Abstract
Widespread infections with community-associated (CA) methicillin-resistant Staphylococcus aureus (MRSA) have occurred in the United States with the dissemination of the USA300 strain beginning in 2000. We examined 105 isolates obtained from children treated at the University of Chicago from 1994 to 1997 (75 methicillin-susceptible S. aureus [MSSA] and 30 MRSA isolates) in order to investigate for possible evidence of USA300 during this period. Infections were defined epidemiologically based on medical record review. The isolates underwent multilocus sequence typing (MLST), as well as assays for the Panton-Valentine leukocidin (PVL) genes, the protein A gene (spa), and arcA and opp3, proxy markers for the arginine catabolic mobile element (ACME), characteristic of USA300 MRSA. MRSA isolates also underwent staphylococcal cassette chromosome mec (SCCmec) typing and pulsed-field gel electrophoresis (PFGE) subtyping. MSSA isolates belonged to 17 sequence type (ST) groups. The 12 epidemiologically defined CA-MRSA infection isolates were either ST1 (n = 4) or ST8 (n = 8). They belonged to 3 different PFGE types: USA100 (n = 1), USA400 (n = 5), and USA500 (n = 6). Among the CA-MRSA infection isolates, 8 (67%) were PVL+. None of the MRSA or MSSA isolates contained arcA or opp3. Only one MRSA isolate was USA300 by PFGE. This was a health care-associated (HA) MRSA isolate, negative for PVL, that carried SCCmec type II. USA300 with its characteristic features was not identified in the collection from the years 1994 to 1997.
INTRODUCTION
In the mid-1990s, there was a major shift in the clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) infections in the United States. One of the first reports of a different epidemiology for MRSA infections was from the children's hospital at the University of Chicago Medical Center (UCMC), in which previously healthy children were found to have MRSA infections (1). Notably, the incidence of MRSA infections among children having no previous exposure to the health care system increased significantly from 1988–1990 to 1993–1995 at the UCMC. Another report documented rapidly fatal infections in four children in North Dakota and Minnesota from 1997 to 1999 (2). A further rapid increase in the incidence of MRSA disease among previously healthy people was documented at the UCMC in 1998 to 1999 (3) and 2004 to 2005 (4), and in other geographic areas in the United States (5–16). These infections were caused by newly emergent strains of MRSA. Because these infections occurred in community settings, the new strains of MRSA causing these infections were called community-associated (CA) MRSA strains, and the infections, as defined by epidemiologic criteria (4, 6), have been called CA-MRSA infections.
CA-MRSA strains have been studied extensively. Pulsed-field gel electrophoresis (PFGE) revealed that the initial strains found in the Midwest, Alaska, and New York belonged to a single clonal group classified as USA400 (17). Since 2000, however, the vast majority of isolates causing CA-MRSA infections have belonged to clonal group USA300 (17–19).
The USA300 and USA400 strains almost always carry the Panton-Valentine leukocidin (PVL) toxin genes and staphylococcal chromosome cassette mec (SCCmec) type IV (18–20). The SCCmec element includes the mecA gene, which is necessary for the MRSA phenotype. Unlike other CA-MRSA strain types, USA300 most often carries another large chromosomally encoded element, the arginine catabolic mobile element (ACME), which likely enhances the ability of USA300 strains to live on the skin and resist antimicrobial peptides, including spermidine (21).
Prior to 2000, when CA-MRSA infections were relatively rare, there were many circulating CA-MRSA strain types (22). There is evidence that before 2000, USA400 was the predominant CA-MRSA strain type in the United States (23–25). However, there are few published data on clinical syndromes or on strain types of MRSA or methicillin-susceptible S. aureus (MSSA) isolates from children in the 1990s. Some have hypothesized that USA300 became common as a cause of MRSA infections after a precursor strain acquired the ACME (26).
We therefore set out to compare the clinical and microbiologic characteristics of infections caused by MRSA and MSSA isolates, including community-associated and health care-associated (HA) S. aureus infections, among children at the UCMC before 2000.
(Some data in this article were presented at the Annual Meeting, Infectious Diseases Society of America, Boston, MA, 20 to 23 October 2011.)
MATERIALS AND METHODS
Samples of MSSA (n = 75) and MRSA (n = 30) isolates were obtained from children treated at the UCMC from 1994 to 1997. All S. aureus isolates obtained from children during this period by the Clinical Microbiology Laboratory at the UCMC were prospectively collected and were frozen at −70°C. Several boxes of these isolates were lost between 1997 and 2011. All the isolates held in the boxes that remained were included in the present study, except that when there was >1 isolate in the collection from a single patient, only the first isolate obtained was included.
The study was approved by the Institutional Review Board (IRB) of the Biological Sciences Division of the University of Chicago. For each S. aureus isolate, the medical record was abstracted by a physician; data included age, gender, date of birth, date of S. aureus isolate collection, place of culture, anatomic site of culture, clinical diagnosis, medical history, antibiotic use in the previous 12 months, information on hospitalization, hemodialysis, indwelling catheters, surgery, day care attendance and stays in long-term-care facilities or rehabilitation centers in the previous 12 months, and history of skin lesions. Antimicrobial susceptibility data from the medical record, determined according to contemporary National Committee for Clinical Laboratory Standards (NCCLS) guidelines, were recorded for each S. aureus isolate. A clinical syndrome was assigned to each patient, and each isolate was categorized as being obtained from a colonized or infected patient. Infectious syndromes included skin and soft tissue infection (SSTI), bacteremia without focus, respiratory infection, urinary infection, bone and joint infection, or other. Each infection was classified as a CA, health care-associated-community onset (HACO), or hospital onset (HO) S. aureus infection by the CDC clinical criteria (27).
Stored isolates underwent multilocus sequence typing (MLST) (28) and PCR for detection of the PVL genes (29), the protein A gene (spa) (30), and arcA and opp3, proxy markers for the ACME (18). MRSA isolates underwent SCCmec typing (31, 32). All MRSA isolates underwent PFGE and were classified as USA100 to USA1200 according to a standard protocol (17). The hypervariable region of the spa gene was sequenced, and the spa type was determined (30) for all MRSA isolates identified as USA300 or USA500 by PFGE.
The presence of putative risk factors for the acquisition of MRSA infection was tabulated for all subjects. Individual univariate risk factors were assessed by a t test, Kruskal-Wallis test, χ2 test, or Fisher exact test, as appropriate. All analyses were performed using Stata, version 12 (StataCorp, College Station, TX).
RESULTS
Genotyping.
The 75 MSSA isolates belonged to 17 sequence types (STs) (Table 1).
TABLE 1.
Molecular characterization of 75 methicillin-susceptible Staphylococcus aureus isolates, University of Chicago Medical Center, 1995 to 1997a
| MLST | Total no. of isolates (%) | PVL | Yr of isolation | No. of isolates |
|---|---|---|---|---|
| 1 | 2 (2.7) | − | 1996 | 1 |
| + | 1996 | 1 | ||
| 5 | 10 (13.3) | − | 1995 | 2 |
| − | 1996 | 7 | ||
| − | 1997 | 1 | ||
| 8 | 4 (5.3) | − | 1996 | 3 |
| − | 1995 | 1 | ||
| 15 | 12 (16) | − | 1995 | 2 |
| − | 1996 | 7 | ||
| − | 1997 | 3 | ||
| 25 | 3 (4) | − | 1995 | 1 |
| − | 1996 | 1 | ||
| − | 1997 | 1 | ||
| 30 | 16 (21.3) | − | 1995 | 2 |
| − | 1996 | 7 | ||
| − | 1997 | 6 | ||
| + | 1996 | 1 | ||
| 39 | 1 (1.3) | − | 1996 | 1 |
| 45 | 13 (17.3) | − | 1995 | 3 |
| − | 1996 | 9 | ||
| − | 1997 | 1 | ||
| 51 | 1 (1.3) | − | 1996 | 1 |
| 59 | 4 (5.3) | − | 1995 | 1 |
| − | 1996 | 3 | ||
| 96 | 1 (1.3) | − | 1997 | 1 |
| 101 | 1 (1.3) | − | 1996 | 1 |
| 109 | 3 (4) | − | 1996 | 2 |
| − | 1995 | 1 | ||
| 121 | 1 (1.3) | + | 1995 | 1 |
| 435 | 1 (1.3) | − | 1996 | 1 |
| 474 | 1 (1.3) | − | 1996 | 1 |
| 508 | 1 (1.3) | − | 1995 | 1 |
| Total | 75 (100) | 75 |
All isolates were negative for opp3 and arcA, markers for the arginine catabolic mobile element (ACME), by PCR. MLST, multilocus sequence typing; PVL, Panton-Valentine leukocidin.
The 30 MRSA isolates belonged to 6 PFGE clonal groups (Table 2). Only 2 MRSA isolates could not be assigned by PFGE to a defined USA type. The MRSA isolates were assigned to 6 MLSTs. The most common were ST8 (12 isolates [40%]) and ST1 (11 isolates [37%]). The majority (24 isolates [80%]) carried SCCmec type IV. Eighteen of the MRSA isolates carried the PVL genes (60%). None carried arcA or opp3. All 8 USA500 MRSA isolates belonged to ST8, were PVL+, carried SCCmec type IV, and belonged to spa type t064. The single USA300 MRSA isolate as determined by PFGE, collected in 1995, was from a bloodstream infection and belonged to ST8. However, this isolate carried SCCmec type II; the ACME and PVL were absent; and it had spa type t064. Eight of 11 (73%) ST1 isolates and 10/12 (83%) ST8 MRSA isolates were PVL+ and carried SCCmec type IV (Table 2), characteristics of CA-MRSA isolates.
TABLE 2.
Molecular characterization of 30 methicillin-resistant Staphylococcus aureus isolates, University of Chicago Medical Center, 1994 to 1997a
| PFGE type (80% similarity cutoff)b | Total no. of isolates (%) | MLST | SCCmec type | Presence of gene |
Yr of acquisition | spa typec | No. of isolates | ||
|---|---|---|---|---|---|---|---|---|---|
| PVL | arcA | opp3 | |||||||
| NT | 2 (6.7) | 8 | IV | + | − | − | 1997 | NA | 1 |
| 12 | IV | − | − | − | 1997 | NA | 1 | ||
| USA100 | 2 (6.7) | 5 | II | − | − | − | 1995 | NA | 1 |
| 5 | II | − | − | − | 1997 | NA | 1 | ||
| USA200 | 3 (10.0) | 5 | II | − | − | − | 1994 | NA | 1 |
| 5 | II | − | − | − | 1995 | NA | 1 | ||
| 36 | II | − | − | − | 1996 | NA | 1 | ||
| USA300 | 1 (3.3) | 8 | II | − | − | − | 1995 | t064 | 1 |
| USA400 | 13 (43.3) | 1 | IV | + | − | − | 1996 | NA | 3 |
| 1 | IV | + | − | − | 1997 | NA | 5 | ||
| 1 | IV | − | − | − | 1995 | NA | 2 | ||
| 1 | IV | − | − | − | 1997 | NA | 1 | ||
| 8 | IV | + | − | − | 1997 | NA | 1 | ||
| 8 | IV | − | − | − | 1994 | NA | 1 | ||
| USA500 | 8 (26.7) | 8 | IV | + | − | − | 1996 | t064 | 6 |
| 8 | IV | + | − | − | 1997 | t064 | 2 | ||
| USA600 | 1 (3.3) | 45 | IV | − | − | − | 1997 | NA | 1 |
| Total | 30 (100) | 30 | |||||||
PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; PVL, Panton-Valentine leukocidin; arcA and opp3, markers for the arginine catabolic mobile element (ACME) detected by PCR.
The isolates fell within the standard criteria used for inclusion in the USA typing scheme (at least 80% similarity to the pattern of a particular USA type). NT, nontypeable; the isolate(s) indicated did not fall into one of the known USA type clonal groups. All isolates showed banding patterns by PFGE. If an isolate had >80% pattern similarity with multiple USA types, the USA type that had the highest percentage of similarity was designated as the type.
Only USA300 and USA500 isolates were genotyped using spa typing. NA, not applicable (testing was not conducted).
The PVL genes were carried by 60% (18/30) of MRSA isolates but only by 4% (3/75) of MSSA isolates (P < 0.001). The PVL+ MSSA isolates belonged to ST121 and ST1 (causing SSTIs) and ST30 (causing a joint infection). All HA- and HACO-MSSA infections were caused by PVL-negative isolates.
Comparing MSSA and MRSA infections.
There were no significant differences between MSSA and MRSA patients in clinical syndrome, age, gender, race, or site of care (inside or outside the hospital) at the time of culture (Table 3). The MSSA and MRSA patients also showed no significant differences in history of chronic skin, lung, liver, cardiac, or renal disease, cystic fibrosis, HIV infection, or use of systemic antibacterial antibiotics in the previous year (data not shown). There were no significant differences in history of hospitalization, surgery, hemodialysis, or stay in a long-term-care facility in the previous year, or in the presence of a central venous catheter at the time of culture. By CDC epidemiologic criteria, 12 MRSA infections (40% of those of known type by CDC epidemiologic criteria) were CA-MRSA, 13 (45%) were HO-MRSA, and 4 (14%) were HACO-MRSA infections; 2 infections were of unknown onset (Table 3).
TABLE 3.
Demographic and clinical characteristics of patients with methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus infections at the University of Chicago Medical Center, 1994 to 1997
| Characteristic | No. (%) of patients with: |
P | |
|---|---|---|---|
| MRSA (n = 30) | MSSA (n = 75) | ||
| Clinical syndrome | 0.5 | ||
| Bacteremia, endocarditis, or sepsis | 8 (27) | 14 (19) | |
| Osteomyelitis or septic arthritis | 0 | 4 (5) | |
| Respiratory | 3 (10) | 2 (3) | |
| Skin and soft tissue infection | 16 (53) | 36 (48) | |
| Urinary tract infection | 0 | 1 (1) | |
| Colonization | 3 (10) | 14 (19) | |
| Othera | 0 | 2 (3) | |
| Age group (yr) | 0.2 | ||
| <1 | 9 (30) | 26 (35) | |
| 1–2 | 7 (23) | 8 (11) | |
| 3–5 | 2 (7) | 13 (17) | |
| 6–12 | 9 (30) | 15 (20) | |
| 13–18 | 3 (10) | 13 (17) | |
| Gender | 0.3 | ||
| Male | 14 (47) | 43 (57) | |
| Female | 16 (53) | 32 (43) | |
| Race | 0.1 | ||
| African-American | 23 (77) | 42 (56) | |
| White | 3 (10) | 15 (20) | |
| Unknown or other | 4 (13) | 18 (24) | |
| Type of insurance | 0.5 | ||
| Public assistance | 23 (77) | 49 (65) | |
| Private | 6 (20) | 24 (32) | |
| Uninsured | 1 (3) | 2 (3) | |
| Presence of risk factors for HA-MRSA | |||
| Inpatient culture obtained >48 h after admission | 13 (46) | 31 (41) | 0.6 |
| Hospital stay, past yr | 8 (27) | 29 (39) | 0.2 |
| Surgery, past yr | 5 (17) | 26 (35) | 0.07 |
| Hemodialysis, past yr | 0 | 0 | NAc |
| Indwelling catheter | 9 (30) | 30 (40) | 0.9 |
| Stay in long-term-care facility, past yr | 0 | 3 (4) | 0.6 |
| Location of careb | 0.1 | ||
| Intensive care units | 9 (30) | 32 (43) | |
| Other inpatient units | 13 (43) | 26 (35) | |
| Emergency department | 4 (13) | 6 (8) | |
| Outpatient | 2 (7) | 11 (15) | |
| Unknown | 2 (7) | 0 | |
| CDC criteria for infection type | 0.2 | ||
| CA | 12 (40) | 21 (28) | |
| HO | 13 (45) | 31 (41) | |
| HACO | 4 (14) | 23 (31) | |
| Previous MRSA isolation, UCMC laboratory report | 5 (17) | 2 (3) | 0.02 |
Other clinical syndromes include sinusitis and liver abscess.
The location of care was unknown for 2 subjects.
NA, not applicable.
Among the MSSA isolates, 21 (28%) were classified as CA-MSSA, 31 (41%) as HO-MSSA, and 23 (31%) as HACO-MSSA infection isolates. There was no significant difference between MSSA and MRSA infections in the percentage of isolates cultured from inpatients >48 h after hospital admission or in the percentage classified as CA or HA S. aureus according to CDC epidemiologic criteria (Table 3).
Resistance to clindamycin and gentamicin, although unusual, was more common among MRSA isolates than among MSSA isolates in our sample (23% versus 5% for clindamycin [P = 0.005] and 17% versus 3% for gentamicin [P = 0.02], respectively). Clindamycin resistance, assessed by single-agent testing (i.e., the D-test for inducible clindamycin resistance was not performed), was identified in 8 MRSA isolates, including 2 USA100, 3 USA200, and 1 USA600 isolate, the USA300 MRSA isolate, and 1 isolate for which no PFGE type could be assigned.
Comparing the genotypes and clinical syndromes of CA, HO, and HACO infections.
All 12 (39%) CA-MRSA infection isolates (as defined by CDC criteria) were either ST1 (n = 4 [33%]) or ST8 (n = 8 [67%]). They belonged to 3 PFGE types: USA100 (n = 1), USA400 (n = 5), and USA500 (n = 6).
Among the CA-MRSA infection isolates, 8 (67%) were PVL+. Among the HO- and HACO-MRSA isolates, 10/19 (53%) were PVL+.
Among the CA-MSSA isolates, 3/21 (14%) were PVL+ and belonged to ST1, ST30, or ST121. No HA-MSSA infections were caused by PVL+ isolates.
The 2 USA100 MRSA isolates were obtained from a CA-MRSA SSTI and from a HA-MRSA bloodstream infection.
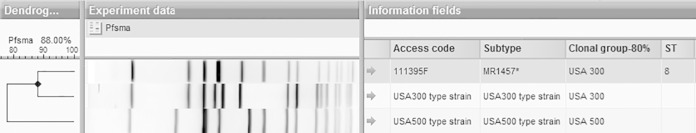
The USA300 MRSA infection isolate was from a case of HA-MRSA bacteremia and was resistant to clindamycin, erythromycin, gentamicin, and trimethoprim-sulfamethoxazole. The PFGE pattern of this isolate is shown in Fig. 1.
FIG 1.
Dendrogram showing the PFGE gel result for the sole USA300 MRSA isolate obtained in this study (isolate 111395F, which belonged to ST8, carried SCCmec type II, did not carry the PVL gene or arcA, and had spa type t064) as well as those for control USA300 and USA500 strains.
USA400 MRSA infections, of which 5/12 (42%) were CA-MRSA infections by CDC criteria, included SSTIs (n = 8), bacteremia (n = 1), and respiratory infections (n = 3). The 5 USA400 CA-MRSA infections were all SSTIs.
Interestingly, USA500 isolates accounted for 8 of the 30 MRSA isolates (27%). These were uniformly PVL+, which is unusual for USA500 isolates. All of them also carried SCCmec type IV, lacked arcA, and belonged to spa type t064. The majority (6/8 [75%]) were from CA-MRSA infections. The 6 USA500 CA-MRSA infections were SSTIs; of the 2 USA500 HA-MRSA isolates, 1 colonized a patient asymptomatically and the other caused a bloodstream infection.
DISCUSSION
USA300 has become the most common genetic background of MRSA causing infections in the United States, but its origins have not been well explored in epidemiologic studies. The emergence of USA300 MRSA was extremely rapid. We found that USA300 was almost absent among the MRSA isolates that we obtained before 1998 at the UCMC despite the early report of CA-MRSA infections in children at our center increasing between 1988 and 1995 (1). The single pre-1998 USA300 isolate identified in the present study differed genotypically from typical USA300 isolates in that it lacked the genes encoding the PVL toxin and arcA from the ACME, and it carried SCCmec type II. At the UCMC in 1994 to 1997, USA400 and USA500 isolates were likely the most common clonal types causing MRSA infections in children.
It has been suggested that USA500 was a precursor to USA300 MRSA, which became common after 2000 (26). The pulsotype of one isolate in our study clustered with USA300, but the genetic characteristics more closely resembled those of USA500, since this isolate lacked arcA and had spa type t064. This is consistent with the possibility that this single USA300 MRSA isolate (by pulsotype) was a progenitor that arose from USA500.
It is noteworthy that we identified 2 isolates that were USA400 but were typed as ST8 by MLST. This was an unexpected finding.
The group of MSSA and MRSA patients in 1994 to 1997 for whom we had isolates did not differ in the demographic or clinical variables assessed, suggesting that the risk factors for MRSA infections were similar to those for MSSA infections in children at our center. This is surprising, because in the era before 2000, it may have been surmised that MRSA infections would still have been most common among children with health care exposure. Furthermore, the MSSA backgrounds identified in 1994 to 1997, with a predominance of ST5, ST15, ST30, and ST45, were similar in distribution to the STs of MSSA isolates identified at our center in adults and children in 2004 to 2005 (33). Also, in 2004 to 2005, 7% of 114 MSSA isolates from children and adults at the UCMC were PVL+, and in our sample from children in 1994 to 1997, 4% of 75 MSSA isolates were PVL+, a nonsignificant difference (33). This suggests that the genotypes of MSSA isolates collected from patients at the UCMC did not change dramatically in the first decade of the CA-MRSA era in the United States (1993 to 2003).
Few MRSA isolates obtained from patients in the United States before 2001 have been genotyped. From a review of the published literature, we identified 9 articles (34–42) that included RIDOM (ribosomal differentiation of medical microorganisms) spa typing, MLST, or USA system PFGE typing data on 438 MRSA isolates, collected mostly from adults in the United States in 1988 to 2000. For 342 isolates, MLST was performed, identifying 27 STs. The 7 commonest STs accounted for 88.6% (303/342) of the isolates; these were ST1 (n = 15), ST5 (n = 66), ST8 (n = 95), ST30 (n = 13), ST36 (n = 72), ST59 (n = 25), and ST105 (n = 17). Among 107 isolates that underwent spa typing, no isolates of type t008, the type most closely associated with USA300, were reported. Many of the spa types reported in references 34 to 42 have been associated with either ST1 or USA400 in later studies (43–49). None was typed as USA300 or carried both the ACME and PVL. Importantly, however, PFGE was not performed on many of the ST8 isolates reported (see Table S1 in the supplemental material).
Our study has certain limitations. We studied the available sample of MRSA and MSSA isolates from 1994 to 1997. There was no known bias in the isolate collection, and we believe that we were able to ascertain the major STs circulating at our center at the time. The study was retrospective, and therefore, we were not able to interview the patients. We may not have identified all risk factors for health care exposure in our medical record review, which may have resulted in the misclassification of CA- and HACO-MRSA patients. Our study may have been underpowered to detect characteristics associated with MRSA and MSSA patients, but we examined all subjects for whom isolates were available.
In summary, we identified only a single USA300 isolate, which lacked the ACME and PVL, from 1994 to 1997, suggesting that the typical USA300 MRSA, prevalent after 2000, was not yet circulating before 1998 in the pediatric population served at the UCMC. In contrast, USA500/ST8-IV-t064 MRSA was commonly identified. We suggest that the USA500 strain type may have been a precursor of USA300. USA400 and USA500 were likely the predominant MRSA strain types circulating among children at our center in 1994 to 1997. Further genomic studies of these strain types may elucidate the relationship among these types and suggest the critical differences that enabled USA300 MRSA to succeed as the predominant CA-MRSA strain type in North America. We found that few clinical characteristics could distinguish pediatric patients with MRSA and MSSA infections during the study period. There was little change in the circulating MSSA strain types at our center between 1995 to 1997 and 2004 to 2005, and PVL+ MSSA strains were exceptional in 1994 to 1997 and remained so in 2004 to 2005.
Supplementary Material
ACKNOWLEDGMENT
M.Z.D. was supported by NIH NIAID grant K23 AI095361.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00096-15.
REFERENCES
- 1.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC). 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. MMWR Morb Mortal Wkly Rep 48:707–710. [PubMed] [Google Scholar]
- 3.Hussain FM, Boyle-Vavra S, Bethel CD, Daum RS. 2000. Current trends in community-acquired methicillin-resistant Staphylococcus aureus at a tertiary care pediatric facility. Pediatr Infect Dis J 19:1163–1166. doi: 10.1097/00006454-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 4.David MZ, Glikman D, Crawford SE, Peng J, King KJ, Hostetler MA, Boyle-Vavra S, Daum RS. 2008. What is community-associated methicillin-resistant Staphylococcus aureus? J Infect Dis 197:1235–1243. doi: 10.1086/533502. [DOI] [PubMed] [Google Scholar]
- 5.Naimi TS, LeDell KH, Boxrud DJ, Groom AV, Steward CD, Johnson SK, Besser JM, O'Boyle C, Danila RN, Cheek JE, Osterholm MT, Moore KA, Smith KE. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin Infect Dis 33:990–996. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- 6.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK; Active Bacterial Core surveillance (ABCs) MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 7.Casey JA, Cosgrove SE, Stewart WF, Pollak J, Schwartz BS. 2013. A population-based study of the epidemiology and clinical features of methicillin-resistant Staphylococcus aureus infection in Pennsylvania, 2001–2010. Epidemiol Infect 141:1166–1179. doi: 10.1017/S0950268812001872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landrum ML, Neumann C, Cook C, Chukwuma U, Ellis MW, Hospenthal DR, Murray CK. 2012. Epidemiology of Staphylococcus aureus blood and skin and soft tissue infections in the US military health system, 2005–2010. JAMA 308:50–59. doi: 10.1001/jama.2012.7139. [DOI] [PubMed] [Google Scholar]
- 9.Purcell K, Fergie JE. 2002. Exponential increase in community-acquired methicillin-resistant Staphylococcus aureus infections in South Texas children. Pediatr Infect Dis J 21:988–989. doi: 10.1097/00006454-200210000-00028. [DOI] [PubMed] [Google Scholar]
- 10.Sattler CA, Mason EO Jr, Kaplan SL. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr Infect Dis J 21:910–917. doi: 10.1097/00006454-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Hultén KG, Kaplan SL, Gonzalez BE, Hammerman WA, Lamberth LB, Versalovic J, Mason EO Jr. 2006. Three-year surveillance of community onset health care-associated Staphylococcus aureus infections in children. Pediatr Infect Dis J 25:349–353. doi: 10.1097/01.inf.0000207404.50143.1e. [DOI] [PubMed] [Google Scholar]
- 12.Ochoa TJ, Mohr J, Wanger A, Murphy JR, Heresi GP. 2005. Community-associated methicillin-resistant Staphylococcus aureus in pediatric patients. Emerg Infect Dis 11:966–968. doi: 10.3201/eid1106.050142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank AL, Marcinak JF, Mangat PD, Schreckenberger PC. 1999. Increase in community-acquired methicillin-resistant Staphylococcus aureus in children. Clin Infect Dis 29:935–936. doi: 10.1086/520463. [DOI] [PubMed] [Google Scholar]
- 14.Buckingham SC, McDougal LK, Cathey LD, Comeaux K, Craig AS, Fridkin SK, Tenover FC. 2004. Emergence of community-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee children's hospital. Pediatr Infect Dis J 23:619–624. doi: 10.1097/01.inf.0000131981.67342.c4. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez K, Halpern MS, Sarnquist C, Soni S, Arroyo AC, Maldonado Y. 2013. Staphylococcal infections in children, California, USA, 1985–2009. Emerg Infect Dis 19:10–20. doi: 10.3201/eid1901.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen AE, Goldstein M, Carroll K, Song X, Perl TM, Siberry GK. 2006. Evolving epidemiology of pediatric Staphylococcus aureus cutaneous infections in a Baltimore hospital. Pediatr Emerg Care 22:717–723. doi: 10.1097/01.pec.0000236832.23947.a0. [DOI] [PubMed] [Google Scholar]
- 17.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 19.Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother 64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 20.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan ES, Diep BA, Carleton HA, Charlebois ED, Sensabaugh GF, Haller BL, Perdreau-Remington F. 2003. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin Infect Dis 37:1384–1388. doi: 10.1086/379019. [DOI] [PubMed] [Google Scholar]
- 23.Furtado N, Mangat PD, Frank AL, Janda W, Boyle S, Daum RS. 2006. Two clindamycin susceptible (community associated) MRSA epidemics in Chicago, abstr 378 Prog Abstr 44th Annu Meet Infectious Diseases Society of America (IDSA), Toronto, Ontario, Canada, 2006 IDSA, Arlington, VA. [Google Scholar]
- 24.David MZ, Rudolph KM, Hennessy TW, Zychowski DL, Asthi K, Boyle-Vavra S, Daum RS. 2012. MRSA USA300 at Alaska Native Medical Center, Anchorage, Alaska, USA, 2000–2006. Emerg Infect Dis 18:105–108. doi: 10.3201/eid1801.110746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tattevin P, Diep BA, Jula M, Perdreau-Remington F. 2008. Long-term follow-up of methicillin-resistant Staphylococcus aureus molecular epidemiology after emergence of clone USA300 in San Francisco jail populations. J Clin Microbiol 46:4056–4057. doi: 10.1128/JCM.01372-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4(6):e00889–13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallen AJ, Mu Y, Bulens S, Reingold A, Petit S, Gershman K, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Patel PR, Fridkin SK; Active Bacterial Core surveillance (ABCs) MRSA Investigators of the Emerging Infections Program. 2010. Health care-associated invasive MRSA infections, 2005–2008. JAMA 304:641–648. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 28.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 30.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J Clin Microbiol 43:4719–4730. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David MZ, Boyle-Vavra S, Zychowski DL, Daum RS. 2011. Methicillin-susceptible Staphylococcus aureus as a predominantly healthcare-associated pathogen: a possible reversal of roles? PLoS One 6(4):e18217. doi: 10.1371/journal.pone.0018217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla SK, Stemper ME, Ramaswamy SV, Conradt JM, Reich R, Graviss EA, Reed KD. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J Clin Microbiol 42:3752–3757. doi: 10.1128/JCM.42.8.3752-3757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla SK, Karow ME, Brady JM, Stemper ME, Kislow J, Moore N, Wroblewski K, Chyou PH, Warshauer DM, Reed KD, Lynfield R, Schwan WR. 2010. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J Clin Microbiol 48:3582–3592. doi: 10.1128/JCM.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray CK, Holmes RL, Ellis MW, Mende K, Wolf SE, McDougal LK, Guymon CH, Hospenthal DR. 2009. Twenty-five year epidemiology of invasive methicillin-resistant Staphylococcus aureus (MRSA) isolates recovered at a burn center. Burns 35:1112–1117. doi: 10.1016/j.burns.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Faria NA, Carrico JA, Oliveira DC, Ramirez M, de Lencastre H. 2008. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol 46:136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crum NF, Lee RU, Thornton SA, Stine OC, Wallace MR, Barrozo C, Keefer-Norris A, Judd S, Russell KL. 2006. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. Am J Med 119:943–951. doi: 10.1016/j.amjmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Chung M, Dickinson G, De Lencastre H, Tomasz A. 2004. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J Clin Microbiol 42:542–547. doi: 10.1128/JCM.42.2.542-547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brady JM, Stemper ME, Weigel A, Chyou PH, Reed KD, Shukla SK. 2007. Sporadic “transitional” community-associated methicillin-resistant Staphylococcus aureus strains from health care facilities in the United States. J Clin Microbiol 45:2654–2661. doi: 10.1128/JCM.02579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlebois ED, Perdreau-Remington F, Kreiswirth B, Bangsberg DR, Ciccarone D, Diep BA, Ng VL, Chansky K, Edlin BR, Chambers HF. 2004. Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 39:47–54. doi: 10.1086/421090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira DC, Milheiriço C, Vinga S, de Lencastre H. 2006. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J Antimicrob Chemother 58:23–30. doi: 10.1093/jac/dkl208. [DOI] [PubMed] [Google Scholar]
- 43.Stemper ME, Brady JM, Qutaishat SS, Borlaug G, Reed J, Reed KD, Shukla SK. 2006. Shift in Staphylococcus aureus clone linked to an infected tattoo. Emerg Infect Dis 12:1444–1446. doi: 10.3201/eid1209.051634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li V, Chui L, Louie L, Simor A, Golding GR, Louie M. 2013. Cost-effectiveness and efficacy of spa, SCCmec, and PVL genotyping of methicillin-resistant Staphylococcus aureus as compared to pulsed-field gel electrophoresis. PLoS One 8(11):e79149. doi: 10.1371/journal.pone.0079149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K, McClure JA, Elsayed S, Tan J, Conly JM. 2008. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J Infect Dis 197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 46.Tristan A, Bes M, Meugnier H, Lina G, Bozdogan B, Courvalin P, Reverdy ME, Enright MC, Vandenesch F, Etienne J. 2007. Global distribution of Panton-Valentine leukocidin–positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis 13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartels MD, Boye K, Rhod Larsen A, Skov R, Westh H. 2007. Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg Infect Dis 13:1533–1540. doi: 10.3201/eid1310.070503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li V, Chui L, Simmonds K, Nguyen T, Golding GR, Yacoub W, Ferrato C, Louie M. 2014. Emergence of new CMRSA7/USA400 methicillin-resistant Staphylococcus aureus spa types in Alberta, Canada, from 2005 to 2012. J Clin Microbiol 52:2439–2446. doi: 10.1128/JCM.00505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williamson DA, Roberts SA, Ritchie SR, Coombs GW, Fraser JD, Heffernan H. 2013. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in New Zealand: rapid emergence of sequence type 5 (ST5)-SCCmec-IV as the dominant community-associated MRSA clone. PLoS One 8(4):e62020. doi: 10.1371/journal.pone.0062020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.