Abstract
This study evaluated the added value of selective preenrichment for the detection of rectal carriage of extended-spectrum-beta-lactamase-producing Enterobacteriaceae (ESBL-E). ESBL-E rectal carriage was identified in 4.8% of hospitalized patients, and 25.9% of ESBL-E rectal carriers were identified with selective preenrichment only.
TEXT
The steady worldwide spread of extended-spectrum-beta-lactamase-producing Enterobacteriaceae (ESBL-E) has affected health care and community settings, as well as livestock farming (1–4). Extended-spectrum beta-lactamase (ESBL) confers resistance to the majority of beta-lactam antibiotics, including third-generation cephalosporins, which limits the options for antimicrobial therapy and results in increased morbidity and mortality rates and health care costs (5, 6). Appropriate antimicrobial therapy and infection control measures depend on rapid sensitive laboratory detection methods. Unfortunately, reliable standardized methods for direct molecular detection of ESBL-E in clinical specimens are currently not available for routine use in medical microbiology. Therefore, the targeted ESBL-E screening of clinical specimens relies on the use of selective screening agars for ESBL, several of which, with comparable high sensitivities, have been described (7–13). It has been shown that preenrichment using a broth improves the performance of selective screening agars for the detection of methicillin-resistant Staphylococcus aureus in clinical specimens (14–17). However, the use of preenrichment for the detection of ESBL-E is still controversial (18) and is not common practice in clinical or research settings. Although several studies on the occurrence of ESBL-E have used nonselective or selective preenrichment (19, 20), comparative data that quantify the added value of preenrichment are limited. A comparative study showed that nonselective preenrichment improved the detection of ESBL-E in throat and rectal surveillance cultures from intensive care unit (ICU) patients (21). That study was performed in a setting in which all patients received selective decontamination of the digestive tract (SDD), and the use of a nonselective medium would hardly have been hampered by overgrowth with nonresistant flora. The detection of ESBL-E in fecal or rectal samples from patients who are not receiving antibiotics, however, is complicated by the presence of nonresistant gastrointestinal flora and would benefit from the use of selective culture media. At present, no comparative data on the use of preenrichment with a selective broth for the detection of ESBL-E rectal carriage are available. This study aimed to determine the added value of selective preenrichment for the detection of rectal carriage of ESBL-E in hospitalized patients.
In October 2011, an ESBL-E prevalence survey was performed in an 850-bed Dutch teaching hospital, where yearly ESBL-E prevalence surveys are part of the routine infection control policy. Rectal swabs were taken from all patients who were hospitalized on the day of the survey and were willing to participate in the survey.
The methods used to detect ESBL-E rectal carriage in this study are in accordance with national and international guidelines (18, 22). Nylon-flocked swabs with Amies liquid (ESwab) (Copan Diagnostics, Murrieta, CA) were used for collection and transport of rectal samples (23). Swabs were kept at 2 to 7°C and were processed within 24 h after sampling, at the local microbiology laboratory. The ESwab system enables the inoculation of multiple culture media with equivalent aliquots of the collected specimen and thus avoids the need to collect more than one sample per patient for comparison of culture methods (23). A blood agar plate and a selective ESBL screening agar (EbSA) plate (Cepheid Benelux, Apeldoorn, The Netherlands) (7) were each inoculated with 100 μl of the ESwab Amies liquid. The remaining Amies liquid was used to inoculate 5 ml of selective tryptic soy broth with vancomycin and cefotaxime (TSB-VC) (Cepheid Benelux, Apeldoorn, The Netherlands). EbSA has high sensitivity for the detection of ESBL-E, which is comparable to that of other selective ESBL screening agars (7, 11). EbSA plates consist of MacConkey agar and are divided into two sections. One section contains cefotaxime at 1 mg/liter and the other ceftazidime at 1 mg/liter. Both sections contain cloxacillin at 400 mg/liter, to inhibit the growth of Enterobacteriaceae that produce AmpC beta-lactamase, and vancomycin at 64 mg/liter, to inhibit the growth of enterococci. TSB-VC contains cefotaxime at 0.25 mg/liter and vancomycin at 8 mg/liter, to inhibit the growth of cefotaxime-susceptible Enterobacteriaceae and enterococci, respectively. Both agar plates and TSB-VC were incubated for 18 to 24 h at 35 to 37°C. Subsequently, 100 μl of the TSB-VC was subcultured on an EbSA plate that was incubated for another 18 to 24 h at 35 to 37°C. The blood agar plate served as a growth control. Cultures were rejected when the blood agar plate showed no bacterial growth, which was judged to be indicative of inappropriate sampling. The Vitek 2 system (bioMérieux, Marcy l'Etoile, France) was used for species identification and susceptibility testing of all isolates that grew on either one of the EbSA agar plates. Enterobacteriaceae with MICs for cefotaxime and/or ceftazidime above the screening breakpoint value of 1 mg/liter were considered suspect for the production of ESBL (18, 22). Production of ESBL was phenotypically confirmed with the combination disk diffusion method with cefotaxime (30 μg), ceftazidime (30 μg), and cefepime (30 μg), both alone and combined with clavulanic acid (10 μg) (Rosco, Taastrup, Denmark). The combination disk diffusion method has straightforward interpretation and is known for its high sensitivity and specificity (24). Test results were considered positive if the inhibition zone around the disk was ≥5 mm larger for the combination with clavulanic acid (18, 22). Finally, genotypic confirmation of the presence of ESBL genes was performed with the Check-MDR CT103 DNA microarray (Check-Points, Wageningen, The Netherlands), which covers a wide range of representatives of the most prevalent ESBL gene families (SHV, TEM, and CTX-M) (25).
On the day of the prevalence survey, 642 patients were hospitalized (Fig. 1). Rectal swabs were obtained from 570 patients (88.8%), of which 564 (98.9%) were evaluable. ESBL-E were detected in rectal swabs from 27 patients (4.8%), by either direct cultures or cultures with preenrichment. No ESBL-E were cultured for 535 patients (94.9%), and data for cultures with preenrichment were missing for 2 patients (0.4%). Escherichia coli was the predominant ESBL-positive species identified (n = 24 [88.9%]). The other ESBL-positive species were Morganella morganii (n = 1), Enterobacter cloacae (n = 1), and Pantoea agglomerans (n = 1). ESBL genes of the CTX-M-1 group, i.e., CTX-M-1-like (n = 10 [37.0%]), CTX-M-3-like (n = 1 [3.7%]), and CTX-M-15-like (n = 9 [33.3%]), were the most prevalent genes identified. Other ESBL genes belonged to the CTX-M-9 group (n = 3 [11.1%]), the SHV family (SHV 238S + 240K) (n = 3 [11.1%]), and the TEM family (TEM 104K + 238S) (n = 1 [3.7%]). The observed overall prevalence of ESBL-E rectal carriage of 4.8% and the distribution of ESBL genes are similar to those found in previous studies in Dutch health care settings (20, 26).
FIG 1.
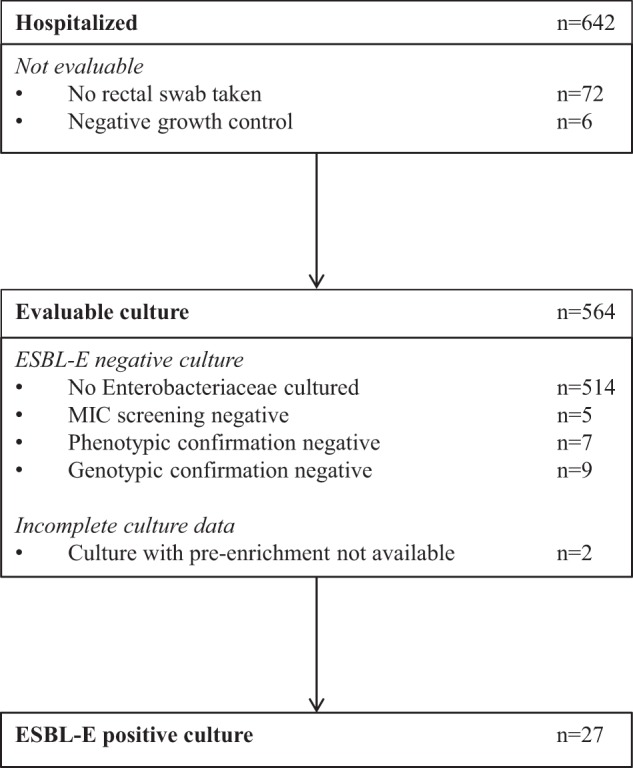
Recruitment of patients.
The performance of the culture methods was compared for patients with complete data for both methods and separately for ESBL-E-positive and ESBL-E-negative cultures (Table 1). The overall percent agreement and the exact conditional McNemar test (SPSS Statistics, version 19.0; IBM Corp., Armonk, NY) were used to compare concordant and discordant results, respectively. Rectal swabs from 20 patients were ESBL-E positive in both the direct cultures and the cultures with preenrichment. For 7 patients, ESBL-E were detected only in the cultures with preenrichment. The overall percent agreement between the methods for patients with ESBL-E-positive cultures was 74.1% (95% confidence interval [CI], 55.1% to 87.1%) (20/27 patients). The direct cultures failed to identify 25.9% (95% CI, 12.9% to 44.9%) (7/27 patients) of ESBL-E rectal carriers (McNemar test, P = 0.016), corresponding to 1.2% (95% CI, 0.5% to 2.6%) of the hospitalized patients (7/562 patients). Rectal swabs from 514 patients were ESBL-E negative in both the direct cultures and the cultures with preenrichment. False-positive growth was observed for 7 patients in the direct cultures and for 18 patients in the cultures with preenrichment. Therefore, the overall percent agreement between methods for patients with ESBL-E-negative cultures was 96.8% (95% CI, 94.9% to 98.0%) (518/535 patients), whereas a 2.1% increase (95% CI, 1.1% to 3.7%) (11/535 patients) in false-positive growth was observed for the cultures with preenrichment, compared to the direct cultures (McNemar test, P = 0.013). The ratio between the increase in the yield of screening (n = 7) and the increase in false-positive growth (n = 11) will most likely increase with increasing prevalence of ESBL-E rectal carriage. A sensitivity analysis in which the cultures with missing data for the cultures with preenrichment were included and their outcomes were varied to the extremes confirmed the findings of the primary analysis (data not shown).
TABLE 1.
Comparison of methods
| Result for direct culturea | Result for preenriched culturea |
|||
|---|---|---|---|---|
| No. of ESBL-E-positive culturesb |
No. of ESBL-E-negative culturesc |
|||
| Growth | No growth | Growth | No growth | |
| Growth | 20 | 0 | 4 | 3 |
| No growth | 7 | 0 | 14 | 514 |
Growth indicates the growth of Enterobacteriaceae. ESBL-E, extended-spectrum-beta-lactamase-producing Enterobacteriaceae.
Overall percent agreement, 74.1% (95% CI, 55.1% to 87.1%); exact conditional McNemar test, P = 0.016.
Overall percent agreement, 96.8% (95% CI, 94.9% to 98.0%); exact conditional McNemar test, P = 0.013.
Patient and strain characteristics were compared for ESBL-E-positive cultures that were detected either by both methods or by only the cultures with preenrichment (Table 2). A lack of growth in the direct cultures was not associated with species, the presence of inducible or derepressed chromosomal AmpC beta-lactamase genes, the type of ESBL gene, age, gender, the length of hospital stay, or antibiotic use.
TABLE 2.
Patient and strain characteristics of ESBL-E-positive cultures
| Characteristic | Both methods (n = 20) | Preenrichment only (n = 7) |
|---|---|---|
| Species (no.) | ||
| Escherichia coli | 19 | 5 |
| Morganella morganii | 1 | 0 |
| Enterobacter cloacae | 0 | 1 |
| Pantoea agglomerans | 0 | 1 |
| Chromosomal AmpC beta-lactamase gene (no.) | ||
| No | 19 | 5 |
| Yes | 1 | 2 |
| ESBL resistance gene (no.) | ||
| CTX-M-1-like | 6 | 4 |
| CTX-M-3-like | 1 | 0 |
| CTX-M-15-like | 8 | 1 |
| CTX-M-9 group | 3 | 0 |
| SHV 238S + 240K | 1 | 2 |
| TEM 104K + 238S | 1 | 0 |
| Age (yr) | ||
| Median | 64 | 73 |
| Range | 5–86 | 58–84 |
| Gender (no.) | ||
| Male | 12 | 4 |
| Female | 8 | 3 |
| Length of hospital stay (days) | ||
| Median | 5 | 4 |
| Range | 1–15 | 1–27 |
| Antibiotic use (no.)a | ||
| No | 16 | 7 |
| Amoxicillin-clavulanic acid, i.v. | 3 | 0 |
| Flucloxacillin, i.v. | 1 | 0 |
i.v., intravenously.
Whether there is a difference in the risk of nosocomial spread between ESBL-E identified in direct cultures and those identified in cultures with preenrichment cannot be concluded from our data. It could be argued that ESBL-E carriage that is not identified in direct cultures reflects low ESBL-E loads, with less potential for transmission to other patients. However, ESBL-E loads may vary over time, and they are known to increase following antibiotic use (21). As ESBL-E-screening is aimed at identifying patients who are at risk of spreading ESBL-E to other patients during hospitalization, and not only at the time of screening, we consider low ESBL-E loads potentially as important as high loads.
A disadvantage of preenrichment is the prolongation of the time to result by 1 day. For optimal rapidity and sensitivity, however, direct cultures and cultures with preenrichment can be used side by side. Whether this approach is cost-effective will depend on the local epidemiology of ESBL-E and the infection control measures applied to contain ESBL-E.
In conclusion, the use of selective preenrichment of rectal swabs improves the detection of ESBL-E rectal carriage. This potentially adds to infection control policies that aim to prevent the nosocomial spread of ESBL-E.
ACKNOWLEDGMENTS
This study was supported by the Netherlands Organization for Health Research and Development (ZonMw) (project 205100010).
We thank K. G. M. Moons and M. S. M. van Mourik for their advice on the analysis and presentation of the data. We are grateful to the infection control practitioners of our hospital for collecting demographic and clinical patient data and to the microbiology technicians of our laboratory for processing the rectal swabs.
REFERENCES
- 1.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control. 2014. Antimicrobial resistance surveillance in Europe 2013: annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control, Stockholm, Sweden: http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2013.pdf. [Google Scholar]
- 3.Dutch Foundation of the Working Party on Antibiotic Policy. 2014. NethMap 2014: consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands in 2013. Dutch Foundation of the Working Party on Antibiotic Policy, Rotterdam, Netherlands: http://www.wageningenur.nl/upload_mm/1/a/1/0704c512-5b42-4cef-8c1b-60e9e3fb2a62_NethMap-MARAN2014.pdf. [Google Scholar]
- 4.Central Veterinary Institute of Wageningen University and Research Centre. 2014. MARAN 2014: monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2013. Central Veterinary Institute of Wageningen University and Research Centre, Wageningen, Netherlands: http://www.wageningenur.nl/upload_mm/1/a/1/0704c512-5b42-4cef-8c1b-60e9e3fb2a62_NethMap-MARAN2014.pdf. [Google Scholar]
- 5.Ammerlaan HSM, Troelstra A, Kruitwagen CLJJ, Kluytmans JAJW, Bonten MJM. 2009. Quantifying changes in incidences of nosocomial bacteraemia caused by antibiotic-susceptible and antibiotic-resistant pathogens. J Antimicrob Chemother 63:1064–1070. doi: 10.1093/jac/dkp036. [DOI] [PubMed] [Google Scholar]
- 6.Rottier WC, Ammerlaan HSM, Bonten MJM. 2012. Effects of confounders and intermediates on the association of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae and patient outcome: a meta-analysis. J Antimicrob Chemother 67:1311–1320. doi: 10.1093/jac/dks065. [DOI] [PubMed] [Google Scholar]
- 7.Al Naiemi N, Murk JL, Savelkoul PHM, Vandenbroucke-Grauls CMJ, Debets-Ossenkopp YJ. 2009. Extended-spectrum beta-lactamases screening agar with AmpC inhibition. Eur J Clin Microbiol Infect Dis 28:989–990. doi: 10.1007/s10096-009-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glupczynski Y, Berhin C, Bauraing C, Bogaerts P. 2007. Evaluation of a new selective chromogenic agar medium for detection of extended-spectrum β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 45:501–505. doi: 10.1128/JCM.02221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stürenburg E, Sobottka I, Laufs R, Mack D. 2005. Evaluation of a new screen agar plate for detection and presumptive identification of Enterobacteriaceae producing extended-spectrum β-lactamases. Diagn Microbiol Infect Dis 51:51–55. doi: 10.1016/j.diagmicrobio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Huang TD, Bogaerts P, Berhin C, Guisset A, Glupczynski Y. 2010. Evaluation of Brilliance ESBL agar, a novel chromogenic medium for detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol 48:2091–2096. doi: 10.1128/JCM.02342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overdevest ITMA, Willemsen I, Elberts S, Verhulst C, Kluytmans JAJW. 2011. Laboratory detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae: evaluation of two screening agar plates and two confirmation techniques. J Clin Microbiol 49:519–522. doi: 10.1128/JCM.01953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Färber J, Moder KA, Layer F, Tammer I, König W, König B. 2008. Extended-spectrum beta-lactamase detection with different panels for automated susceptibility testing and with a chromogenic medium. J Clin Microbiol 46:3721–3727. doi: 10.1128/JCM.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paniagua R, Valverde A, Coque TM, Baquero F, Cantón R. 2010. Assessment of prevalence and changing epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae fecal carriers using a chromogenic medium. Diagn Microbiol Infect Dis 67:376–379. doi: 10.1016/j.diagmicrobio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Böcher S, Smyth R, Kahlmeter G, Kerremans J, Vos MC, Skov R. 2008. Evaluation of four selective agars and two enrichment broths in screening for methicillin-resistant Staphylococcus aureus. J Clin Microbiol 46:3136–3138. doi: 10.1128/JCM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böcher S, Middendorf B, Westh H, Mellmann A, Becker K, Skov R, Friedrich AW. 2010. Semi-selective broth improves screening for methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 65:717–720. doi: 10.1093/jac/dkq001. [DOI] [PubMed] [Google Scholar]
- 16.McAllister SK, Albrecht VS, Fosheim GE, Lowery HK, Peters PJ, Gorwitz R, Guest JL, Hageman J, Mindley R, McDougal LK, Rimland D, Limbago B. 2011. Evaluation of the impact of direct plating, broth enrichment, and specimen source on recovery and diversity of methicillin-resistant Staphylococcus aureus isolates among HIV-infected outpatients. J Clin Microbiol 49:4126–4130. doi: 10.1128/JCM.05323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkade E, Ferket M, Kluytmans J. 2011. Clinical evaluation of Oxoid Brilliance MRSA agar in comparison with bioMérieux MRSA ID medium for detection of livestock-associated meticillin-resistant Staphylococcus aureus. J Med Microbiol 60:905–908. doi: 10.1099/jmm.0.021964-0. [DOI] [PubMed] [Google Scholar]
- 18.Netherlands Society for Medical Microbiology. 2012. NVMM guideline: laboratory detection of highly resistant microorganisms (HRMO), version 2.0. Netherlands Society for Medical Microbiology, Leeuwarden, Netherlands: http://www.nvmm.nl/system/files/2012.11.15%20richtlijn%20BRMO%20%28version%202.0%29%20-%20RICHTLIJN.pdf. [Google Scholar]
- 19.Reuland EA, Overdevest ITMA, al Naiemi N, Kalpoe JS, Rijnsburger MC, Raadsen SA, Ligtenberg-Burgman I, van der Zwaluw KW, Heck M, Savelkoul PHM, Kluytmans JAJW, Vandenbroucke-Grauls CMJE. 2013. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin Microbiol Infect 19:542–549. doi: 10.1111/j.1469-0691.2012.03947.x. [DOI] [PubMed] [Google Scholar]
- 20.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis 17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murk JLAN, Heddema ER, Hess DLJ, Bogaards JA, Vandenbroucke-Grauls CMJE, Debets-Ossenkopp YJ. 2009. Enrichment broth improved detection of extended-spectrum-beta-lactamase-producing bacteria in throat and rectal surveillance cultures of samples from patients in intensive care units. J Clin Microbiol 47:1885–1887. doi: 10.1128/JCM.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing. 2013. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, version 1.0. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf. [Google Scholar]
- 23.Van Horn KG, Audette CD, Sebeck D, Tuckert KA. 2008. Comparison of the Copan ESwab system with two Amies agar swab transport systems for maintenance of microorganism viability. J Clin Microbiol 46:1655–1658. doi: 10.1128/JCM.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drieux L, Brossier F, Sougakoff W, Jarlier V. 2008. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14(Suppl 1):90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 25.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. 2012. Evaluation of a DNA microarray for the rapid detection of extended-spectrum β-lactamases (TEM, SHV and CTX-M), plasmid mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J Antimicrob Chemother 67:1865–1869. doi: 10.1093/jac/dks156. [DOI] [PubMed] [Google Scholar]
- 26.Hoogendoorn M, Smalbrugge M, Stobberingh EE, van Rossum SV, Vlaminckx BJ, Thijsen SF. 2013. Prevalence of antibiotic resistance of the commensal flora in Dutch nursing homes. J Am Med Dir Assoc 14:336–339. doi: 10.1016/j.jamda.2012.11.001. [DOI] [PubMed] [Google Scholar]


