcox MH, Shetty N, Fawley WN, Shemko M, Coen P, Birtles A, Cairns M, Curran MD, Dodgson KJ, Green SM, Hardy KJ, Hawkey PM, Magee JG, Sails AD, Wren MW. 2012. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin Infect Dis 55:1056–1063. doi: 10.1093/cid/cis614. [DOI] [PubMed] [Google Scholar]
- 9.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, Lambert ML, Drudy D, Fitzpatrick F, Wiuff C, Brown DJ, Coia JE, Pituch H, Reichert P, Even J, Mossong J, Widmer AF, Olsen KE, Allerberger F, Notermans DW, Delmée M, Coignard B, Wilcox M, Patel B, Frei R, Nagy E, Bouza E, Marin M, Akerlund T, Virolainen-Julkunen A, Lyytikäinen O, Kotila S, Ingebretsen A, Smyth B, Rooney P, Poxton IR, Monnet DL. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe. Euro Surveill 13:pii=18942 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=18942. [PubMed] [Google Scholar]
- 10.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 11.Smits WK. 2013. Hype or hypervirulence: a reflection on problematic C. difficile strains. Virulence 4:592–596. doi: 10.4161/viru.26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Lee Y, Lee K, Riley TV, Kim H. 2014. The changes of PCR ribotype and antimicrobial resistance of Clostridium difficile in a tertiary care hospital over 10 years. J Med Microbiol 63:819–823. doi: 10.1099/jmm.0.072082-0. [DOI] [PubMed] [Google Scholar]
- 13.Arvand M, Hauri AM, Zaiss NH, Witte W, Bettge-Weller G. 2009. Clostridium difficile ribotypes 001, 017, and 027 are associated with lethal C. difficile infection in Hesse, Germany. Euro Surveill 14:pii=19403 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=19403. [DOI] [PubMed] [Google Scholar]
- 14.Taori SK, Wroe A, Poxton IR. 2013. Clostridium difficile infections in southeast Scotland: mortality and recurrence in a region without PCR ribotype 027. J Med Microbiol 62:1468–1477. doi: 10.1099/jmm.0.061093-0. [DOI] [PubMed] [Google Scholar]
- 15.Borgmann S, Kist M, Jakobiak T, Reil M, Scholz E, von Eichel-Streiber C, Gruber H, Brazier JS, Schulte B. 2008. Increased number of Clostridium difficile infections and prevalence of Clostridium difficile PCR ribotype 001 in southern Germany. Euro Surveill 13:pii=19057 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=19057. [PubMed] [Google Scholar]
- 16.Baldan R, Cavallerio P, Tuscano A, Parlato C, Fossati L, Moro M, Serra R, Cirillo DM. 2010. First report of hypervirulent strains polymerase chain reaction ribotypes 027 and 078 causing severe Clostridium difficile infection in Italy. Clin Infect Dis 50:126–127. doi: 10.1086/649011. [DOI] [PubMed] [Google Scholar]
- 17.Di Bella S, Paglia MG, Johnson E, Petrosillo N. 2012. Clostridium difficile 027 infection in Central Italy. BMC Infect Dis 12:370. doi: 10.1186/1471-2334-12-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, study group ECDIS. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 19.Guastalegname M, Grieco S, Giuliano S, Falcone M, Caccese R, Carfagna P, D'ambrosio M, Taliani G, Venditti M. 2014. A cluster of fulminant Clostridium difficile colitis in an intensive care unit in Italy. Infection 42:585–589. doi: 10.1007/s15010-014-0597-1. [DOI] [PubMed] [Google Scholar]
- 20.Spigaglia P, Barbanti F, Dionisi AM, Mastrantonio P. 2010. Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J Clin Microbiol 48:2892–2896. doi: 10.1128/JCM.02482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furci L, Baldan R, Bianchini V, Trovato A, Ossi C, Cichero P, Cirillo DM. 2015. A new role for human α-defensin 5 in the fight against Clostridium difficile hypervirulent strains. Infect Immun 83:986–995. doi: 10.1128/IAI.02955-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer MP, Kuijper EJ, van Dissel JT, European Society of Clinical Microbiology and Infectious Diseases. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI). Clin Microbiol Infect 15:1067–1079. doi: 10.1111/j.1469-0691.2009.03099.x. [DOI] [PubMed] [Google Scholar]
- 23.McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK; Ad Hoc. Clostridium difficile Surveillance Working group. 2007. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol 28:140–145. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 24.Langlois-Klassen D, Senthilselvan A, Chui L, Kunimoto D, Saunders LD, Menzies D, Long R. 2013. Transmission of Mycobacterium tuberculosis Beijing strains, Alberta, Canada, 1991 to 2007. Emerg Infect Dis 19:701–711. doi: 10.3201/eid1905.121578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson S, Torpdahl M, Olsen KE. 2008. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 14:1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt C, Löffler B, Ackermann G. 2007. Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn Microbiol Infect Dis 59:1–5. doi: 10.1016/j.diagmicrobio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40:2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carman RJ, Genheimer CW, Rafii F, Park M, Hiltonsmith HF, Lyerly DM. 2009. Diversity of moxifloxacin resistance during a nosocomial outbreak of a predominantly ribotype ARU 027 Clostridium difficile diarrhea. Anaerobe 15:244–248. doi: 10.1016/j.anaerobe.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Di Bella S, Musso M, Cataldo MA, Meledandri M, Bordi E, Capozzi D, Cava MC, Chiaradonna P, Prignano G, Petrosillo N. 2013. Clostridium difficile infection in Italian urban hospitals: data from 2006 through 2011. BMC Infect Dis 13:146. doi: 10.1186/1471-2334-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernaz N, Hill K, Leggeat S, Nathwani D, Philips G, Bonnabry P, Davey P. 2009. Temporal effects of antibiotic use and Clostridium difficile infections. J Antimicrob Chemother 63:1272–1275. doi: 10.1093/jac/dkp128. [DOI] [PubMed] [Google Scholar]
- 31.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 32.Kaier K, Hagist C, Frank U, Conrad A, Meyer E. 2009. Two time-series analyses of the impact of antibiotic consumption and alcohol-based hand disinfection on the incidences of nosocomial methicillin-resistant Staphylococcus aureus infection and Clostridium difficile infection. Infect Control Hosp Epidemiol 30:346–353. doi: 10.1086/596605. [DOI] [PubMed] [Google Scholar]
- 33.Hensgens MP, Goorhuis A, Notermans DW, van Benthem BH, Kuijper EJ. 2009. Decrease of hypervirulent Clostridium difficile PCR ribotype 027 in the Netherlands. Euro Surveill 14:pii=19402 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=19402. [DOI] [PubMed] [Google Scholar]
- 34.Belmares J, Johnson S, Parada JP, Olson MM, Clabots CR, Bettin KM, Peterson LR, Gerding DN. 2009. Molecular epidemiology of Clostridium difficile over the course of 10 years in a tertiary care hospital. Clin Infect Dis 49:1141–1147. doi: 10.1086/605638. [DOI] [PubMed] [Google Scholar]
- 35.Brazier JS, Raybould R, Patel B, Duckworth G, Pearson A, Charlett A, Duerden BI, HPA Regional Microbiology Network. 2008. Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007 to 2008. Euro Surveill 13:pii=19000 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=19000. [DOI] [PubMed] [Google Scholar]
- 36.Terhes G, Brazier JS, Urban E, Soki J, Nagy E. 2006. Distribution of Clostridium difficile PCR ribotypes in regions of Hungary. J Med Microbiol 55:279–282. doi: 10.1099/jmm.0.46141-0. [DOI] [PubMed] [Google Scholar]
- 37.Sawabe E, Kato H, Osawa K, Chida T, Tojo N, Arakawa Y, Okamura N. 2007. Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a 5-year period. Eur J Clin Microbiol Infect Dis 26:695–703. doi: 10.1007/s10096-007-0355-8. [DOI] [PubMed] [Google Scholar]
- 38.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, Rogers MA, Washer L, Newton DW, Malani PN, Young VB, Aronoff DM. 2012. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis 55:1661–1668. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cloud J, Noddin L, Pressman A, Hu M, Kelly C. 2009. Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin Gastroenterol Hepatol 7:868–873. doi: 10.1016/j.cgh.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Barbut F, Rupnik M. 2012. Editorial commentary: 027, 078, and others: going beyond the numbers (and away from the hypervirulence). Clin Infect Dis 55:1669–1672. doi: 10.1093/cid/cis790. [DOI] [PubMed] [Google Scholar]
- 41.Pépin J, Valiquette L, Cossette B. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173:1037–1042. doi: 10.1503/cmaj.050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A. 2005. A predominantly clonal multiinstitutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 43.Collins DA, Hawkey PM, Riley TV. 2013. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2:21. doi: 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han SH, Kim H, Lee K, Jeong SJ, Park KH, Song JY, Seo YB, Choi JY, Woo JH, Kim WJ, Kim JM. 2014. Epidemiology and clinical features of toxigenic culture-confirmed hospital-onset Clostridium difficile infection: a multicentre prospective study in tertiary hospitals of South Korea. J Med Microbiol 63:1542–1551. doi: 10.1099/jmm.0.070672-0. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Seo MR, Kang JO, Kim Y, Hong SP, Pai H. 2014. Clinical characteristics of relapses and reinfections in Clostridium difficile infection. Clin Microbiol Infect 20:1198–1204. doi: 10.1111/1469-0691.12704. [DOI] [PubMed] [Google Scholar]