Abstract
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) is an emerging technology for rapid identification of bacterial and fungal isolates. In comparison to conventional methods, this technology is much less labor intensive and can provide accurate and reliable results in minutes from a single isolated colony. We compared the cost of performing the bioMérieux Vitek MALDI-TOF MS with conventional microbiological methods to determine the amount saved by the laboratory by converting to the new technology. Identification costs for 21,930 isolates collected between April 1, 2013, and March 31, 2014, were directly compared for MALDI-TOF MS and conventional methodologies. These isolates were composed of commonly isolated organisms, including commonly encountered aerobic and facultative bacteria and yeast but excluding anaerobes and filamentous fungi. Mycobacterium tuberculosis complex and rapidly growing mycobacteria were also evaluated for a 5-month period during the study. Reagent costs and a total cost analysis that included technologist time in addition to reagent expenses and maintenance service agreement costs were analyzed as part of this study. The use of MALDI-TOF MS equated to a net savings of $69,108.61, or 87.8%, in reagent costs annually compared to traditional methods. When total costs are calculated to include technologist time and maintenance costs, traditional identification would have cost $142,532.69, versus $68,886.51 with the MALDI-TOF MS method, resulting in a laboratory savings of $73,646.18, or 51.7%, annually by adopting the new technology. The initial cost of the instrument at our usage level would be offset in about 3 years. MALDI-TOF MS not only represents an innovative technology for the rapid and accurate identification of bacterial and fungal isolates, it also provides a significant cost savings for the laboratory.
INTRODUCTION
Mass spectrometry (MS) has been traditionally utilized for chemical analysis, although its utility was limited to low-molecular-weight organic compounds (1). Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS expanded the field and allowed for the analysis of biological molecules with no theoretical upper limit of mass (2). Previously employed to determine the mass of peptides and proteins, this emerging technology has been adapted for rapid identification of bacterial and fungal isolates in the clinical microbiology laboratory. In an increasing number of settings, MALDI-TOF MS has replaced traditional identification methods, including microscopy and determination of phenotypic characteristics, which typically require multiple steps (3, 4).
The performance of MALDI-TOF MS for the identification of microorganisms was examined in numerous studies and shown to be accurate and reliable (5–8). In comparison to conventional methods, this technology is much less labor intensive and can provide accurate and reliable results in minutes from a single isolated colony. The only reagents utilized are the target slides that contain sample spots used for the identification of microorganisms, an organic matrix solution, formic acid for yeast isolates, pipette tips, and disposable loops or toothpicks for sample application. Therefore, reagent rental programs are usually not available from the two manufacturers of MALDI-TOF MS, so the instrument is purchased, which is a large capital cost to the laboratory. In addition, fairly expensive annual maintenance contracts of $25,000 to $30,000 are needed to ensure limited downtime of this technically complex equipment. Causes of downtime are multifaceted and consist of various issues, including time required for remote access to software for manipulation or tuning, and hardware maintenance/repair, reducing the time the instrument is available on a given day for identification of isolates. Sufficient protocols need to be in place for times when the MALDI-TOF MS will be unavailable. Although the initial instrumentation price is high and maintenance expenses are significant, the cost of identifying an isolate can be very low. Based on this, use of MALDI-TOF MS has the potential to improve laboratory efficiency, reduce turnaround times, and lower costs (9).
We compared the cost of performing the bioMérieux Vitek MALDI-TOF MS (Durham, NC, USA) with that of conventional microbiological methods to determine the amount saved by the laboratory after converting to the new technology. This study examined the cost savings encountered when reagents and total cost were calculated.
MATERIALS AND METHODS
Study design.
We conducted a 12-month retrospective analysis of potential cost savings incurred by the Clinical Microbiology and Immunology Laboratory (CMIL) at the University of North Carolina Hospitals (UNCH) after implementation of MALDI-TOF MS for routine identification of bacteria and yeasts. UNCH is an 803-bed academic medical center composed of acute care, cancer, children's, neuroscience, and women's hospitals located in Chapel Hill, North Carolina. Identification costs for 21,930 isolates collected during a 1-year period between April 1, 2013, and March 31, 2014, associated with MALDI-TOF MS and conventional methodologies were directly compared. Specimens consisted of the most commonly isolated organisms in the clinical microbiology laboratory, including Enterobacteriaceae, enterococci, Gram-negative glucose nonfermenters (GNFs), staphylococci, streptococci, and yeast. A comprehensive and structured review of the laboratory information system (SCC Soft Computer, Clearwater, FL) was conducted to determine the number of isolates identified during the study period.
Analysis of costs.
Three cost calculations were analyzed as part of this study: reagent cost, a direct cost analysis that included technologist time in addition to reagent expenses, and a total cost analysis that included reagent, technologist, and maintenance costs. Reagent expenditures associated with traditional microbiological methods and MALDI-TOF MS were directly calculated for each specimen in the study. A standardized formula was used to determine total technologist cost: technologist cost = (identification time per isolate [minutes]) × (number of isolates identified) × (mean base technologist salary [per minute]). This was then added to reagent costs to derive the direct cost: direct cost = technologist cost + reagent cost. Lastly, we calculated the total cost with the addition of the maintenance service agreement contracts to provide a full picture of the total savings incurred over the life of the instrument: total cost = direct cost + maintenance cost/isolate.
Since the maintenance contract was a fixed rather than a variable cost, we determined the maintenance cost based on our actual maintenance contract amount for fiscal year 2014 and divided that by the total number of isolates identified. Our maintenance cost for the instrument and software for fiscal year 2014 was $29,700, which included the annual service agreement covering the instrument hardware and Myla software.
Identification time was calculated based on prior experiences with traditional and MALDI-TOF MS methods. Traditional microbiological methods utilized by the UNCH CMIL include the Vitek 2 microbial identification system (bioMérieux, Durham, NC) for identification of Enterobacteriaceae. Escherichia coli isolates were an exception, as identification was based mainly on colony morphology and spot indole (Remel, Lenexa, KS). Vitek 2, pyrrolidonyl arylamidase (PYR) disks, 6.5% NaCl broth, and bile esculin agar (Remel, Lenexa, KS) were utilized for identification of enterococci, while oxidase (Remel), Vitek 2, and molecular sequencing were used for detection of GNFs. Staphylococci and streptococci were identified with various agglutination and presumptive (e.g., CAMP, A disk, P disk) tests. API strips (bioMérieux), CHROMagar Candida media (Becton, Dickinson and Company, Franklin Lakes, NJ), and rapid assimilation of trehalose (RAT) testing (Remel) for Candida glabrata were used for yeast identifications (Fig. 1).
FIG 1.
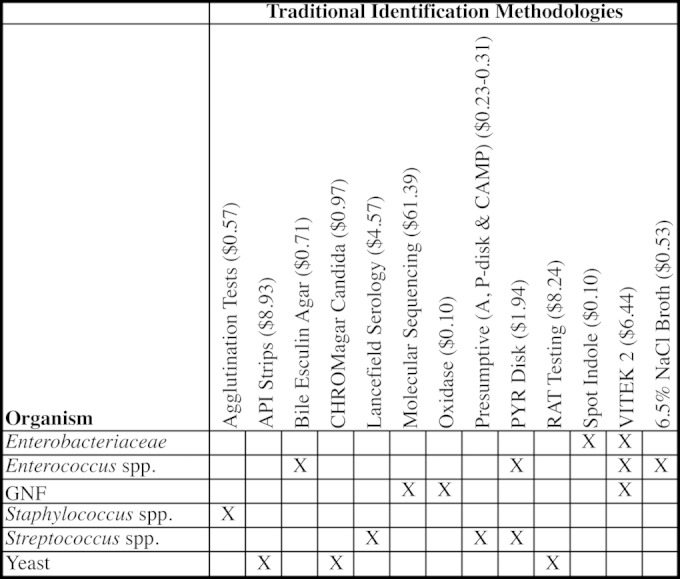
Traditional methods required for identification, by organism category. GNF, Gram-negative glucose nonfermenters.
For these traditional methods, we provided an average of 6 minutes of technologist time for a positive identification of all bacterial organisms except staphylococci. Staphylococcus aureus were traditionally identified via BactiStaph (Remel, Lenexa, KS), while other Staphylococcus spp. were detected with a combination of a tube coagulase test and BactiStaph, both simple agglutination methodologies, and were therefore assigned 3 minutes per detection. Yeast isolates were allotted between 3 and 20 minutes per positive identification, depending on the genus and species of the yeast isolated. Our laboratory has been able to essentially eliminate the routine use of biochemical batteries, API strips for yeast identification, RAT testing, and Vitek 2 bacterial identification cards, which significantly cut reagent and quality control costs. The UNCH CMIL mean base technologist salary utilized for this study was $27.00/hour, with an additional 20% for fringe benefits, totaling $0.54/minute.
The UNCH CMIL laboratory staff went through extensive training and internal performance monitoring to properly operate the bioMérieux Vitek MS and enhance workflow. This included exercises on how to apply the proper amount of colony growth onto the disposable target slides (i.e., spotting) to achieve a quality result, troubleshoot identification issues as they arise, and know when to reanalyze (i.e., refire) a spot. The technologists were taught to apply one colony per isolate onto a spot on the target slide, add 1 μl of the matrix solution, and allow it to dry. We urge one sample spot per isolate, no duplicates. An exception would be if various colony morphologies exist on a plate and the technologist wants to rule out another organism, then other morphologies would be sampled. For yeast isolates, 0.5 μl of formic acid was applied to the sample and allowed to dry prior to addition of the organic matrix solution. For workflow, each technologist evaluated his or her culture, and suspicious colonies were marked and put aside for testing on MALDI-TOF MS. When a sufficient batch of cultures is ready to be analyzed, the technologists each spotted their own isolates, since they were the most familiar with the culture they just evaluated. The bioMérieux Vitek MALDI-TOF MS utilizes disposable target slides with three acquisition groups (AGs) per slide. Each acquisition group has a maximum of 16 spots for identification of isolates, for a total of 48 sample spots per disposable target slide. Staff were encouraged to utilize as much of the AG as possible to maximize slide usage. Once an AG was filled, the slide was inserted into the MALDI-TOF MS for analysis.
Analysis takes only minutes, and the result is usually an organism identification that is reported out in our laboratory information system. However, there are a number of reasons that a reportable identification may not be generated. The first is due to a failure, where no identification is generated. Spots prepared too thickly or too thinly account for the largest number of identification failures, underscoring the importance of proper training. Further reasons for failures include analysis of isolates that are not included in the current MALDI-TOF MS database and are ultimately identified by 16S rRNA gene sequencing, isolates that fail to meet our own internal standards (UNCH CMIL accepts ≥80% probability for valid identification), and isolates that generate identifications that do not match with colony morphology and are subject to further review. Additionally, some identifications reveal other morphologies of organisms that have already been identified or were not otherwise reported independently (e.g., oropharyngeal flora) and as such do not generate a reportable identification for that particular sample. One drawback of the instrument is that it cannot distinguish between Escherichia coli and Shigella sp., so testing of these isolates will result in a warning. Isolates that are non-lactose fermenting and non-beta-hemolytic require further testing on the Vitek 2 to differentiate.
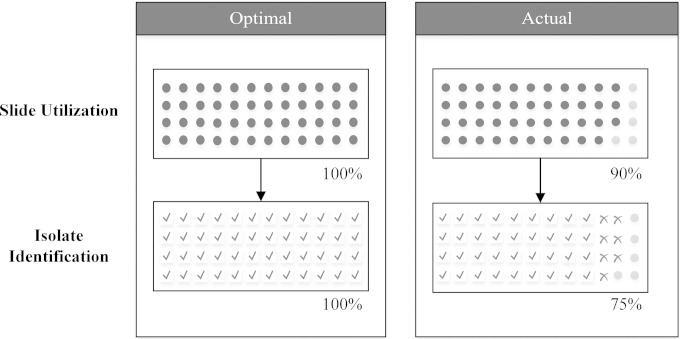
Costs for MALDI-TOF MS were calculated by tallying the total expenditures required to perform the test. This included the commercial organic matrix solution for all organisms plus formic acid for yeasts, Vitek MS target slides, pipette tips for application of the matrix and formic acid solutions, and toothpicks for applying the colonies to the target slide. To properly calculate the cost of the disposable target slides, we estimated the total efficiency of reportable MALDI-TOF MS identifications per slide. This was estimated at 75%. In other words, three out of every four sample spots available on the target slide provided an organism identification that was reportable (Fig. 2). This estimate included unused sample spots and the inability to acquire reportable identifications. Technologist time was estimated at 2.5 minutes per isolate for spotting specimens and operating the instrument, which also included the resampling of failed spots. Additional technologist and reagent costs associated with instrument reanalysis were not included because they were negligible, as the technologist only needed to select the sample spot to be reanalyzed and the instrument automatically retested the isolate. No further technologist time was required.
FIG 2.
Diagram of Vitek disposable target slide utilization and isolate identification with an optimal and actual scenario.
With these assumptions, the total reagent cost per isolate was $0.43. This included $0.10 for the organic matrix solution, $0.01 for the toothpick, $0.06 for the pipette tip, and $0.26 for each sample spot. All cost estimates were rounded to the nearest cent. The technologist time was then multiplied by the average technologist salary at UNCH CMIL and added to the reagent cost to account for direct costs. Calculations were performed for reagent and direct costs utilizing the aforementioned formulas. Differences in costs between MALDI-TOF MS and conventional methods are reported in gross and percent savings.
Acid-fast bacilli.
As an addendum to the main study, we analyzed costs over a 5-month period to determine the cost savings of implementing MALDI-TOF MS for the prompt identification of rapidly growing mycobacteria (RGM) and Mycobacterium tuberculosis complex (MTC). From May 5 to October 4, 2014, 40 isolates of RGM and MTC were identified by the CMIL. The process of preparing RGM and MTC isolates for identification on MALDI-TOF MS is complex and time-consuming, requiring properly trained laboratory personnel to achieve accurate results. This procedure involves inactivation of an acid-fast bacillus (AFB) isolate with 70% ethanol along with mechanical disruption of the organisms with glass beads, centrifugation to concentrate the sample, resuspension of the pellet with 70% formic acid and acetonitrile, application of the end product onto the target slide, and covering the specimen with matrix. These isolates were evaluated using the Saramis database v4.12. This total process was allotted 23 minutes of technologist time per identification of RGM and MTC isolates.
In contrast, traditional identification involves molecular sequencing of the 16S rRNA gene for the determination of most AFB isolates (10). In our laboratory, where sequencing is done weekly, this typically adds an extra week or more to the identification. Additionally, differentiation between two species of RGM, Mycobacterium abscessus and Mycobacterium chelonae, requires further molecular sequencing, in our case of the hsp65 region, which adds additional cost and time, as this methodology was also only performed once a week in our laboratory (11). Each molecular sequencing method is allotted 25 min of technologist time to complete.
Calculations for RGM and MTC were performed for reagent costs and total cost utilizing a specialized formula. Reagent costs were directly calculated. For 16S molecular sequencing, reagents equated to $61.39 and doubled to $122.78 for hsp65 sequencing per test performed. Reagent costs for MALDI-TOF MS were calculated to be $11.33 with all of the aforementioned required materials and consumables. Total costs were then calculated utilizing the direct cost calculation previously mentioned, where technologist time required for each isolate is added to the direct reagent cost. Of note, the MALDI-TOF MS total cost was calculated with testing and identification of only two isolates performed per AG and per run. This reflected our laboratory workflow, as we batch specimens and perform MALDI-TOF MS runs for AFB once or twice weekly. Differences in costs between MALDI-TOF MS and conventional methods are reported in gross and percent savings.
RESULTS
Over the 12-month study, from April 1, 2013, to March 31, 2014, a total of 21,930 isolates were identified in the UNCH CMIL.
Reagent cost analysis.
Traditional identification of the isolates would have cost $78,689.62 in reagents alone, compared to $9,581.01 for identification with MALDI-TOF MS. The use of MALDI-TOF MS equated to a substantial net savings of $69,108.61, or 87.8%, in reagent costs compared to the use of traditional methods (Table 1). When analyzed by organism type, the largest savings were realized among Gram-negative GNFs, with a 92.9% reduction in reagent costs with MALDI-TOF MS. Traditional costs for identifying these organisms mainly involved Vitek 2 and 16S molecular sequencing. This large percentage of savings was expected, since the use of MALDI-TOF MS for identification of bacteria and yeasts from culture utilizes a minimal amount of reagents.
TABLE 1.
Reagent cost comparison between traditional and MALDI-TOF MS methods
| Organism | No. of samples | Reagent costs ($) |
Cost savings |
||
|---|---|---|---|---|---|
| Traditional | MALDI-TOF | $ | % | ||
| Enterobacteriaceae | 7,503 | 27,407.46 | 3,226.29 | 24,181.17 | 88.2 |
| Enterococcus spp. | 1,454 | 8,361.20 | 625.22 | 7,735.98 | 92.5 |
| GNFa | 3,489 | 21,154.03 | 1,501.56 | 19,652.47 | 92.9 |
| Staphylococcus spp. | 5,790 | 8,003.24 | 2,489.70 | 5,513.54 | 68.9 |
| Streptococcus spp. | 2,332 | 10,149.14 | 1,002.76 | 9,146.38 | 90.1 |
| Yeast | 1,362 | 3,614.55 | 735.48 | 2,879.07 | 79.7 |
| Total | 21,930 | 78,689.62 | 9,581.01 | 69,108.61 | 87.8 |
Gram-negative glucose nonfermenters.
Direct cost analysis.
If total costs were calculated to include technologist time, traditional identification would have cost $142,532.69, compared to $39,186.51 for identification with MALDI-TOF MS (Table 2). Therefore, the implementation of MS for identification of bacteria and yeasts netted a savings of $103,346.18, or 72.5%. Again, Gram-negative GNFs yielded the largest savings with MALDI-TOF MS, where 80.9% total cost savings was observed.
TABLE 2.
Direct cost comparison between traditional and MALDI-TOF MS methods
| Organism | No. of samples | Total cost ($) |
Cost savings |
||
|---|---|---|---|---|---|
| Traditional | MALDI-TOF | $ | % | ||
| Enterobacteriaceae | 7,503 | 51,717.18 | 13,355.34 | 38,361.84 | 74.2 |
| Enterococcus spp. | 1,454 | 13,072.16 | 2,588.12 | 10,484.04 | 80.2 |
| GNFa | 3,489 | 32,458.39 | 6,211.71 | 26,246.68 | 80.9 |
| Staphylococcus spp. | 5,790 | 17,383.04 | 10,306.20 | 7,076.84 | 40.7 |
| Streptococcus spp. | 2,332 | 17,704.82 | 4,150.96 | 13,553.86 | 76.6 |
| Yeast | 1,362 | 10,197.09 | 2,574.18 | 7,622.91 | 74.8 |
| Total | 21,930 | 142,532.69 | 39,186.51 | 103,346.18 | 72.5 |
Gram-negative glucose nonfermenters.
The above calculations represent an estimation of the annual savings without the maintenance agreement contracts, which would apply to the first year after purchase of the instrument, since maintenance is included during the initial year of operation. To properly calculate the cost savings of each subsequent year, beginning with year 2, the annual budget of the maintenance contract needs to be added in. With the additional service contracts added, the laboratory savings fell to $73,646.18, or 51.7% (Table 3). Interestingly, there was a loss of 4.4% in savings for the identification of Staphylococcus spp. when maintenance agreement contracts were added to the calculation. There were no other remarkable differences among the other categories.
TABLE 3.
Total cost comparison between traditional and MALDI-TOF MS methods, including maintenance agreement costs
| Organism | No. of samples | Total cost ($) |
Cost savings |
||
|---|---|---|---|---|---|
| Traditional | MALDI-TOF | $ | % | ||
| Enterobacteriaceae | 7,503 | 51,717.18 | 23,516.72 | 28,200.46 | 54.5 |
| Enterococcus spp. | 1,454 | 13,072.16 | 4,557.29 | 8,514.87 | 65.1 |
| GNFa | 3,489 | 32,458.39 | 10,936.89 | 21,521.50 | 66.3 |
| Staphylococcus spp. | 5,790 | 17,383.04 | 18,147.65 | −764.61 | −4.4 |
| Streptococcus spp. | 2,332 | 17,704.82 | 7,309.21 | 10,395.61 | 58.7 |
| Yeast | 1,362 | 10,197.09 | 4,418.75 | 5,778.34 | 56.7 |
| Total | 21,930 | 142,532.69 | 68,886.51 | 73,646.18 | 51.7 |
Gram-negative glucose nonfermenters.
Acid-fast bacilli.
Through traditional methods utilizing molecular sequencing, it would have cost the CMIL $5,392.08 to properly identify the 40 RGM and MTC specimens, or $134.80 per isolate. The direct cost from MALDI-TOF MS was only $1,403.20, or $35.08 per isolate, which equates to a 74.0% savings. These savings were reflected by the reduced cost of reagents required for MALDI-TOF MS versus molecular sequencing methods, since technologist times were similar between the two methods (23 and 25 min, respectively). Each isolate that required 16S sequencing cost the UNCH CMIL $61.39 in reagents, while use of hsp65 doubled that cost. Even with the additional reagents required for performing MALDI-TOF MS, reagent costs accounted for only $11.33 per isolate, or 89.7% less than molecular sequencing. Therefore, even during a short period of 5 months, the laboratory was able to save nearly $4,000, or $100 per identification, by eliminating the need for molecular sequencing.
Cost per sample.
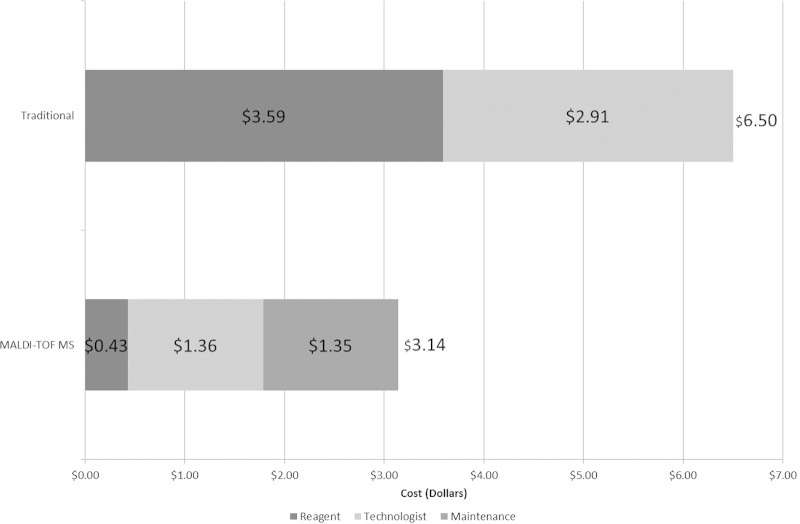
We also calculated the average cost per sample identified for the various scenarios described. This simply represents the mean of the total costs, including reagent, technologist, and maintenance costs, for all isolates identified during the study period. Reagent costs for the traditional methods utilized averaged $3.59 per isolate and those for MALDI-TOF MS were only $0.43 (Fig. 3). Total costs with traditional methods, including reagent, technologist time, and maintenance agreement contracts, were determined to be $6.50 per isolate reported, compared to $3.14 for with MALDI-TOF MS.
FIG 3.
Mean reagent and total costs (with and without maintenance costs) per identification of isolates by traditional and MALDI-TOF MS methods.
DISCUSSION
The transition from conventional microbiological methods to MALDI-TOF MS technology resulted in significant cost savings. The laboratory savings estimate was $73,646.18 (51.7%) in total costs during the 12-month study (Table 3). This included reagent, technologist, and maintenance agreement costs. Workflow, which affects the amount of time a technologist spends on identifying an isolate, is an important aspect that has direct implications for total costs. This includes but is not limited to the number of touches the technologist performs during the process of isolate determination. The UNCH CMIL underwent a Lean assessment that was conducted by bioMérieux as part of the purchase of the MALDI-TOF MS, which helped the laboratory refine protocols to maximize time and efforts when identifying specimens with the MALDI-TOF MS.
Laboratories can maximize resources and minimize costs by batching runs and waiting for all of the sample spots within an acquisition group on the target slide to be filled prior to loading the slides onto the MS; the remaining AGs would be accessible for additional runs at a later time. This helps boost slide usage efficiency and provides more savings. This philosophy should be possible for most runs throughout the day without impacting turnaround time significantly. An optimal scenario assumes maximum utilization of reagents and technologist time; i.e., all sample spots on the disposable slide are utilized, and 100% of the spots result in a reportable identification (Fig. 2). If this scenario were true, then the total cost savings would be $80,894.12, or 56.8%, annually instead of $73,646.18. Realistically, we believe the actual cost savings are between this optimal scenario and our study calculations, since it is not feasible to expect that every spot will be utilized on each slide and result in a reportable identification.
Not surprisingly, implementation of MALDI-TOF MS reduced reagent costs by 88% compared to the costs of conventional methods. This is because the only consumables utilized in our laboratory for MALDI-TOF MS were the disposable target slides, matrix, formic acid (mainly for yeasts), pipette tips, and toothpicks for spotting organisms.
Interestingly, there was only one category where it was less expensive to perform traditional identification than identification with MALDI-TOF MS. Identification of Staphylococcus spp. by MS cost $764.61 more than by the traditional latex agglutination methods. Although the technologist time and reagent costs were similar for both methods, addition of the maintenance contract increased the cost of MALDI-TOF MS identification above that for conventional identification of Staphylococcus species. However, traditional methods of identification of Staphylococcus saprophyticus and Staphylococcus lugdunensis required more technologist time and biochemical tests, which delayed identification of these organisms for up to 48 h. Additionally, the Vitek MS provided our laboratory with the ability to identify coagulase-negative Staphylococcus isolates to the species level, which was not done previously. This is beneficial for blood and sterile body fluid isolates to help differentiate between true infection and contamination of blood and sterile body fluid cultures when multiple colonial morphologies were detected (12–14). For us, these benefits far outweigh the added costs of MALDI-TOF MS. Additionally, some rapid biochemical tests, such as spot indole for Escherichia coli and PYR for Streptococcus pyogenes, may be less expensive than MS. The inclusion and exclusion of certain organisms in this new methodology is something that each laboratory should consider and incorporate into implementation protocols, because MALDI-TOF MS reagent costs, workflow, and maintenance contracts vary significantly.
Verification studies.
Verification studies for implementation of MALDI-TOF MS began in June 2012, and the laboratory went live with the first set of organisms in October 2012. For this validation, we included 731 organisms across 3,411 sample spots. Through June 2013, the verifications cost an estimated $4,357.78, and more organisms continue to be verified as we encounter them in the laboratory.
Acid-fast bacilli.
Because of the expense of identifying rapidly growing mycobacteria and Mycobacterium tuberculosis by molecular means, this group of AFB is likely to be more efficiently identified by MALDI-TOF MS. Our laboratory cultures AFB through traditional mycobacterial methods utilizing liquid (Bactec MGIT 960 mycobacterial detection system) and solid (Lowenstein-Jensen medium; Becton, Dickinson and Company, Franklin Lakes, NJ) media. Once positive growth is verified via a Kinyoun stain, the organism is subcultured to 7H11 medium. After sufficient growth, the organisms are prepared for identification by MALDI-TOF MS. The rate of growth determines whether the organism is an RGM, and colony morphology helps determine whether MTC is in the differential.
In February 2014, RGM and MTC were added to the menu of approved organisms for identification on MALDI-TOF MS. These identifications utilize the research-use-only (RUO) Saramis database v4.12 and incur a higher total cost due to a time-consuming extraction technique required to process and inactivate the AFB prior to organism identification by MALDI-TOF MS. These costs still represent a significant 74% reduction in direct costs after moving to MALDI-TOF MS from traditional molecular sequencing methods while reducing the turnaround time.
Capital costs.
The main stumbling block with implementing MALDI-TOF MS in the clinical microbiology setting is the capital cost of acquiring the instrument. With an approximate $270,000 price tag for the instrument and associated in vitro diagnostics and RUO software databases, the initial financial hurdle may be too high for some laboratories to overcome. Moreover, laboratories must be mindful of the maintenance that is associated with the MALDI-TOF MS. The UNCH CMIL pays an additional $29,700 annually for instrument and database/software maintenance. Our laboratory requires MALDI-TOF MS fine-tuning every 3 to 4 weeks and has undergone a full laser and three linear detector replacements in the <3 years that the instrument has been in use. The fine-tuning process, which takes several hours, does not interrupt workflow or increase cost in our laboratory, as we batch specimens and test them once the instrument is ready for operation again. We estimate that the MALDI-TOF MS was nonfunctional for extended periods (i.e., >18 h) for a total of 4 days during the study period. During these times, we reverted to traditional methods of identification. Maintenance is an integral part of the cost calculation for MALDI-TOF MS and needs to be strongly considered by every laboratory planning on implementing this technology. It should be noted that maintenance costs begin in year 2, since the first year is covered by the manufacturer warranty.
Therefore, understanding the cost savings after implementation of MALDI-TOF MS is important because of its large initial fiscal investment. With an annual savings estimate of $73,646.18, coupled with the initial year's savings of $103,346.18, we anticipate that the capital cost of the instrument would be offset in just >3 years. The net savings will only increase as we continue to validate more organisms by MALDI-TOF MS that will ultimately require less molecular sequencing and labor-intensive technologist time for identification.
Limitations.
There are several limitations to the study. We were limited by database inquiries and our financial test cost analyses. The laboratory information system has limited querying capabilities. While we were able to identify the number of isolates reported during the study period, we were unable to identify exactly how many sample spots were utilized for identification of the organisms that were reported. The only way to track this is to prospectively audit the number of spots that were utilized and cross-reference that number with the number of isolate reports that were generated. This was not possible for us due to resource constraints. We were, however, able to audit all of the spots generated over a 7-day period and found that 79.6% of the spots generated a reportable identification. Similarly, a 2-month audit of our slide usage efficiency revealed that 89.8% of sample spots were utilized. These surveys showed that our estimate of 75% total slide efficiency was relatively close to what actually occurs in our laboratory. However, without prospective monitoring of slide consumption and efficiency throughout the study period, it is impossible to accurately calculate this value.
It should be noted that this was a cost-savings study that estimated the amount saved by the laboratory after routine implementation of MALDI-TOF MS for identification of the more common organisms encountered in the clinical microbiology laboratory. It is not a true cost-effectiveness study that examines costs compared to a defined clinical or natural outcome to improve health (15). A cost-effectiveness study would require evaluation of the turnaround time to results and how it impacts patient care, which is beyond the scope of this paper. Further and more in-depth studies, such as that by Tan et al. (9), are needed to determine the cost-effectiveness of this new technology.
MALDI-TOF MS not only represents an innovative technology for the rapid and accurate identification of bacterial and fungal isolates, it also provides significant cost savings for the laboratory. Despite the high capital cost of the instrument, the ease of performance, the rapid turnaround time to results, and the modest cost of testing for each sample make this new methodology a paradigm shift in the field of clinical microbiology.
ACKNOWLEDGMENTS
We thank the laboratory staff at the UNCH CMIL for their contributions, continued diligence, and dedication to their craft, which helped tremendously with our cost savings.
REFERENCES
- 1.Shah HN, Gharbia S. 15 June 2010. Mass spectrometry for microbial proteomics. John Wiley & Sons, Inc., Chichester, United Kingdom. doi: 10.1002/9780470665497. [DOI] [Google Scholar]
- 2.Karas M, Hillenkamp F. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem 60:2299–2301. [DOI] [PubMed] [Google Scholar]
- 3.Buchan BW, Ledeboer NA. 2013. Advances in identification of clinical yeast isolates by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:1359–1366. doi: 10.1128/JCM.03105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Carolis E, Vella A, Vaccaro L, Torelli R, Spanu T, Fiori B, Posteraro B, Sanguinetti M. 2014. Application of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. J Infect Dev Ctries 8:1081–1088. doi: 10.3855/jidc.3623. [DOI] [PubMed] [Google Scholar]
- 5.Chao QT, Lee TF, Teng SH, Peng LY, Chen PH, Teng LJ, Hsueh PR. 2014. Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS One 9:e109376. doi: 10.1371/journal.pone.0109376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol 48:1169–1175. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr AM. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin Lab 55:289–296. [PubMed] [Google Scholar]
- 8.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 9.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol 50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kothavade RJ, Dhurat RS, Mishra SN, Kothavade UR. 2013. Clinical and laboratory aspects of the diagnosis and management of cutaneous and subcutaneous infections caused by rapidly growing mycobacteria. Eur J Clin Microbiol Infect Dis 32:161–188. doi: 10.1007/s10096-012-1766-8. [DOI] [PubMed] [Google Scholar]
- 12.Abdul Rahman Z, Hamzah SH, Hassan SA, Osman S, Md Noor SS. 2013. The significance of coagulase-negative staphylococci bacteremia in a low resource setting. J Infect Dev Ctries 7:448–452. doi: 10.3855/jidc.2535. [DOI] [PubMed] [Google Scholar]
- 13.Al Wohoush I, Rivera J, Cairo J, Hachem R, Raad I. 2011. Comparing clinical and microbiological methods for the diagnosis of true bacteraemia among patients with multiple blood cultures positive for coagulase-negative staphylococci. Clin Microbiol Infect 17:569–571. doi: 10.1111/j.1469-0691.2010.03372.x. [DOI] [PubMed] [Google Scholar]
- 14.Usha MG, Shwetha DC, Vishwanath G. 2013. Speciation of coagulase negative Staphylococcal isolates from clinically significant specimens and their antibiogram. Indian J Pathol Microbiol 56:258–260. doi: 10.4103/0377-4929.120383. [DOI] [PubMed] [Google Scholar]
- 15.Gold MR. 1996. Cost-effectiveness in health and medicine. Oxford University Press, New York, NY. [Google Scholar]