Abstract
Dysregulation of microRNAs (miRNAs) is involved in the initiation and progression of several human cancers, including breast cancer (BC), as strong evidence has been found that miRNAs can act as oncogenes or tumor suppressor genes. This review presents the state of the art on the role of miRNAs in the diagnosis, prognosis, and therapy of BC. Based on the results obtained in the last decade, some miRNAs are emerging as biomarkers of BC for diagnosis (i.e., miR-9, miR-10b, and miR-17-5p), prognosis (i.e., miR-148a and miR-335), and prediction of therapeutic outcomes (i.e., miR-30c, miR-187, and miR-339-5p) and have important roles in the control of BC hallmark functions such as invasion, metastasis, proliferation, resting death, apoptosis, and genomic instability. Other miRNAs are of interest as new, easily accessible, affordable, non-invasive tools for the personalized management of patients with BC because they are circulating in body fluids (e.g., miR-155 and miR-210). In particular, circulating multiple miRNA profiles are showing better diagnostic and prognostic performance as well as better sensitivity than individual miRNAs in BC. New miRNA-based drugs are also promising therapy for BC (e.g., miR-9, miR-21, miR34a, miR145, and miR150), and other miRNAs are showing a fundamental role in modulation of the response to other non-miRNA treatments, being able to increase their efficacy (e.g., miR-21, miR34a, miR195, miR200c, and miR203 in combination with chemotherapy).
Keywords: Breast cancer, microRNA/miRNA, circulating biomarker, theranostic, diagnosis, prognosis, prediction and therapy.
1. Introduction
In 1993, Lee et al. 1 described that a small non-coding RNA in Caenorhabditis elegans was able to regulate the expression and function of another protein-coding mRNA. The discovery of microRNAs (miRNAs or miRs) had a profound impact on the understanding of many gene regulation processes in the following years. Since they were first discovered, the physiological relevance of miRNAs in regulating plant and animal gene expression has been established.
The primary repository for miRNA sequences and annotations, miRBase (www.mirbase.org), debuted in 2006 with just 218 miRNA loci 2-4. Since then, novel high-throughput sequencing techniques applied to miRNA analysis have allowed the discovery of more than 28000 mature miRNAs (miRBase release June 21, 2014). MiRNAs participate in the post-transcriptional regulation of gene expression in almost all key cellular processes 5, such as regulation of cell proliferation, differentiation, angiogenesis, migration, and apoptosis.
Significant evidence has accumulated in the last few years, showing a fundamental role of miRNAs in the development of many diseases 6-9. In particular, in cancer, aberrations in miRNA expression levels have been linked to the onset and progression of various types of cancer 10.
Breast cancer (BC) is the second most common cancer in the world, and by far, the most frequent cancer among women, contributing to an estimated 25% of all new cancers or cases diagnosed in 2012 11. Several biological features are routinely used for the diagnosis and prognosis of patients with BC and for determining the therapy, e.g., histological grade 12, lymph node status, hormone receptor status, and human epidermal growth factor receptor type 2 (HER2) status 13. Some of these factors have been associated with the survival rate of patients and their clinical outcome after treatment 14. However, some patients, with a similar combination of BC features, have been found to have different clinical outcomes. Thus, the role of these factors in determining diagnosis and prognosis and in predicting therapeutic outcomes in BC remains limited 15.
New affordable methods are therefore needed to help diagnosis and prognosis and to suggest the most appropriate treatment for patients with BC on an individual basis. As a solution, miRNAs have been proposed as promising biomarkers of BC because they can be readily detected in tumor biopsies (non-circulating miRNAs) 16, 17 and are also stably found in body fluids (circulating miRNAs), particularly in blood, plasma, serum, and saliva 18, 19. These circulating miRNAs are highly reliable and protected from endogenous RNAse activity, being bound to lipoproteins such as HDL, associated with Argonaute 2 (Ago2) protein 20, or packaged into microparticles (such as exosome-like particles, microvescicles, and apoptotic bodies.) 19, 20.
Recently, miRNA profiling has been assessed to improve BC classification and to differentiate patients with BC as responding or not responding to therapies, with promising results 21. It is now clear that these tools have the potential to provide new diagnostic, prognostic, and predictive biomarkers for BC, with a great impact on the clinical management of patients with BC 15.
In this review, we focused on the recent findings related to the role of miRNAs in BC and on how miRNAs have the potential to answer actual clinical needs, such as identification of biomarkers for early and differential diagnosis, prognosis, and prediction of response to specific therapies. New therapeutic strategies represented by miRNA-based theranostic approaches in BC are also introduced and could become a starting point for the future development of novel therapeutic tools.
2. miRNA biogenesis and mechanisms of action
2a. miRNA biogenesis
miRNAs are small, evolutionarily conserved, non-coding RNAs that are approximately 18-25 nucleotides in length and constitute the dominating class of small RNAs in most somatic tissues. Other small RNAs in animals include silencing RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), which are typical of germinal cells 22, and non-coding mitochondrial RNAs (ncmRNAs) 23. Although many aspects of the miRNA biogenesis pathway and repressive mechanisms are still obscure, the key processes have been fully characterized.
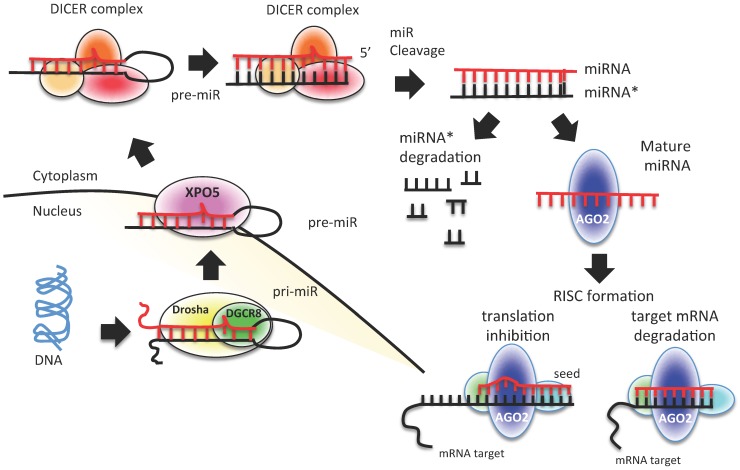
miRNAs are transcribed from individual genes containing their own promoter, or intragenically from spliced portions of protein-coding genes 24. Like protein-coding genes, miRNAs with their own promoters are almost exclusively transcribed by RNA polymerase II in a primary transcript called pri-miRNA 24 (Figure 1). This long transcript contains a 7-methylguanosine cap at the 5′ end, a 3′ poly-(A) tail, and sometimes also introns. To be processed, pri-miRNAs are recognized by Drosha ribonuclease and its partner, the double-stranded RNA binding protein DGCR8, through interaction with a stem-loop structure within the miRNA in which the sequences are not perfectly complementary 25, 26. Processing of pri-miRNAs gives rise to precursor miRNAs (pre-miRNAs) of approximately 70 nucleotides 24 (Figure 1). Some intronic miRNAs, called mirtrons, could bypass Drosha processing and use the splicing machinery to generate pre-miRNAs 24. The generated pre-miRNAs are then exported from the nucleus to the cytoplasm by exportin 5 (XPO5) 27-29, where they are cleaved by the RNase III enzyme Dicer 1 in union with transactivation-responsive RNA-binding protein 2 (TARBP2) and AGO2 (DICER complex). The processing generates a double-stranded miRNA-miRNA* duplex 30. The 2 strands are then separated: the mature miRNA (the guide strand) is incorporated into the RNA-induced silencing complex (RISC), whereas the passage miRNA* strand can be loaded in the RISC as well or degraded 31-33. The mature miRNA guides the AGO protein of the RISC to the complementary mRNA sequence on the target to repress its expression 24 (Figure 1).
Figure 1.
miRNA biogenesis process. A schematic representation of canonical miRNA biogenesis pathway. Each miRNA is transcribed by RNA polymerase II (pri-miRNA) from genomic DNA within the nucleus; pri-miRNA is recognized by Drosha-DGCR8 and processed to pre-miRNA. Pre-miRNA is exported to the cytoplasm by exportin 5 (XPO5), where it is processed and cleaved by DICER complex to a double strand miRNA (miRNA*-miRNA). The duplex is cleaved, and only the mature miRNA is loaded into the RISC complex. The degree of homology of the miRNA “seed” to the 3′ UTR target sequence of the mRNA determines the mRNA translational repression or degradation.
2b. miRNA mechanisms of action
The major determinant for miRNA binding to its target mRNA is a 6-8-nucleotide sequence at the 5′ end of the miRNA, the “seed” sequence 24. Any sequence complementarity between the loaded miRNA and the seed region triggers a detectable decrease in target mRNA expression levels. Seed matches can occur in any region of the mRNA but are more likely to be present in the 3′ untranslated region (3′ UTR) of a mRNA 34, 35. Several lines of evidence indicate that miRNAs can also bind to other regions in the target mRNA 36. Depending on the degree of homology to the 3′ UTR target sequence, miRNAs can induce the translational repression or degradation of mRNAs. Given that each miRNA is capable of regulating the expression of many genes, each miRNA can simultaneously regulate multiple cellular signaling pathways.
Apart from the “traditional” mechanism of action of miRNAs described above, other “non-canonical” mechanisms have been proposed recently. Some evidence indicates that miRNAs could increase the translation of a target mRNA by recruiting protein complexes at the AU-rich region of the target mRNA or they could indirectly increase target mRNA levels by interacting and modulating repressor proteins that block the translation of the target mRNA 37. Other evidence suggests that miRNAs could enhance ribosome biogenesis, thereby modulating protein synthesis, or skip cell cycle arrest, thereby activating target gene repression 34, 38.
3. Methods for miRNA target prediction and miRNA-target interaction validation
3a. Methods for miRNA target prediction
Uncovering of miRNA-regulated networks needs large-scale and unbiased methods for miRNA target identification. For instance, the differential expression of a single miRNA would be followed by downstream gene or proteome-wide analysis. A single miRNA could regulate a set of genes responsible for a particular malignant phenotype. The silencing of that single miRNA can alter the entire set of genes.
To overcome this complexity and to predict the target genes, several algorithms have been developed. The main difficulty in miRNA target prediction is to detect the specific sequences within genes where one miRNA is fully or partially complementary 39, considering the small size of miRNAs and their low specificity.
A collection of tools is available, each with a distinct approach to miRNA target prediction and different features 40. The suitable tool can be decided depending on the requirements 12.
The major features of computational target prediction are as follows: sequence composition (e.g., seed match), conservation, and thermodynamic stability (e.g., free energy).
i) Seed match is the start of many computational methods for miRNA target prediction. A seed match usually consists of Watson-Crick (WC) complementarity between the miRNA and miRNA target nucleotides. WC complementarity occurs when adenosine (A) pairs with uracil (U) and guanine (G) pairs with cytosine (C). The seed is a sequence from the 1st to the 8th nucleotide at the 5′ end of the miRNA. However, algorithms based only on WC complementarity show low accuracy and a high number of false-positive results 26.
Other sequence compositions can be used as features for miRNA target prediction tools. Bartel et al. 31 showed that the AU residues in target sites improve the accessibility of miRNAs to form duplexes. Recent studies have suggested that coding regions of mRNAs can also include target sites for miRNAs 41. In addition, it has been demonstrated that a transcript can contain multiple target sites for a single miRNA; however, when the target sites show overlapping sequences, miRNA-mRNA pairing can be compromised 42.
ii) Conservation analysis was introduced in order to reduce false-positive results. Conservation refers to the maintenance of sequence homology across species 40. In general, there is higher conservation in the miRNA seed region than in the non-seed region 43. However, the limit of this approach was demonstrated by Bentwich et al., who showed that several non-conserved miRNAs were missing 43.
iii) Free energy (or Gibbs free energy) can be used as a feature for miRNA target prediction 44, 45. The thermodynamic stability of the miRNA-mRNA duplex shows the strength of the binding between a miRNA and its target by predicting how the miRNA and its candidate target will hybridize. The free energy is related to duplex formation between the miRNA and its target site. In particular, pairing can be determined by removing existing secondary structures 46. The free energy is established by the difference between the energy expended in opening the target site structure and that gained by forming the duplex 46, 47.
Many computational algorithms have been developed and implemented as software tools for miRNA target prediction using some of the described features. These packages are very useful to select putative miRNA targets for further biological validation. The most common classifiers are based on machine learning algorithms, e.g., support vector machine (SVM), neural networks, hidden Markov model (HMM), and Naive Bayes (NB). These machine learning methods are trained on a so-called “training” dataset that contains a set of known miRNA sequences (positive training dataset) and a set of sequences that do not contain miRNAs, such as mRNAs, tRNAs, and rRNAs (negative training dataset), which represent the limit of this approach 47. Several studies have tried to overcome this problem with the use of only true/positive models 48-50. However, the results are worse than those obtained with approaches that utilize both positive and negative training sets 49. Many tools of machine learning-based approaches for miRNA target prediction are currently available, e.g., HHMMiR 51, PicTar 52, MiRFinder 53, RNAmicro 50, 54, ProMiR 55, MiRRim 56, BayesMiRNAFind 57, and SSCprofiler 58.
Other computational algorithms use approaches different from machine learning. The TargetScan algorithm was the first miRNA target prediction tool for human genome 40. It searches for perfect complementarity in the seed region, and all seed sequences outside complementarity are filtered out. Predictions are ranked by a combinatorial score on the basis of sequence composition (seed sequence), conservation, and thermodynamic stability (free energy).
Diana-microT uses a larger frame for scanning complementarity. It focuses on orthologous human and mouse 3′ UTRs from the mRNA Reference Sequences (RefSeq) database and 94 miRNAs conserved in human and mouse. It applies a modified dynamic programming algorithm to calculate the minimum free energy for each segment with a miRNA 59.
The miRanda algorithm gives scores for seed complementary regions. The results are evaluated for free energy. Each target that has a predicted free energy below a threshold is then passed to the last step, i.e., conservation 60.
These algorithms are summarized in Table 1, together with their main characteristic approaches and features.
Table 1.
The main algorithms for computational miRNA-target prediction
| Algorithm | Features | Approach | References |
|---|---|---|---|
| HHMMiR | Seed match, and conservation | HMM | 51 |
| PicTar | Seed match | HMM | 52 |
| MiRFinder | Seed match, and conservation | SVM | 53 |
| RNAmicro | Sequence composition, conservation, and thermodynamic stability | SVM | 54 |
| ProMir | Sequence composition, conservation and thermodynamic stability. | HMM | 55 |
| MiRRim | Sequence composition, conservation, and free energy. | HMM | 56 |
| BayesMiRNAFind | Sequence composition and free energy. | Naïve Bayes Classifier | 57 |
| SSCprofiler | Sequence composition, conservation and free energy. | HMM | 58 |
| Diana-microT | Seed match, conservation, and free energy | Dynamic programming algorithm | 59 |
| TargetScan | Seed match, conservation, and free energy | Combinatorial score | 40 |
| MiRanda | Seed match, conservation, and free energy | Score | 60 |
3b. miRNA-target interaction validation
Many experimental technologies for validating miRNA-mRNA interactions have been developed 61, 62. In general, the effects of differential miRNA expression on the target gene obtained through transfection of miRNA mimic or miRNA inhibitor oligonucleotides or constructs 63 are established at the protein level by western blotting and at the mRNA level by quantitative real-time PCR (qRT-PCR), with a specific probe for the target gene 61, 62. The most important disadvantage of these techniques is that they are not able to distinguish between direct and secondary miRNA-target interactions.
3b.1 Luciferase assay
Reporter assays are commonly used to study gene expression coupled with other cellular events, such as receptor activity or intracellular signal transduction of protein-protein interactions. To analyze direct miRNA-mRNA interactions, the firefly luciferase-based assay is widely used because the reporter activity is available immediately upon translation, the assay is very rapid and sensitive, and no background luminescence is found in the host cells (Figure 2). To be used as a reporter assay for validation of the interaction of a miRNA with the 3′ UTR of a gene of interest (GOI), the luciferase-based assay needs cloning of the 3′ UTR of the GOI, where the miRNA-recognized sequence is supposed to be present, downstream of the luciferase gene in the reporter vector (Figure 2). The cells are then transfected with this construct in the presence or absence of the miRNA mimic oligonucleotide. If the miRNA is able to recognize the seed in the 3′ UTR of the GOI, the level of luciferase expression is decreased, thus causing a diminished bioluminescence emission (Figure 2B); on the other hand, if the miRNA does not interact with the 3′ UTR, the emission of light is unaffected (Figure 2A). The disadvantages of this type of reporter assays are that they are laborious, expensive, sensitive only for the 3′ UTR chosen for cloning, and difficult to use for transfection 62, 63.
Figure 2.
In vitro validation of miRNA-target direct interaction. Cultured cell lines are transfected with a reporter vector containing firefly (FIR) luciferase gene and the 3′ UTR of the gene of interest (GOI). The level of expression of FIR luciferase is measured in a luminometric assay. Cells are then exposed to the mimic miRNA, which is supposed to enter within the cell and to interact with the 3′ UTR of the GOI. If no interaction between miRNA and the 3′ UTR of GOI happens (a), we could observe no alteration in the level of expression of luciferase, thus no alteration in the emitted chemoluminescence, as FIR gene produced an active, luminescent protein. The complete interaction between the miRNA and the 3′ UTR of the GOI (b) leads to reduced FIR luciferase expression, with a decrease of luminescence levels. Other luminescent genes, such as Renilla (REN) luciferase, are usually used as reference genes for luminescence normalization.
3b.2 RISC immunoprecipitation
Another biochemical method to identify and isolate direct miRNA-target complexes is based on the immunoprecipitation of RISC components (such as AGO and TNRC6). This method is able to capture low-abundant and transient miRNA-mRNA pairs. Target mRNAs undergoing direct miRNA regulation are co-immunoprecipitated along with the RISC and are identified by qRT-PCR, microarray, or deep sequencing 64. The successful pull-down of the entire complex relies on the strong interaction between the miRNA-target complex and RISC and on the ability of the used antibody to precipitate AGO2, the core RISC protein usually used for complex immunoprecipitation. Some companies have developed a dominant negative mutant of an RISC protein subunit to trap the miRNA-target complex into the RISC, thus limiting further processing 65. This strategy allows the recovery of transient and low-abundance mRNA targets that would otherwise be lost. A FLAG epitope is then used for the capture of the entire complex 65. qRT-PCR or next-generation sequencing techniques are used to confirm the interaction between the miRNA and the target mRNA.
4. miRNAs and BC
Advanced technologies, such as microarray expression data, have shown that aberrant miRNA expression is the rule rather than the exception in BC 66, 67. The tight integration of miRNAs in physiological circuits could become a problem, because the dysregulation of a small number of miRNAs could profoundly affect the expression profile of the cells, driving them toward transformation 68. BC miRNAs, which have an important role in the pathophysiology of the disease, facilitating invasion, metastasis, epithelial to mesenchymal transition (EMT), and maintenance of BC stem cells, have become an interesting topic in BC management.
4a. Mechanisms altering miRNA expression levels
Because of amplification, each miRNA can increase the control over its target gene. If the target gene is an oncogene, the cancer does not develop (oncosuppressor-miRs); if the target gene is a tumor suppressor, the cancer develops (oncomiRs). Due to deletion, each miRNA can reduce the control over its target gene. If the target gene is an oncogene, the cancer develops (oncomiRs); if the target gene is a tumor suppressor, the cancer does not develop (oncosuppressor-miRs).
Several mechanisms can influence miRNA expression levels (Figure 3). Tumors often present altered levels of mature miRNAs 101 as a consequence of the following:
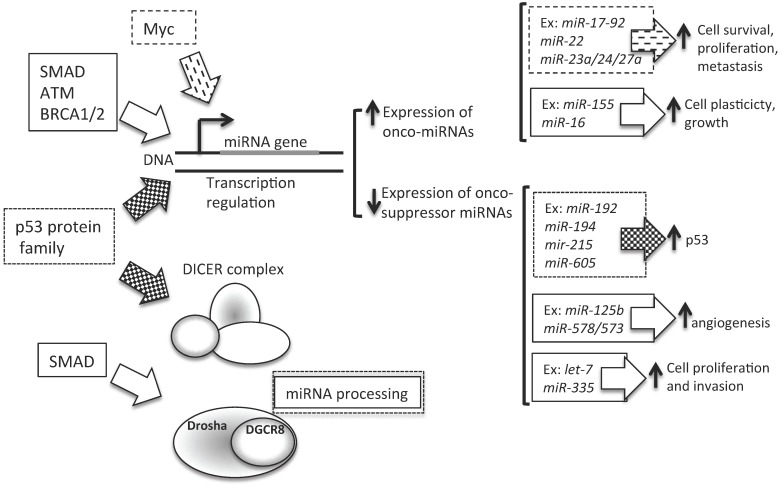
Figure 3.
Altered steps in miRNA biogenesis lead to cancer. A schematic representation of altered steps of the miRNA biogenesis pathway, commonly deregulated in cancer: 1. miRNA genes contain upstream regulator elements (enhancers/repressors) and promoter regions, indicating that miRNAs are subjected to CpG methylation (CpG promoter met); 2. The alteration in the copy number of miRNA (due to genomic amplification or deletion, activating or repressing mutation, loss of epigenetic silencing and transcriptional activation) could increase the oncogenic miRNAs or decrease the tumor suppressor miRNAs; 3. Alteration in the miRNA processing machinery, i.e. downregulation of Drosha, could decrease the cropping of pri-miR to pre-miR; 4. XPO5 mutation could prevent pre-miR export to the cytoplasm; 5. Mutation of TARBP2 or downregulation of DICER1 decrease mature miRNA levels, causing finally a loss on tumor suppressor miRNAs; 6 and 7. Accumulation of oncogenic miRNAs or loss of tumor suppressor miRNAs could finally lead to cancer development.
1. Epigenetic mechanisms (Figure 3, section 1). A large proportion of miRNA loci on the genome are associated with CpG islands, giving strong bases for their regulation by methylation (Figure 3, section 1) 69. A recent critical review on aberrant DNA methylation of miRNAs in BC showed that although aberrant DNA methylation is a well-described mechanism for gene silencing, an actual demonstration of the link between miRNA expression and gene methylation was still missing in several of the analyzed studies 70. However, Castilla et al. have clearly demonstrated in 70 BC cases that a relationship exists between miR-200 family expression, gene methylation, and metastatic potential of the tumors 71. A mapping-based study has identified miRNA promoters silenced in BC 72, and different patterns of methylation have been observed in the miR-200b cluster promoter in different BC sub-types 72. Aure et al., focusing their attention on let7e-3p miRNA, found that the genomic region that encodes for this miRNA belongs to a hypomethylated, and thus silenced, chromosome 73. The researchers have associated let-7e-3p downregulation with poorer BC prognosis 73. Another epigenetic phenomenon altered in BC is histone acetylation. Studies with deacetylase inhibitors have revealed that the reduction of acetylated histones could diminish the expression of anti-oncogenic miRNAs 74, 75.
2. A genetic alteration (Figure 3, sections 1 and 2), i.e., frameshift mutations resulting from microsatellite instability. Such genetic alternations can affect the expression of several mRNAs, e.g., the mRNA of TARBP2 (Figure 3, section 5), the Dicer stabilizing protein. This has been found, for example, in colorectal and gastric cancer 76 and in BC 77. Moreover, more than half of the known miRNAs are located in cancer-associated region, such as fragile sites, minimal regions of loss of heterozygosity, minimal regions of amplification (minimal amplicons), or common breakpoint regions 78. In the literature, some miRNA families emerge to be overall more involved in tumor development 79, such as the let-7 miRNA family. In BC, several let-7 family members, together with miR-125b, miR100, and miR34a, have been found to be located at fragile sites of human chromosomes (11q23-q24D), potentially contributing to aberrant miRNA expression 78.
3. Defects in the miRNA biogenesis pathway (Figure 3, sections 3-5): each step of miRNA biogenesis could be affected, thus altering miRNA expression levels and making the cell suitable for oncogenic changes. Reduced Dicer and Drosha expression (Figure 3, sections 3 and 5) have been associated with high-grade BC and shorter metastasis-free survival or with higher-grade BC and shorter disease-free survival 80-83. Reduced Dicer expression (Figure 3, section 5) has been also found in many other human tumors 84, e.g., in prostate 85, gastric 86, or squamous cell carcinoma 87. In BC, reduced Dicer expression has been associated with the triple-negative phenotype 83, 88. Moreover, in BC, nucleolin (NCL), a component of the Drosha/DGCR8 microprocessor complex, has been demonstrated to promote the maturation of a set of metastasis-promoting miRNAs (miR-221/222 cluster, miR-21, miR-103, and miR-15a/16) 89, 90. Furthermore, XPO5, a key protein for pre-miRNA export to the cytosol, has been suggested as a possible prognostic biomarker for BC 91 (Figure 3, section 4).
4. Transcriptional repression by other upstream proteins (Figure 4). A plethora of transcription factors can influence the expression levels of a single miRNA. Several lines of evidence suggest that miRNAs and transcription factors work cooperatively. miRNAs are involved in the functional feedback loop, in which transcription factors influence miRNA expression levels and vice versa 92-94. Thus, tumorigenic miRNA expression alterations could be due to the activity of tumor-related transcription factors, such as SMAD 90, 95, p53 protein family (p53, p63, and p73) 96, ataxia telangiectasia mutated (ATM) 97, and Myc 98. In BC, the BC 1, early onset (BRCA1) transcription factor 99 and the epidermal growth factor receptor (EGFR/HER1), a hypoxic transcription factor involved in the regulation of the RISC 100, are able to inhibit miRNA maturation, thus enhancing cell survival and invasiveness.
Figure 4.
Contribution of transcription to miRNA level alteration in cancer. Several transcription factors are able to control the level of expression of miRNAs. In particular, as described in the text, SMAD, Myc, ATM, BRCA1/2 and p53 influence miRNA transcription. P53 can regulate onco-suppressor miRNAs, which are involved in the control of p53 turnover. SMAD, ATM, BRCA1/2 and Myc could influence the transcription levels of miRNAs involved in cell plasticity, cell proliferation and survival, and cell invasion control. Moreover, SMAD is also involved in miRNA processing, by Drosha expression levels control. Ex: example of miRNA regulated by transcription factors.
4b. miRNAs and BC progression models
Modeling cancer disease is not easy, because cancer encompasses several histopathologies, involving genetic and genomic variations, and distinct clinical outcomes. A major challenge in advancing the knowledge of cancer is the availability of a single experimental model system that recapitulates the complex biology of the disease. Because of this complexity, no single model would be expected to mimic all features of the disease. The existing experimental models include two-dimensional (2D) and three-dimensional (3D) cell line cultures, xenografted mice, and engineered mice.
1. 2D cell culture. 2D cell culture studies in the oncogenic field have played a pivotal role in furthering our understanding of the disease mechanisms and drug discovery. The majority of scientific studies on miRNAs use 2D cell cultures for modulation of single miRNA expression and validation of the interaction between a single miRNA and its predicted targets via protein or gene expression analyses. This culture condition is easy to be manipulated, less expensive than the other approaches, and particularly suitable when a small number of miRNAs have to be studied.
Recently, particular attention has been given to emerging inadequacies associated with 2D culture systems, such as their inability to fully emulate in vivo tumor growth conditions and to provide physiological relevance. In fact, in the body, nearly all cells reside in an extracellular matrix (ECM) consisting of a complex 3D architecture, and interact with neighboring cells through biochemical and mechanical cues. These features cannot be obtained in 2D culture conditions. Cell-cell and cell-ECM interactions establish a 3D communication network that maintains the specificity and homeostasis of the tissue and influences tumor growth and its interaction with the whole organ. This approach has been extensively used in many works on the assessment of miRNAs in BC 101-103.
2. 3D cell culture. To overcome some shortcomings of 2D cultures, 3D cell cultures have been developed, with the use of specific matrix (such as natural ECM-based hydrogels, 3D spheroids, and trans-well inserts) that are able to support the growth of tumor cells for the establishment of physiological cell-cell and cell-ECM interactions of the native tissues. These matrix supports can mimic the environmental conditions in which the tumor cells grow with greater physiological relevance than conventional 2D cultures. The development of new biological supports is further fueled by the optimism that 3D models may significantly accelerate translational research in cancer biology. For example, use of 3D tumor cell culture is emerging as an important tool to characterize the morphogenesis of mammary epithelial cells and to elucidate the tumor-modulating actions of ECM. Focusing on miRNAs, the comparative analysis of 2D and 3D cell cultures has revealed a profound difference in miRNA profiles between the 2 culture conditions, particularly for BC cells and lung adenocarcinoma 104, 105. In particular, the miRNA profiles in 2D and 3D cultures of 2 BC cell lines were compared. The findings revealed that the 3D culture exhibited a greater discrimination between the miRNA profiles than the 2D culture 105. For example, the lower expression of miR-429 was highlighted in the 3D culture-specific miRNA profile better than that in the 2D culture-specific profile, correlating with the 3D invasive capacity of the MDA-MB-231 BC cell line.
3. Xenografted mouse models. This approach takes advantage of the injection of cancer cells from human immortalized cancer cell lines or tumor cells from patients into mouse tissue to study the development of the tumor in its native environment. This method can be used to obtain a miRNA profile during ongoing tumor development. Otherwise, tumor-xenografted mice are used for the therapeutic study of miRNA modulation. In fact, several reports have examined the effect of miRNA modulation by treating xenografted animals with oligonucleotides that increase (miRNA mimic) or decrease (antagomiR) the expression levels of a specific, single miRNA or by using an expression vector for miRNA level modulation 106. Thus, the effects of miRNA modulation are analyzed by measuring the growth of the tumor, its invasive capacity, presence of metastatic masses, and vascularization of the tumor 107.
Several models to study miRNA involvement in BC progression are based on BC cell lines or patient specimens implanted into mouse mammary fat pads. For example, use of the triple-negative MDA-MB-231 BC cell line to generate a xenografted BC model allows identification of miR-124 as the key regulator of the myc/p27/phospho-Rb pathway, which is usually altered in BC and ovarian cancer 108. Another BC model, obtained by the orthotopic implantation of xenografted human BC specimens into NOD/SCID mouse mammary fat pads (called patient-derived human-in-mouse breast tumor xenograft model or PDX model), was used to study the spontaneous generation of BC-derived lung cancer metastasis. With a combined approach of gene expression arrays and global miRNA analysis, miR-138 was demonstrated to be a key regulator of tumor invasion in lung, targeting the EMT process of BC cells 109. Another xenografted mouse model was used to demonstrate the anti-metastatic potential of the peptide nucleic acid (PNA)-modified antagomiR-21 oligonucleotide on BC cells. Yan et al. 110 demonstrated that use of the antagomiR-21 oligonucleotide is able to block proliferation, cell migration, and in vivo tumor growth of 2 BC cell lines (MCF7 and MDA-MB-231) implanted in BALB/c-nude mice, proposing the use of this oligonucleotide for potential therapeutic applications in BC treatment.
4. Engineered mouse models have been widely used. These models could be of 2 types. The first type, the genetically engineered mouse model, obtained by oncogene amplification or tumor suppressor gene deletion to characterize a specific cancer, is used to generate an expression profile of miRNAs to clarify which miRNAs are involved in the development of that specific tumor. In BC, this model has been used to identify a miRNA profile associated with 8 different mammary-engineered mouse models 111. In the second type, knocking-in or -out of specific miRNAs in the mouse germline allows to study the influence of miRNAs on tumor progression 112. This approach also involves using genetic constructs to induce miRNA overexpression or downregulation in a particular tissue, at a particular development stage, or under pharmacological control. For example, several strains of mice lacking or overexpressing cancer-associated miRNAs have been developed and characterized. These include germline transgenic or knockout mice for the following: miR-155, which, if overexpressed in B cell lineage, induces B cell malignancy 113; miR-21, which leads to lung tumorigenesis if ubiquitously deleted 114; miR-17-92 and its paralogs, whose overexpression in lymphocytes induces lymphoproliferative disease and autoimmunity 115; miR-15 and miR-16, whose deletion induces lymphoproliferative disorders 116; miR-146, which causes myeloid sarcomas and lymphomas when deleted 117; and miR-29, whose deletion causes B-cell lymphoma 118.
To our knowledge, no germline transgenic miRNA-engineered mouse models have been proposed yet to study BC onset and development.
5. miRNAs and cancer stem cells (CSCs)
5a. miRNAs involved in CSCs
The most recent definition of CSCs or tumor-initiating cells (TICs) identifies these cells as “a small subset of the cancerous population responsible for tumor initiation and growth, which also possesses the characteristic properties of quiescence, indefinite self-renewal, intrinsic resistance to chemotherapy and radiotherapy, and the capability to give rise to differentiated progeny” 119. Chemotherapeutic agents kill differentiated tumor cells, but CSCs are generally unharmed. The existence of CSCs propels resistance to chemotherapy, disease progression, and disease relapse. The origin of CSCs is still an ambiguous issue. The main hypotheses regarding the origin of CSCs include the following: i) malignant transformation of normal stem cells, ii) de-differentiation of mature cancer cells through EMT, and iii) induction of pluripotent cancer cells. Leukemia stem cells (LSCs) in acute myeloid leukemia (AML) are the first and best characterized CSCs, often providing a working template for other types of cancers 120. Similar to normal hematopoietic stem cells (HSCs), LSCs have extensive self-renewal property and are responsible for the maintenance of the bulk of leukemia blasts 120. The somatic stem cell hypothesis describes the process where a dormant stem cell present in the adult organism could be transformed to a CSC because of a mutation or inappropriate regulation of stem cell pathways. The transformation of an adult stem cell (ASC) into a CSC could be due to epigenetic reprogramming processes (alteration in the DNA methylation of CpG islands, leading to the expression or silencing of specific genomic regions; alteration in chromatin remodeling, which controls the accessibility of chromatin to transcription factors; and alteration in specific miRNA expression levels) 121.
The miRNA profile of CSCs is remarkably different from that of non-stem cancer cells, and many miRNAs have been shown to regulate the self-renewal and differentiation properties of CSCs 122, 123, such as the let-7 miRNA family 124. Being a tumor suppressor, the tumorigenic potential of the let-7 family is due to its downregulation in many tumors, such as lung cancer or BC 125. Cancer initiation, progression, and aggressiveness are driven by CSCs 126-128. The let-7 miRNA family appears to play a substantial role in the CSC phenotype. In fact, it seems that each tumor, being either hematologic or solid, includes a minor population of CSCs, capable of tumor initiation 129. These TICs have downregulated let-7 expression and, having tumor stem cell properties, can also undergo asymmetric division, thereby sustaining differentiated tumor proliferation 130. In BC, let-7 is found to be downregulated. In normal tissue, it plays the role of a regulator of self-renewal, acting as a pro-differentiation miRNA, whereas in BC it is repressed by the Wnt/β-catenin pathway 124. Thus, its loss in BC leads to an increase in the CSC population.
In addition to let-7, miR-34 has been described as a regulator of the Notch signaling pathway, necessary for stem cell maintenance, in colon CSCs 131. Asymmetric cell division, a characteristic of CSCs required for self-renewal, is directed toward symmetry by the presence of miR-146a, which targets Numb to stabilize β-catenin expression and leads to symmetrical division 132. In BC, expression of miR-34 leads to cell cycle arrest 133, whereas its downregulation increases the invasive capacity and metastatic potential of BC cell lines in vitro and in vivo 134. All the discussed miRNA as summarized in Table 2.
Table 2.
Examples of miRNAs involved in CSC phenotype, and EMT process. This table focuses on few examples of miRNAs described in the text, with a particular attention on their function and the type of cancer where they have been found.
| miRNA annotation | Function | Tumor | Ref. |
|---|---|---|---|
| let-7 | Regulator of self-renewal, cell proliferation and EMT | Lung, BC | 124 |
| miR-34, miR-146a | Symmetric and asymmetric division of CSCs | colon | 132 |
| miR-34, | Cell cycle control, invasion capacity and metastatic potential | BC | 133, 134 |
| let-7, miR-10, miR-34, miR-200, miR-205; miR-30 | Stemness and EMT regulation | BC | 139, 141, 142 |
| miR-155, miR-21 | EMT, cell migration and invasion control | BC | 133, 140 |
5b. miRNAs and EMT
Emerging evidence demonstrates that miRNAs play an essential role in controlling stem cell properties, such as self-renewal and differentiation, by regulating the expression of certain key stem cell regulatory genes 135-137 and by regulating EMT 138, 139. EMT refers to the process in which tumor epithelial cells acquire mesenchymal features, with high invasiveness and metastatic abilities. In fact, EMT is associated with the loss of intracellular junctions and epithelial polarity and increase in cell motility, which are fundamental characteristics for tumorigenesis, invasion, and metastasis that allow cancer cells to infiltrate adjacent stroma and metastasize to distant sites. These phenotypic changes appear to be induced by several miRNAs, such as let-7, miR-10, miR-34, miR-200, and miR-205 139. In BC, miR-155 and miR-21, described as oncomiRs, are implicated in EMT, cell migration, and invasion control. A well-known target of miR-21 is PTEN, a tumor suppressor, which negatively regulates the PI3K pathway 133, 140. Growing evidence suggests that BC cell plasticity, necessary for the spread of a tumor, arises because of partial reactivation of EMT in a mature cancer cell in order to give the cell pluripotency and a stem-like phenotype.
All the discussed miRNAs are summarized in Table 2.
6. Potential of miRNAs as BC biomarkers
BC is a heterogeneous disease with several morphological appearances, molecular features, behaviors, and response to therapy 143, 144. Therapeutic management of BC is based on the availability of strong diagnostic, prognostic, and predictive factors to guide the decision and the choice of different treatment options 145-147.
The current in vivo diagnostic tools for BC, e.g., mammography and ultrasound, are used for the detection of early-stage BC. However, several technical limitations exist for these techniques, such as breast density or calcification detection. Other imaging modalities, e.g., magnetic resonance imaging (MRI), have been proposed as complementary diagnostic modalities, with limited sensitivity.
Some proteins have been associated with BC by the analysis of expression levels of specific mRNAs, e.g., carcinoembryonic antigen (CEA) and CA-125 148. For BC diagnosis and prognosis, several mRNA-based genetic tests are currently available, such as the PAM50 assay (based on the NanoString technology), MammaTyper assay (based on the qRT-PCR technology), MammaPrint test (based on the microarray technology), Oncotype DX test (based on the qRT-PCR technology), Endopredict (based on the qRT-PCR technology), and Genomic Grade Index (based on the microarray technology) 149. Use of independent cores for gene expression testing in BC, coming from different gene signatures, may be a successful strategy to overcome tumor heterogeneity and sampling error.
Although direct measurements of tissue gene biomarkers have greatly improved BC diagnosis, the invasive and unpleasant nature of the diagnostic procedures limits their application. Isolation and subsequent characterization of circulating miRNAs provide the opportunity to bypass the problems associated with tissue biopsy, which is required as per the currently available genetic tests. In fact, circulating miRNAs are small molecules, found in body fluids (blood, plasma, serum, saliva, urine…). Being important regulators of gene expression and being dysregulated in several types of cancer diseases 150, circulating miRNAs have become interesting in new cancer biomarker research. They have been found to be stably and specifically expressed in mammary tissues and in body fluids when the disease is ongoing 151, 152. These features enable them to respond to the current clinical needs, allowing them to be used as easy, affordable, and clinically accessible molecular biomarkers in the retrospective analysis of large tissue collections and for diagnosis, prognosis, and prediction of therapeutic outcomes in BC.
6a. miRNAs dysregulated in BC
Several studies have looked at possible specific miRNAs dysregulated in BC with a diagnostic purpose 153, 154. Dysregulated miRNAs could be divided into 2 groups, being either upregulated or downregulated (Table 3).
Table 3.
BC deregulated miRNAs: an overview.
| miRNA annotations | Samples type | Ref. | |
|---|---|---|---|
|
Up-regulated miRNAs |
miR-21 | BC cell lines | 141, 155-157, 163 |
| miR-221/222 cluster | BC cell lines | 161 | |
| miR-9, miR10b, miR-29a, miR-96, miR-146a, miR-181, miR-373, miR-375, miR-520c, miR589 | BC cell lines | 162-165, 167, 198, 199 | |
| miR-10b | BC cell lines | 167 | |
| miR-155 | BC cell lines | 158, 163, 200 | |
| miR-210 | BC cell lines | 197 | |
|
Down-regulated miRNAs |
miR-30, miR-31, miR-34, miR-93, miR-126, miR-146a, miR-195, miR-205, miR-206, miR-503 | BC cell line; TNBC cell lines |
163, 170-175, 201 |
| let-7 family | BC cell lines | 163, 175, 182 | |
| miR-92a cluster | BC cell lines; TNBC cell lines | 187-191 | |
| miR-200 family | BC cell lines | 163, 197 | |
| miR-15/16 cluster, miR-103/107, miR-145, miR-335, miR-128b | BC cell lines | 137 | |
| miR-10b* | BC cell lines and xenografted tumor | 168, 169 |
TNBC= triple negative BC.
Increased expression of miR-21 has been found in vitro in human BC cell lines and tissues, playing a key role in all phases of BC pathogenesis 141, 155, although it also appears to be able to monitor early BC onset 156. miR-21 activity controls cell proliferation, G2/M check point, and metastasis diffusion 157-159 and the expression of many anti-oncogenes, including TPM1, programmed cell death 4, maspin, and Bcl-2, to support the metastasis and hyperplasia of BC cells 160.
Several other miRNAs have been validated to be overexpressed in BC; these include the miR-221/222 cluster 161, miR-9, miR10b, miR-29a, miR-96, miR-146a, miR-181, miR-373, miR-375, miR-520c, and miR589 162, highlighting their potential use for BC diagnosis, prognosis, and therapeutic studies 80, 137, 163-165.
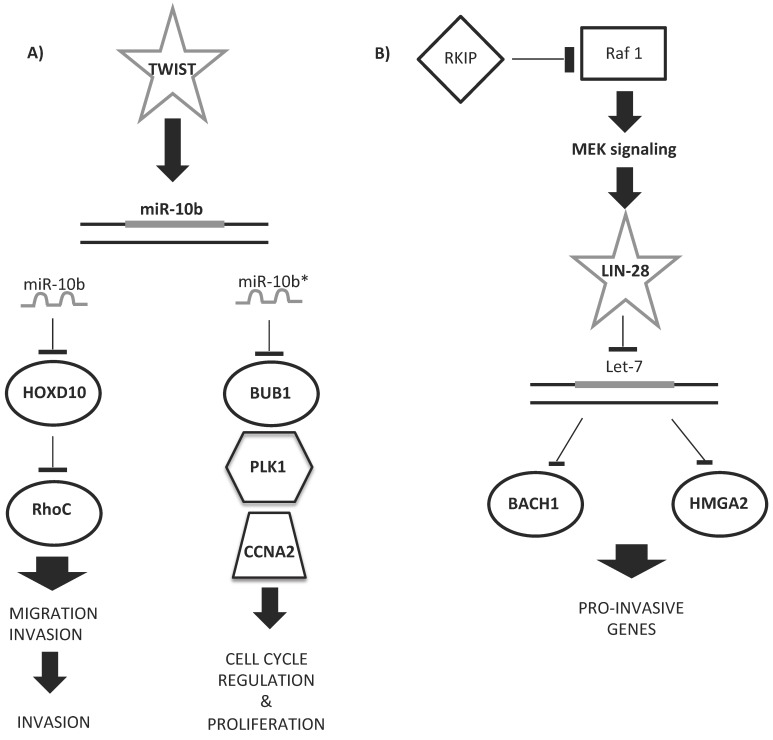
Some upregulated miRNAs could cooperate in controlling a network of functional genes to help tumor development or metastasis. Figure 5 shows examples of miRNA regulatory networks in BC that promote metastasis through their ability to coordinately target multiple genes 166. Ma et al. 167 proved the role of miR-10b as a driver of metastasis: miR-10b, under the control of the TWIST transcription factor, binds HOXD10 gene, enhancing cell migration and invasion. HOXD10, in turn, inhibits the Ras homolog gene family, member C (RHOC) protein, favoring metastatic diffusion of the tumor (Figure 5A). The miR-10b locus also encodes for miR-10b*/miR10b-3p. miR-10b*, although considered functionally irrelevant, was very recently demonstrated to be important for BC insurgence and development 168. Hence, if miR-10b-5p upregulation leads to the induction of ECM remodeling factors for metastatic invasion, miR-10b-3p downregulation is involved in primary BC onset and development, as its overexpression inhibits the proliferation of BC cell lines by targeting cell cycle regulator proteins (BUB1, PLK1, and CCNA2) 168, 169 (Figure 5A).
Figure 5.
Examples of miRNA regulatory networks in BC that promote metastasis. A) Two examples of the role of miR-10b/10b* in the regulation of either migration and invasion (left side) or cell cycle and proliferation (right side) processes. B) Example of let-7 regulatory role in the pro-invasive gene network control.
Among the downregulated miRNAs, miR-30a, miR-31, miR34, miR-93, miR-125, miR-126, mR-146a, miR-195, miR-200, miR-205, miR-206, miR-503, and let-7 170-174 have been shown to have a role in BC pathogenesis through the loss of their tumor suppressor properties. Overall, the main mechanisms affected by downregulated miRNAs are cell cycle, proliferation, and metastasis diffusion. Among all, the aforementioned members of the let-7 family are particularly relevant for BC development, as they function physiologically as tumor suppressors and are often inactivated in cancer 175, 176. let-7 miRNAs have been demonstrated to regulate multiple oncogenes, such as RAS, high-mobility group AT-hook 2 (HMGA2), c-Myc, and caspase-3 177-180, and several genes involved in the maintenance of stem cell phenotype 181. Thus, downregulation of let-7 family members could be one of the key events in the initiation of cancer owing to the acquisition of stem cell-like properties 182. Members of the let-7 family are among the more reproduced biomarkers identified in the in silico research of a miRNA BC signature. In fact, the let-7 family is present in several miRNA expression signatures of BC tissues 183, 184 and also in other types of tumor 183, 185, 186. In our recent publication, we proposed let-7c as a possible biomarker of a 4-miRNA signature, capable of distinguishing between grade 1 and grade 3 BC samples 21.
miR-92a is another possibly downregulated miRNA. This miRNA belongs to the miR-17-92 family, which can promote tumor proliferation by controlling the PI3K/Akt/mTOR pathway 187. Moreover, this miRNA has Bcl-2 interacting mediator of cell death (Bim) and p53 proteins as targets, thus inhibiting tumor cell apoptosis and cell cycle arrest and promoting tumorigenesis 98, 188, 189. In addition, it is involved in promoting tumor invasion and metastasis by modulating the TGF-β signaling pathway 190, 191.
Another downregulated miRNA, miR-206, has been found to be underexpressed in estrogen receptor (ER)α-positive BC, both in patient samples and BC cell lines 192, 193, and in lymph node metastatic BC 194, 195. With regard to its functions, it has been recently demonstrated to regulate the 3′ UTR of cyclin D1, inducing G1 arrest and a decrease in cell proliferation in BC cells 196, suggesting a potential role as a tumor suppressor. It has been shown that miR-206 regulates ERα via interaction with its 3′ UTR 193, demonstrating a specific role in most aggressive types of BC.
Other downregulated miRNAs, typical of BC tissues, are a group of miRNAs usually expressed in stem cells. This group includes the miR-200 family 197, miR-15/16, miR-103/107, miR-128b, miR-145, and miR-335 137. All these miRNAs are downregulated in CSCs, targeting common genes (Bmi1 and Suz12 component, Zeb1/2, and Klf4), all belonging to a regulatory circuit that sustains the breast CSC state 137.
6b. miRNAs as biomarkers of diagnosis, prognosis, and therapy prediction in BC
Several attempts have been made to identify affordable BC signatures for diagnosis, prognosis, and prediction of the therapeutic response (Table 4).
Table 4.
Circulating and non-circulating miRNAs as BC biomarkers. All the reported miRNAs have been validated on BC patients. For each miRNA, we indicated whether they have a role in diagnosis, prognosis or in prediction of therapy response in BC. For all groups we indicated the biological samples used for the validation, the validation assay, the cohort of data, the main results and the references. This table focused on few examples of single miRNA or miRNA signatures described in the text. GGI=gene expression grade index: TAM=tamoxifen; H= herceptin; N= normal tissue; T= tumor tissues; TNBC= triple negative breast cancer.
| Type of miRNAs | miRNA annotation | Role | Biological Samples | Technique/ cohort |
Results | Ref. |
|---|---|---|---|---|---|---|
| Non-circulating miRNAs | 13 miRNAs (miR-9-1, miR-10b, miR-21, miR-34, miR-29b/102, miR126, miR125a/b1/b2, miR-140as, miR-145, miR-155, miR-194, miR-204, miR-213) | Diagnosis | Tissue | Microarray and northern blot/ 76 BC vs 10 N |
4/13 are downregulated (5 miRNAs are the most constantly deregulated in BC) | 164 |
| 133 miRNAs | Diagnosis | Tissue | Microarray / 99 BC vs 5 N and 33 BC cell lines | 31 miRNAs are associated with tumor subtype or clinical pathological fators | 202 | |
| 15 miRNAs miR-342, miR-299, miR-217, miR-190, miR-135b, miR-218.; miR-520g, miR-377, miR-527-518a, miR-520f-520c; miR-520d, miR-181c, miR-302c, miR-376b, miR-30e |
Diagnosis | Tissue | Microarray / 95 BC vs 17 N | ER+; PR+; HER2/neu+: | 203 | |
| 6 BC-miRNAs signature miR-21 miR-17-5p miR-29b-2 miR-146 miR-155 miR-181b-1 |
Diagnosis | Tissue | Microarray/ 363 T vs 177 N | 31% of the total miRNAs varied among T and N tissues; they identified the most commontly altered miRNAs in solid tumors | 184 | |
|
miR-7, miR-128a, miR-210, and miR-516-3p; miR-210 |
Diagnosis | Tissue | TaqMan / 185 ER+ vs 114 ER- BC |
4-miRNA signature associated with tumor aggressiveness in ER+ BC and miR-210 associated with early relapse in ER- | 204 | |
| let-7c, miR320dmiR567, miR139-5p | Diagnosis | Tissue | Microarray / 42 BC G1 vs 42 BC G3 |
4 miRNA signature | 21 | |
|
let-7a miR-335 |
Diagnosis and prognosis | Tissue | TaqMan/ 60 BC vs 60 N |
Both miRNAs are decreased in BRCA mutant; miR-335 could be used as prognostic marker | 66 | |
| miR-155, miR-493, miR-30e, miR-27a | Diagnosis and prognosis | Tissue | Microarray/ 80 high risk vs 80 low risk | 2 upregulated, 'protective' miRNAs (miR-155, miR-493); 2 downregulated risk-associated miRNAs (miR-30e , miR-27a ) | 205 | |
|
miR-210 miR-148a |
Prognosis | Tissue | qRT-PCR/ 89 ER+ BC +TAM vs 56 N |
miR-210 and miR-148a are associated with relapse free survival; | 206 | |
|
miR-190b miR-339-5p miR-520c-3p /g/h miR-139-3p miR-204 miR-502-5p miR-365 miR-363 miR-7 |
Prediction of therapy response (TAM) | Tissue | Microarray/ 26 patients with recurrence vs 26 patients without recurrence |
miR-7 correlates with tumor grade | 212 | |
|
miR-30c miR-422a miR-30a-3p miR-187 miR-182 |
Prediction of therapy response (TAM) | Tissue | qRT-PCR/ 38 ER+ BC vs 15 BC+TAM |
Higher miR-30a-3p, miR-30c, and miR-182 are associated with treatment benefits | 213 | |
|
miR-21, miR-181b, miR-26a/26b, miR-27b miR-23b, let-7 family, miR-125a-5p/b-5p |
Prediction of therapy response (TAM) | Cell lines and tissue | qRT-PCR/ i) BC cell lines ± ER activation ii) 15 ER+ BC + exemestane and TAM |
All miRNAs are upregulated upon anti-estrogen treatment | 214 | |
| miR-26a, miR-30b, let-7 family, miR-125a/b | Prediction of therapy response (H) | Tissue | Microarray/ 83 BC + H vs adiacent stromal microdissected cells | With SVM technique, they developed a 35 miRNA signature for H treatment response | 216 | |
| Circulating miRNAs | miR-155 | Diagnosis | Serum | qRT-PCR/ 89 BC vs 29 N |
miR-155 is increased, both in primary and metastatic BC | 208 |
| miR-195 | Diagnosis | Blood | qRT-PCR/ 83 BC patients vs 44 N |
miR-195 is increased +19.25 fold | 209 | |
|
miR-29a miR-21 |
Diagnosis and prognosis | Serum | qRT-PCR/ 20 BC sera vs 20 N |
both miRNAs are increased; miR29a correlates with tumor stage | 210 | |
|
miR-16 miR-25 miR-222 miR-324-3 p |
Diagnosis and prognosis | Serum | TaqMan / 48 BC vs 48 N |
All are increased in high risk patients | 211 | |
|
let-7 miR-21 miR-202 |
Diagnosis and prognosis | Blood and serum | RT-PCR/ 136 BC vs 60 non BC |
All miRNAs are increased in BC patients; | 224 | |
|
miR-18b miR-103 miR-107 miR-652 |
Diagnosis, prognosis | Serum | RT-PCR/ 33 primary TNBC vs 33 N | 4 miRNA signature predict tumor relapse and overall survival | 225 | |
| miR-210 | Prediction of therapy response (H) | Plasma | TaqMan / 18 BC+ H vs 11 not responding BC |
miR-210 is higher in patients with residual BC (+2 fold) | 222 | |
| miR-155 | Prediction of therapy response (taxane) | Serum | RT-PCR/ 103 BC+taxane vs 55 N |
miR-155 expression correlates with the treatment course | 223 |
With respect to diagnosis, Iorio et al. 164 identified a 13-miRNA signature that could differentiate BC from normal breast tissues with 100% accuracy. Blenkiron et al. 202 identified 133 miRNAs that displayed aberrant expression levels in breast tumor tissues compared with normal breast tissues. Despite the identification of miRNA with aberrant expression in BC tissues, there remain discrepancies among the different reported miRNA signatures. This is probably because of the intrinsic heterogeneity in BC and because of clinicopathological variables such as the tumor stage, vascular invasion, proliferation index, and expression of HER2, ER, or progesterone receptor (PR). Thus, an attempt has been made to develop a miRNA signature that reflects the histopathological features of the tumor. At the simplest level, BC comprises 3 different histological subtypes: hormone receptor-positive (ER+, PR+) tumors, which cover approximately 60%-70% of diagnosed BCs; HER2+ tumors, which cover 15%-20% of diagnosed BCs; and triple-negative (ER-, PR-, HER2-) tumors 66. Genomic mRNA profiling has subdivided BC into 4 different classes: luminal A (ER+ and low grade), luminal B (ER+ and high grade), HER2+, and basal like (mainly triple negative) 202.
Blenkiron et al. 202 tried to associate a miRNA profile with each of these genomic classes. Among the 309 miRNAs identified in 93 BCs, 9 miRNAs (miR-15b, miR-99a, miR-100, miR-103, miR-107, miR-126*, miR-130a, miR-136, and miR-146b) could discriminate luminal A from luminal B BC 202.
Subsequently, Lowery et al. 203 identified a 15-miRNA predictive signature corresponding to the expression of ER (miR-135b, miR-190, miR-217, miR-218, miR-299, and miR-342), PR (miR-377, miR-520f, miR-520g, and miR-527-518a,), and the HER2 receptor (miR-30e, miR-181c, miR-320c, miR-376b, and miR-520d). The same approach was used by Volinia et al. 184, corresponding to the identification of a 17-miRNA signature for the status of ER (miR-30d and miR-30e), PR (miR-19a, miR-29c, miR-30a-5p, and miR-106b), HER2+ (let-7f, let-7g, miR-10b, miR-107, miR-126, miR-154, and miR-159), and ER/PR (miR-25, miR-142-5p, miR-200a, and miR-205,).
Finally, Foekens et al. 204 described a subset of miRNAs significantly associated with an ER+ luminal signature, identifying particularly 4 miRNAs associated with BC aggressiveness. The discrepancies among the different miRNA signature studies could result from the fact that miRNAs identified in each study were not examined in the others, besides other variables such as clinicopathological parameters of the tumor (tumor size, grade, etc.) or the use of different detection platforms (RT-PCR, next generation sequencing, etc.).
Hence, in our recent publication, starting from public BC databases containing gene expression profiles, copy number information, and miRNA profiles, we have described new 4 miRNA-based signatures, identifying a small group of miRNAs typical of BC, which could distinguish BC with different grades 21.
Several other small signatures or single miRNAs have been proposed with a diagnostic or prognostic aim 66, 205, 206.
All the described miRNAs are summarized in Table 4, section “Non-circulating miRNAs.”
The observation that miRNAs could be secreted by a solid tumor into the surrounding environment and that they are stable in body fluids make miRNAs promising targets easily found in blood, plasma, and serum 207. Some attempts have been made to identify single circulating miRNA or small circulating miRNA signatures with a diagnostic or prognostic purpose (Table 4, section “Circulating miRNAs”). Few examples of single circulating miRNAs proposed as diagnostic or prognostic tools have been suggested by Roth et al. 208, who found miR-155 in the serum of patients with BC and not in healthy controls, and by Heneghan et al. 209 who found elevated miR-195 expression in the blood of only patients with BC. Other miRNAs have been detected in the serum of patients with BC, such as miR-29a and miR-21 210 or the 4-miRNA signature of Hu et al. (miR-16, miR-25, miR-222, and miR-324-3p) 211 (Table 4).
The leading strategy for BC treatment is the use of surgery in combination with or followed by chemotherapy. The most common chemotherapeutics are antracyclines [doxorubicin (DOXO), adriamycin (ADR), and epirubicin], selective ER modulators [tamoxifen (TAM)], taxanes (taxol or paclitaxel and docetaxel), 5-fluorouracil (5-FU), and cyclophosphamide. Despite advances in treatment achieved by the combination of some of these compounds, a large number of patients do not respond to chemotherapy. In this context, miRNAs that are able to predict the therapeutic response of a given patient could help clinicians in the choice of the correct therapeutic approach. Some signatures have been developed in search for miRNAs able to predict the therapeutic response of patients with BC 206, 212-214 (Table 4). TAM, one of the main molecules used in BC treatment, is a drug that reduces or eliminates circulating estrogen or blocks the interaction of ER with genomic targets. In some studies, it was evaluated whether TAM could be a successful treatment for ER+ BC. The analyzed population had already developed metastasis prior to the onset of treatment and the benefit of treatment was measured as an objective response according to the REMARK criteria 215.
Other miRNA signatures have been studied for predicting the response of BC to Herceptin (or trastuzumab, H) 216. This molecule is a recombinant humanized monoclonal antibody against HER2 proteins that blocks the HER2-mediated activation of intracellular kinases and effectors 66. Although H treatment prolongs survival in adjuvant and metastatic settings, a majority of women with HER2+ metastatic disease will develop resistance to the therapy within a year of treatment. Identification of a miRNA signature that predicts patient risk, disease outcome, and tolerability to H therapy would greatly improve the personalized management of HER2+ BC. A few studies have successfully identified a prognostic miRNA signature for the response of BC tissue samples to H therapy 217. Combining the results of non-circulating miRNA signatures of BC cell lines treated with TAM or H, in order to identify miRNAs with predictive ability, only the let-7 family and miR-125a-5p/b-5p emerge as important predictors of therapeutic response 214, 217. miR-125, whose expression correlates with the HER2 status 218, has been found to be significantly downregulated in patients with BC 183, 219. Experimentally, the overexpression of miR-125 reduces ERBB2 and ERBB3, decreasing cell motility and the invasiveness of numerous cancers, including BC 219. The let-7 regulatory network suppresses metastasis by directly targeting the chromatin-remodeling protein HMGA2 and the transcription factor BACH1 180, 220 (Figure 5B). Both targets promote the transcription of pro-invasive genes that suppress cell invasion and metastasis to the bone 180, 220.
Almost all publications of circulating miRNA profile and HER2+ BC response to therapy have used BC cell lines to identify single miRNAs or groups of miRNAs whose expression is altered after prolonged H treatment 217, 221. One miRNA possibly involved in the response to H therapy is miR-210, which is present both in tissue and in body fluids of patients with BC 222. Circulating miR-210 has been associated with H sensitivity, tumor presence, and lymph node metastasis, suggesting a possible use of miR-210 to monitor the response of HER2+ BC to H-based therapies 222. Another miRNA, miR-155, has been used to monitor the effect of taxane treatment on BC. Sun et al. observed the decreased expression of miR-155 in serum after chemotherapy, which reached levels comparable to those of healthy subjects 223.
6c. miRNAs and hallmarks of BC
We have depicted an overview of miRNAs that can already be considered as BC biomarkers. This is outlined in Table 4. We have tried to classify all circulating and non-circulating miRNAs with diagnostic, prognostic, and predictive capacity in relation to their function as described in the literature. In particular, we have related them to altered pathways, the hallmarks of BC 226, generating a daisy-shaped figure (Figure 6) in which each of the petal represents one of the hallmark function altered in BC. The major group of miRNAs (27 miRNAs) affects genes belonging to the proliferation pathway, although some of these miRNAs (miR-155, miR-210, miR-21, and let7 family) are also involved in directing the BC invasion and metastatic pathways, controlled by other 24 miRNAs. Resting cell death and apoptosis are the targets of the third larger 14-miRNA group. Five miRNAs are responsible for the control of angiogenesis, whereas 2 miRNAs control genomic instability. miR-210 is the miRNA with a wider activity, being involved in energy metabolism, angiogenesis, and genomic instability beside the already described role in invasion and proliferation. Other miRNAs, such as miR-21, miR-27a/b, and miR-155, have been demonstrated to have multiple functions. This feature may be partially explained by the fact that the action of certain miRNAs is dependent upon the cellular model or environmental context in which they have been studied. Only 5 miRNAs have not yet been characterized in vitro for their function in BC development (miR-213, miR-299, miR-422a, miR-493, and miR-527).
Figure 6.
miRNA biomarkers and BC hallmarks. miRNAs have a role as diagnostic miRNA, prognostic miRNAs (italics), miRNAs predictive of the BC response to therapy (*), or miRNAs with multiple functions (diagnosis, prognosis, prediction of therapy outcome; underlined). Circulating (red) and non-circulating (black) miRNAs of Table 4 are included.
With respect to the clinical use of the depicted miRNAs, the majority of them (33/59) can be used as diagnostic tools; a small number of them (7/59) (indicated in italics in Figure 6) also have prognostic ability (members of the let-7 family, miR-27a, miR-30, miR-148a, miR-155, miR-210, and miR-335). Some of them (19/59) have the capacity to predict the response of BC to therapy (indicated with * in Figure 6; let-7, miR-7, miR-21, miR-23b, miR-26a/b, miR-27b, miR-30b/c, miR-125 a/b, miR-139-3p, miR-181b, miR-182, miR-187, miR-204, miR-210, miR-339-5p, miR-363, miR-365, miR-502-5p, and miR-520 family); 10/59 (marked in red in Figure 6) are circulating miRNAs, showing different functions in BC, and 2/59 (miR-155 and miR-210) are circulating miRNAs with both diagnostic, prognostic, and predictive role in BC.
7. miRNA for therapeutic use in cancer
Use of miRNAs for the development of new therapeutic strategies is based on 2 approaches: 1) use of miRNAs as drug molecules, based on the synthesis and delivery of specific oligonucleotides, able to increase or decrease miRNA levels in BC or 2) modulation of miRNAs in combination with non-miRNA-based therapies to increase the efficacy of the conventional treatments.
7a. Methods for miRNA modulation
There are 2 main approaches for developing miRNA-based therapies: antagonist and mimic oligonucleotides. MicroRNA antagonists or antagomiRs are generated to inhibit miRNAs that acquire a gain of function in human disease.
The most common strategy to ablate the function of miRNAs is achieved by single-stranded oligonucleotides with miRNA complementary sequences. In contrast, miRNA mimics are used to restore miRNAs that show a loss of function, as in the traditional gene therapy. This approach, also known as miRNA replacement therapy, has attracted much interest as it provides a new opportunity to therapeutically exploit tumor suppressors. The low molecular weight of miRNAs permits the delivery of therapeutic miRNAs as short double-stranded oligonucleotides 227. To improve the efficiency of miRNA/anti-miRNA delivery in vivo, modified miRNA molecules, both miRNA mimics and antagomiRs, with longer half-lives and increased efficiency have been developed, such as anti-miRNA oligonucleotides (AMOs) 228, locked nucleic acid (LNA)-modified oligonucleotides 229, cholesterol-conjugated antagomiRs 230, and the recently developed 2′-O-methoxyethyl-4′-thioRNA (MOE-SRNA) 231.
In recent years, a method has been described to inhibit miRNA function using synthetic mRNAs containing multiple binding sites for a specific miRNA, called miRNA sponges 232, 233. In bladder cancer cell lines it has been demonstrated that the forced expression of a miRNA sponge designed to inhibit miR-21 leads to a reduction in tumor aerobic glycolysis, i.e., the ability of the cells to metabolize glucose even under aerobic conditions 234. miRNA sponges have been validated even in an SUM149-epithelial BC cell xenografted mouse model, where inhibition of Myc-driven miRNA-9 using a synthetic mRNA containing several miR-9 binding sites reduced the development of lung metastases 198. Inhibition of the BC cell proliferation effect has been observed even in another xenografted mouse model implanted with the MDA-MB-231 BC cell line, where a miR-150 sponge-based inhibition led to a reduction in tumor mass proliferation via increase in the P2X7 receptor 235.
7b. miRNA-targeted therapies
Use of miRNAs alone in anti-cancer therapy to inhibit BC proliferation and development is still a challenge, although some promising results have already been obtained in both ex vivo and in vivo experiments. For instance, miR-145 has been chosen as a target therapy in BC cells because it is usually found to be downregulated in BC 236. Use of mimic or inhibitor miRNA oligonucleotides has been exploited in in vivo experiments. For instance, miR-21 has been found to be of particular interest in BC because chemically modified anti-miRNA oligonucleotides have already been developed for the in vivo treatment of xenografted BC mouse models 110.
To avoid rapid degradation and excretion of miRNAs, study of new delivery systems, which could enhance the stability and the delivery to target tissues, is necessary.
Recently, some studies focused on the potential use of nanomaterials to facilitate the delivery of biomolecules inside tumors. In particular, gold nanoparticles, with their high affinity for biomolecules, reduced cytotoxicity, easy size control, and well-developed surface chemistry, have been modified to increase their complementarity for nucleic acids 237, allowing the effective delivery of silencing RNAs inside cells 238, 239. The same approach has been used for the delivery of miR-145 into BC cells 236. Therefore, gold nanoparticles have been successfully used for the delivery of miR-145 oligonucleotides inside BC cell lines 240.
The question regarding the effect of the delivery on non-target organs and systemic toxicity of the compound remains unanswered. An obstacle for the use of miRNAs in therapy is the fact that miRNA modulation can affect hundreds of transcripts in different tissues, being potentially capable of shutting down entire pathways. Thus, till date, few companies have used miRNAs to develop a new class of cancer therapeutics. MRX34, a miR-34a mimic compound, is probably one of the first miRNA replacement agents to undergo clinical trials. At the time of writing this review, Mirna Therapeutics is recruiting participants to a phase I study of MRX34 (NCT01829971) (http://clinicaltrials.gov/ct2/show/NCT01829971). Similarly, Regulus Therapeutics is developing antago-miR-221 for hepatocellular carcinoma treatment and antagomiR-10b for glioblastoma treatment (http://www.regulusrx.com/therapeutic-areas/#Oncology). All these companies suggest that antago-miR oligonucleotides can be easily administered through local or parenteral injection routes with sufficient uptake of the agent to achieve sustained target inhibition in tissues and organs without the need for a formulation. Nevertheless, miRNA features, such as their stability and widespread activity on several targets, lead us to think that before using miRNA in therapies, much work is required to obtain more detailed and comprehensive knowledge about miRNA therapeutic potential, such as miRNA tissue distribution and systemic toxicity.
7c. miRNAs and chemoresistance
miRNAs can potentially be used to increase the response of BC to a therapeutic intervention. As an example, BC has been shown to be chemoresistant when some miRNAs are dysregulated (e.g., miR-125b, 239). miRNA mimic oligonucleotides, which increase the levels of a given miRNA in a BC in which this miRNA is lost, can be used in combination with conventional therapy to obtain an increased benefit for patient outcomes. An example of this approach has been used in the study by Yang et al. 241, where the upregulation of miR-195, obtained by mimic oligonucleotides, supplied to ADR-resistant MCF7 BC cell lines increased the sensitivity of the cells to the treatment, leading to apoptosis through downregulation of Raf-1 and Bcl-2. In addition, the combined treatment of a patient with BC using antago-miR oligonucleotides to shut down the increased miRNA expression levels in a specific patient could potentially increase the effect of conventional therapy. This approach has been used, for example, to demonstrate in vitro that miR-21 antisense oligonucleotides could be used in combination with H to kill resistant BC cells in xenografted mouse models 242.
For a more detail review on the role of miRNAs in chemoresistance modulation, see 243.
7d. miRNAs, CSCs, and chemoresistance
The ability of miRNAs to regulate the CSC phenotype and EMT is critical in the management of therapy resistance. In fact, CSCs in tumors are resting populations. The existing common chemoradiotherapy can target only rapidly proliferating cells. Thus, CSCs will escape from being killed and will thus be able to become resistant, to metastasize, or to generate relapses. miRNA modulation is therefore fundamental to increase the response of the tumor to the therapy. It has been demonstrated, for example, that a leukemia cell line resistant to daunorubicin shows increased levels of miR-21, the miRNA responsible of the control of EMT, whereas the suppression of this miRNA in the same cell line enhances daunorubicin cytotoxicity 244. Furthermore, in BC, therapy resistance is clearly influenced by CSC miRNAs. For example, miR-34a expression, necessary for the maintenance of CSC, is downregulated in BC ADR-resistant cell lines, and its upregulation leads to increased sensitivity to therapy, both in vitro and in vivo 245. Moreover, a close cross-talk among miR-200c, miR-203, and a stem cell transcription factor, Bmi1, has been described in BC; in particular, Yin et al. 246 demonstrated that Bmi1, often upregulated in BC and involved in stem cell maintenance, is regulated by miR-200c and miR-203. Bmi1 expression, and the parallel miR-200c and miR-203 downregulation are accompanied by the reversion of resistance to chemotherapy treatment in different BC cell lines 246.
In all these examples, it is clear that modulation of miRNAs has a critical impact on CSC maintenance, EMT, and, in general, on the response of the disease to therapy. Because CSCs are involved in the relapse of BC, modulation of miRNA in combination with therapy could decrease the possibility of next BC recidivism.
8. Conclusions and future perspective
In this review, we have focused on the recent advances related to miRNAs involved in BC and on their capability to respond to the actual clinical needs for the diagnosis, prognosis, and treatment of patients with BC.
Several lines of evidence have proven that in cancer, including BC, alterations in the levels of miRNAs are not only due to alterations in the miRNA biogenesis mechanisms but also depend on several upstream steps, such as epigenetic control, transcription factors, or mutated protein controls. As a consequence, prolonged aberrant expression of miRNAs, which could act either as tumor suppressors or as oncogenes, may contribute to the onset, progression, and diffusion of BC.
In the recent years, considerable progress has been made to understand the mechanisms responsible for aberrant miRNA expression in BC and several miRNAs or miRNA families have been found as key regulators of BC hallmarks (Figure 6).
Because early diagnosis of BC is essential for better prognosis of patients and because the currently available diagnostic methods show some limitations, miRNAs are emerging as novel diagnostic and prognostic biomarkers for BC. In particular, several miRNAs have shown BC diagnostic potential (i.e., miR-9, miR-10b, and miR-17-5p) and other miRNAs have shown a role in BC prognosis (i.e., miR-148a and miR-335). BC miRNAs have also been proposed as predictors of therapeutic outcomes, because their expression levels enable the prediction of patients' response to specific treatment (i.e., miR-30c, miR-187, and miR-339-5p). A significant number of these miRNAs, found as BC biomarkers, have an important role in the control of BC hallmark functions such as invasion, metastasis, proliferation, resting death, apoptosis, and genome instability.
A few miRNAs have multiple roles in diagnosis, prognosis, and prediction of therapeutic response in BC. Among these miRNAs, some are circulating (e.g., miR-155 and miR-210), and owing to their stability in body fluids, could really become new, easily accessible, affordable, non-invasive, and promising testing tools for the personalized management of patients with BC. Nevertheless, recent evidence suggest that circulating, multiple miRNAs-based profiles have better diagnostic and prognostic performance as well as better sensitivity than individual miRNA assays 225, 247 because the combination of several miRNAs, controlling multiple target genes, can better clarify how each of them contributes to tumor development and better represent the global biological effect of miRNA regulation on the multistep process leading to BC.
However, the full potential of miRNAs should not be exhausted in their use as biomarkers of BC. Future research should be directed to the development and delivery of miRNA-based drugs in BC; in this respect, some miRNAs have already shown promising results (e.g., miR-9, miR-21, miR34a, miR145, and miR150). Thus, particular attention should be given to the optimization of miRNA-based drug stability, improvement of miRNA delivery, and control of the off-target effects of miRNA therapeutics. Moreover, to increase the efficacy of currently used non-miRNA treatments for BC (e.g., chemotherapy), great efforts should be directed to the use of such non-miRNA treatments in combination with miRNAs showing a fundamental role in the modulation of the response to treatment (e.g., miR-21, miR34a, miR195, miR200c, and miR203 in combination with chemotherapy).
Acknowledgments
This work was supported partially by INTEROMICS flagship project (url: http://www.interomics.eu/it/home), National Research Council CUP grant B91J12000190001.
Author contributions
GB and CC wrote the manuscript, and collaborate in figure management and technical editing of the manuscript; GB, CC, and IC critically revised the manuscript.
Abbreviations
- BC
breast cancer
- miRs or miRNAs
microRNAs
- TAM
tamoxifen
- H
herceptin or trastuzumab
- ADR
adriamycin
- CSC
cancer stem cells
- EMT
epithelial-mesenchimal-transition
- MET
mesenchymal-epithelial-transition
- TICs
tumor initiating cells
- N
normal tissue
- T
tumor tissue
- ER
estrogen receptor
- PR
progesterone receptor
- ERBB2/HER2
the receptor tyrosine-protein kinase erbB2.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 3.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S. et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Femminella GD, Ferrara N, Rengo G. The emerging role of microRNAs in Alzheimer's disease. Front Physiol. 2015;6:40. doi: 10.3389/fphys.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondkar AA, Abu-Amero KK. Utility of Circulating MicroRNAs as Clinical Biomarkers for Cardiovascular Diseases. Biomed Res Int. 2015;2015:821823. doi: 10.1155/2015/821823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClelland AD, Kantharidis P. microRNA in the development of diabetic complications. Clin Sci (Lond) 2014;126:95–110. doi: 10.1042/CS20130079. [DOI] [PubMed] [Google Scholar]
- 9.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 10.Palmero EI, de Campos SG, Campos M. et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34:363–370. doi: 10.1590/S1415-47572011000300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 12.Rakha EA, El-Sayed ME, Menon S. et al. Histologic grading is an independent prognostic factor in invasive lobular carcinoma of the breast. Breast Cancer Res Treat. 2008;111:121–127. doi: 10.1007/s10549-007-9768-4. [DOI] [PubMed] [Google Scholar]
- 13.Viale G. The current state of breast cancer classification. Ann Oncol. 2012;23(Suppl 10):x207–210. doi: 10.1093/annonc/mds326. [DOI] [PubMed] [Google Scholar]
- 14.Singletary SE, Allred C, Ashley P. et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20:3628–3636. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 16.Jiang D, Zhao N. A clinical prognostic prediction of lymph node-negative breast cancer by gene expression profiles. J Cancer Res Clin Oncol. 2006;132:579–587. doi: 10.1007/s00432-006-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu H, Xu W, Huang Y, Yang S. Circulating miRNAs: promising biomarkers of human cancer. Asian Pac J Cancer Prev. 2011;12:1117–1125. [PubMed] [Google Scholar]
- 19.Ashby J, Flack K, Jimenez LA. et al. Distribution profiling of circulating microRNAs in serum. Anal Chem. 2014;86:9343–9349. doi: 10.1021/ac5028929. [DOI] [PubMed] [Google Scholar]
- 20.Arroyo JD, Chevillet JR, Kroh EM. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cava C, Bertoli G, Ripamonti M. et al. Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS One. 2014;9:e97681. doi: 10.1371/journal.pone.0097681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 23.Vidaurre S, Fitzpatrick C, Burzio VA. et al. Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy. J Biol Chem. 2014;289:27182–27198. doi: 10.1074/jbc.M114.558841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y, Ahn C, Han J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Denli AM, Tops BB, Plasterk RH. et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 27.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund E, Guttinger S, Calado A. et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 30.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghildiyal M, Xu J, Seitz H. et al. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Qin W, Shi Y, Zhao B. et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eiring AM, Harb JG, Neviani P. et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 39.Ekimler S, Sahin K. Computational Methods for MicroRNA Target Prediction. Genes (Basel) 2014;5:671–683. doi: 10.3390/genes5030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis BP, Shih IH, Jones-Rhoades MW. et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 41.Hafner M, Landthaler M, Burger L. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broderick JA, Salomon WE, Ryder SP. et al. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentwich I, Avniel A, Karov Y. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 44.Yue D, Liu H, Huang Y. Survey of Computational Algorithms for MicroRNA Target Prediction. Curr Genomics. 2009;10:478–492. doi: 10.2174/138920209789208219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofacker IL. How microRNAs choose their targets. Nat Genet. 2007;39:1191–1192. doi: 10.1038/ng1007-1191. [DOI] [PubMed] [Google Scholar]
- 46.Kertesz M, Iovino N, Unnerstall U. et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 47.Lindow M, Gorodkin J. Principles and limitations of computational microRNA gene and target finding. DNA Cell Biol. 2007;26:339–351. doi: 10.1089/dna.2006.0551. [DOI] [PubMed] [Google Scholar]
- 48.Allmer J, Yousef M. Computational methods for ab initio detection of microRNAs. Front Genet. 2012;3:209. doi: 10.3389/fgene.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Ding C, Meraz RF, Holbrook SR. PSoL: a positive sample only learning algorithm for finding non-coding RNA genes. Bioinformatics. 2006;22:2590–2596. doi: 10.1093/bioinformatics/btl441. [DOI] [PubMed] [Google Scholar]
- 50.Yousef M, Jung S, Showe LC, Showe MK. Learning from positive examples when the negative class is undetermined--microRNA gene identification. Algorithms Mol Biol. 2008;3:2. doi: 10.1186/1748-7188-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadri S, Hinman V, Benos PV. HHMMiR: efficient de novo prediction of microRNAs using hierarchical hidden Markov models. BMC Bioinformatics. 2009;10(Suppl 1):S35. doi: 10.1186/1471-2105-10-S1-S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krek A, Grun D, Poy MN. et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 53.Huang TH, Fan B, Rothschild MF. et al. MiRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics. 2007;8:341. doi: 10.1186/1471-2105-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertel J, Stadler PF. Hairpins in a Haystack: recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
- 55.Nam JW, Shin KR, Han J. et al. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terai G, Komori T, Asai K, Kin T. miRRim: a novel system to find conserved miRNAs with high sensitivity and specificity. RNA. 2007;13:2081–2090. doi: 10.1261/rna.655107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yousef M, Nebozhyn M, Shatkay H. et al. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics. 2006;22:1325–1334. doi: 10.1093/bioinformatics/btl094. [DOI] [PubMed] [Google Scholar]
- 58.Oulas A, Boutla A, Gkirtzou K. et al. Prediction of novel microRNA genes in cancer-associated genomic regions--a combined computational and experimental approach. Nucleic Acids Res. 2009;37:3276–3287. doi: 10.1093/nar/gkp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maragkakis M, Alexiou P, Papadopoulos GL. et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enright AJ, John B, Gaul U. et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuhn DE, Martin MM, Feldman DS. et al. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muniategui A, Pey J, Planes FJ, Rubio A. Joint analysis of miRNA and mRNA expression data. Brief Bioinform. 2013;14:263–278. doi: 10.1093/bib/bbs028. [DOI] [PubMed] [Google Scholar]
- 64.Karginov FV, Conaco C, Xuan Z. et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cambronne XA, Shen R, Auer PL, Goodman RH. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci U S A. 2012;109:20473–20478. doi: 10.1073/pnas.1218887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17:313–319. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Shi M, Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. 2009;35:328–334. doi: 10.1016/j.ctrv.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 70.Lehmann U. Aberrant DNA methylation of microRNA genes in human breast cancer - a critical appraisal. Cell Tissue Res. 2014;356:657–664. doi: 10.1007/s00441-014-1793-0. [DOI] [PubMed] [Google Scholar]
- 71.Castilla MA, Diaz-Martin J, Sarrio D. et al. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS One. 2012;7:e47709. doi: 10.1371/journal.pone.0047709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wee EJ, Peters K, Nair SS. et al. Mapping the regulatory sequences controlling 93 breast cancer-associated miRNA genes leads to the identification of two functional promoters of the Hsa-mir-200b cluster, methylation of which is associated with metastasis or hormone receptor status in advanced breast cancer. Oncogene. 2012;31:4182–4195. doi: 10.1038/onc.2011.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aure MR, Leivonen SK, Fleischer T. et al. Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013;14:R126. doi: 10.1186/gb-2013-14-11-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saito Y, Liang G, Egger G. et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 75.Scott GK, Mattie MD, Berger CE. et al. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 76.Yamamoto H, Adachi Y, Taniguchi H. et al. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J Gastroenterol. 2012;18:2745–2755. doi: 10.3748/wjg.v18.i22.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin X, Wu M, Liu P. et al. Up-regulation and worse prognostic marker of cytoplasmic TARBP2 expression in obstinate breast cancer. Med Oncol. 2014;31:868. doi: 10.1007/s12032-014-0868-9. [DOI] [PubMed] [Google Scholar]
- 78.Calin GA, Sevignani C, Dumitru CD. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinhart BJ, Slack FJ, Basson M. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 80.Khoshnaw SM, Rakha EA, Abdel-Fatah T. et al. The microRNA maturation regulator Drosha is an independent predictor of outcome in breast cancer patients. Breast Cancer Res Treat. 2013;137:139–153. doi: 10.1007/s10549-012-2358-0. [DOI] [PubMed] [Google Scholar]
- 81.Khoshnaw SM, Rakha EA, Abdel-Fatah TM. et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135:403–413. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- 82.Yan M, Huang HY, Wang T. et al. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res. 2012;18:343–348. doi: 10.1007/s12253-011-9450-3. [DOI] [PubMed] [Google Scholar]
- 83.Dedes KJ, Natrajan R, Lambros MB. et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer. 2011;47:138–150. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 84.Han Y, Liu Y, Gui Y, Cai Z. Inducing cell proliferation inhibition and apoptosis via silencing Dicer, Drosha, and Exportin 5 in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:201–205. doi: 10.1002/jso.23214. [DOI] [PubMed] [Google Scholar]
- 85.Bian XJ, Zhang GM, Gu CY. et al. Down-regulation of Dicer and Ago2 is associated with cell proliferation and apoptosis in prostate cancer. Tumour Biol. 2014;35:11571–11578. doi: 10.1007/s13277-014-2462-3. [DOI] [PubMed] [Google Scholar]
- 86.Zhang J, Zhang XH, Wang CX. et al. Dysregulation of microRNA biosynthesis enzyme Dicer plays an important role in gastric cancer progression. Int J Clin Exp Pathol. 2014;7:1702–1707. [PMC free article] [PubMed] [Google Scholar]
- 87.Gao C, Li X, Tong B. et al. Up-regulated expression of Dicer reveals poor prognosis in laryngeal squamous cell carcinoma. Acta Otolaryngol. 2014;134:959–963. doi: 10.3109/00016489.2014.920962. [DOI] [PubMed] [Google Scholar]
- 88.Passon N, Gerometta A, Puppin C. et al. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012;65:320–326. doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- 89.Pickering BF, Yu D, Van Dyke MW. Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J Biol Chem. 2011;286:44095–44103. doi: 10.1074/jbc.M111.265439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pichiorri F, Palmieri D, De Luca L. et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J Exp Med. 2013;210:951–968. doi: 10.1084/jem.20120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu X, Lin J, Zack DJ. et al. Analysis of regulatory network topology reveals functionally distinct classes of microRNAs. Nucleic Acids Res. 2008;36:6494–6503. doi: 10.1093/nar/gkn712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fazi F, Rosa A, Fatica A. et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 95.Davis BN, Hilyard AC, Nguyen PH. et al. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suzuki HI, Yamagata K, Sugimoto K. et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 97.Zhang X, Wan G, Berger FG. et al. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Y, Choi PS, Casey SC. et al. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen J, Xia W, Khotskaya YB. et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo QJ, Mills JN, Bandurraga SG. et al. MicroRNA-510 promotes cell and tumor growth by targeting peroxiredoxin1 in breast cancer. Breast Cancer Res. 2013;15:R70. doi: 10.1186/bcr3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jurmeister S, Baumann M, Balwierz A. et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633–651. doi: 10.1128/MCB.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yao Y, Hu J, Shen Z, MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li C, Nguyen HT, Zhuang Y. et al. Comparative profiling of miRNA expression of lung adenocarcinoma cells in two-dimensional and three-dimensional cultures. Gene. 2012;511:143–150. doi: 10.1016/j.gene.2012.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nguyen HT, Li C, Lin Z. et al. The microRNA expression associated with morphogenesis of breast cancer cells in three-dimensional organotypic culture. Oncol Rep. 2012;28:117–126. doi: 10.3892/or.2012.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pramanik D, Campbell NR, Karikari C. et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Q, Jiang Q, Pu Q. et al. MicroRNA-143 inhibits migration and invasion of human non-small-cell lung cancer and its relative mechanism. Int J Biol Sci. 2013;9:680–692. doi: 10.7150/ijbs.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seviour EG, Sehgal V, Lu Y, Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bockhorn J, Prat A, Chang YF. et al. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Cancer Res. 2014;74:7406–7417. doi: 10.1158/0008-5472.CAN-14-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan LX, Wu QN, Zhang Y. et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:R2. doi: 10.1186/bcr2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu M, Yi M, Kim CH. et al. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol. 2011;12:R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trang P, Medina PP, Wiggins JF. et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Costinean S, Zanesi N, Pekarsky Y. et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hatley ME, Patrick DM, Garcia MR. et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xiao C, Srinivasan L, Calado DP. et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klein U, Lia M, Crespo M. et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 117.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santanam U, Zanesi N, Efanov A. et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O'Flaherty JD, Barr M, Fennell D. et al. The cancer stem-cell hypothesis: its emerging role in lung cancer biology and its relevance for future therapy. J Thorac Oncol. 2012;7:1880–1890. doi: 10.1097/JTO.0b013e31826bfbc6. [DOI] [PubMed] [Google Scholar]
- 120.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 121.Shukla S, Meeran SM. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta. 2014;1840:3494–3502. doi: 10.1016/j.bbagen.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 122.Si H, Sun X, Chen Y. et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chhabra R, Saini N. MicroRNAs in cancer stem cells: current status and future directions. Tumour Biol. 2014;35:8395–8405. doi: 10.1007/s13277-014-2264-7. [DOI] [PubMed] [Google Scholar]
- 124.Cai WY, Wei TZ, Luo QC. et al. The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013;126:2877–2889. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 125.Jakopovic M, Thomas A, Balasubramaniam S. et al. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol. 2013;3:261. doi: 10.3389/fonc.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rutnam ZJ, Yang BB. The involvement of microRNAs in malignant transformation. Histol Histopathol. 2012;27:1263–1270. doi: 10.14670/HH-27.1263. [DOI] [PubMed] [Google Scholar]
- 127.Liu C, Kelnar K, Vlassov AV. et al. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–3404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bao B, Ali S, Ahmad A. et al. Differentially expressed miRNAs in cancer-stem-like cells: markers for tumor cell aggressiveness of pancreatic cancer. Stem Cells Dev. 2014;23:1947–1958. doi: 10.1089/scd.2013.0551. [DOI] [PubMed] [Google Scholar]
- 129.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 130.Yu F, Yao H, Zhu P. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 131.Bu P, Chen KY, Chen JH. et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hwang WL, Jiang JK, Yang SH. et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 133.Han M, Liu M, Wang Y. et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One. 2012;7:e39520. doi: 10.1371/journal.pone.0039520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang S, Li Y, Gao J. et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–4303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 135.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iliopoulos D, Lindahl-Allen M, Polytarchou C. et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012;109:14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wellner U, Schubert J, Burk UC. et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 139.Sun X, Jiao X, Pestell TG. et al. MicroRNAs and cancer stem cells: the sword and the shield. Oncogene. 2014;33:4967–4977. doi: 10.1038/onc.2013.492. [DOI] [PubMed] [Google Scholar]
- 140.Han M, Liu M, Wang Y. et al. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol Cell Biochem. 2012;363:427–436. doi: 10.1007/s11010-011-1195-5. [DOI] [PubMed] [Google Scholar]
- 141.Zhang ZJ, Ma SL. miRNAs in breast cancer tumorigenesis (Review) Oncol Rep. 2012;27:903–910. doi: 10.3892/or.2011.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schwarzenbacher D, Balic M, Pichler M. The role of microRNAs in breast cancer stem cells. Int J Mol Sci. 2013;14:14712–14723. doi: 10.3390/ijms140714712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perou CM, Sorlie T, Eisen MB. et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 145.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–392. doi: 10.1023/a:1014778713034. [DOI] [PubMed] [Google Scholar]
- 146.Ravdin PM, Siminoff LA, Davis GJ. et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19:980–991. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 147.Carlson RW, Allred DC, Anderson BO. et al. Invasive breast cancer. J Natl Compr Canc Netw. 2011;9:136–222. doi: 10.6004/jnccn.2011.0016. [DOI] [PubMed] [Google Scholar]
- 148.Opstal-van Winden AW, Rodenburg W, Pennings JL. et al. A bead-based multiplexed immunoassay to evaluate breast cancer biomarkers for early detection in pre-diagnostic serum. Int J Mol Sci. 2012;13:13587–13604. doi: 10.3390/ijms131013587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sinn P, Aulmann S, Wirtz R. et al. Multigene Assays for Classification, Prognosis, and Prediction in Breast Cancer: a Critical Review on the Background and Clinical Utility. Geburtshilfe Frauenheilkd. 2013;73:932–940. doi: 10.1055/s-0033-1350831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mar-Aguilar F, Rodriguez-Padilla C, Resendez-Perez D. Use of serum-circulating miRNA profiling for the identification of breast cancer biomarkers. Methods Mol Biol. 2014;1165:71–80. doi: 10.1007/978-1-4939-0856-1_6. [DOI] [PubMed] [Google Scholar]
- 152.Chan M, Liaw CS, Ji SM. et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 153.Nassar FJ, El Sabban M, Zgheib NK. et al. miRNA as potential biomarkers of breast cancer in the Lebanese population and in young women: a pilot study. PLoS One. 2014;9:e107566. doi: 10.1371/journal.pone.0107566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumour Biol. 2014;35:6235–6244. doi: 10.1007/s13277-014-2202-8. [DOI] [PubMed] [Google Scholar]
- 155.Corcoran C, Friel AM, Duffy MJ. et al. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 156.Ozgun A, Karagoz B, Bilgi O. et al. MicroRNA-21 as an indicator of aggressive phenotype in breast cancer. Onkologie. 2013;36:115–118. doi: 10.1159/000348678. [DOI] [PubMed] [Google Scholar]
- 157.Dong G, Liang X, Wang D. et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31:57. doi: 10.1007/s12032-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 158.Anastasov N, Hofig I, Vasconcellos IG. et al. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol. 2012;7:206. doi: 10.1186/1748-717X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Min W, Wang B, Li J. et al. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97–104. doi: 10.12659/MSMBR.891246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li J, Zhang Y, Zhang W. et al. Genetic heterogeneity of breast cancer metastasis may be related to miR-21 regulation of TIMP-3 in translation. Int J Surg Oncol. 2013;2013:875078. doi: 10.1155/2013/875078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (Review) Int J Oncol. 2013;43:985–994. doi: 10.3892/ijo.2013.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Christodoulatos GS, Dalamaga M. Micro-RNAs as clinical biomarkers and therapeutic targets in breast cancer: Quo vadis? World J Clin Oncol. 2014;5:71–81. doi: 10.5306/wjco.v5.i2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13:13414–13437. doi: 10.3390/ijms131013414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Iorio MV, Ferracin M, Liu CG. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 165.Sandhu R, Rein J, D'Arcy M. et al. Overexpression of miR-146a in basal-like breast cancer cells confers enhanced tumorigenic potential in association with altered p53 status. Carcinogenesis. 2014;35:2567–2575. doi: 10.1093/carcin/bgu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 168.Biagioni F, Bossel Ben-Moshe N, Fontemaggi G. et al. The locus of microRNA-10b: a critical target for breast cancer insurgence and dissemination. Cell Cycle. 2013;12:2371–2375. doi: 10.4161/cc.25380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Biagioni F, Bossel Ben-Moshe N, Fontemaggi G. et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–1229. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hu J, Xu J, Wu Y. et al. Identification of microRNA-93 as a functional dysregulated miRNA in triple-negative breast cancer. Tumour Biol. 2015;36:251–258. doi: 10.1007/s13277-014-2611-8. [DOI] [PubMed] [Google Scholar]
- 171.Zhao FL, Dou YC, Wang XF. et al. Serum microRNA-195 is down-regulated in breast cancer: a potential marker for the diagnosis of breast cancer. Mol Biol Rep. 2014;41:5913–5922. doi: 10.1007/s11033-014-3466-1. [DOI] [PubMed] [Google Scholar]
- 172.Kim SJ, Shin JY, Lee KD. et al. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shukla K, Sharma AK, Ward A, MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol Oncol; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jang K, Ahn H, Sim J. et al. Loss of microRNA-200a expression correlates with tumor progression in breast cancer. Transl Res. 2014;163:242–251. doi: 10.1016/j.trsl.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 175.Yan Y, Zhang F, Fan Q. et al. Breast cancer-specific TRAIL expression mediated by miRNA response elements of let-7 and miR-122. Neoplasma. 2014;61:672–679. doi: 10.4149/neo_2014_082. [DOI] [PubMed] [Google Scholar]
- 176.Xie R, Wang Y, Nie W. et al. Lin28B expression correlates with aggressive clinicopathological characteristics in breast invasive ductal carcinoma. Cancer Biother Radiopharm. 2014;29:215–220. doi: 10.1089/cbr.2014.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 178.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 179.Su JL, Chen PS, Johansson G, Kuo ML. Function and regulation of let-7 family microRNAs. Microrna. 2012;1:34–39. doi: 10.2174/2211536611201010034. [DOI] [PubMed] [Google Scholar]
- 180.Yun J, Frankenberger CA, Kuo WL. et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Worringer KA, Rand TA, Hayashi Y. et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jiang R, Li Y, Zhang A. et al. The acquisition of cancer stem cell-like properties and neoplastic transformation of human keratinocytes induced by arsenite involves epigenetic silencing of let-7c via Ras/NF-kappaB. Toxicol Lett. 2014;227:91–98. doi: 10.1016/j.toxlet.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 183.Mattie MD, Benz CC, Bowers J. et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gowrishankar B, Ibragimova I, Zhou Y. et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15:329–341. doi: 10.4161/cbt.27314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tang Z, Ow GS, Thiery JP. et al. Meta-analysis of transcriptome reveals let-7b as an unfavorable prognostic biomarker and predicts molecular and clinical subclasses in high-grade serous ovarian carcinoma. Int J Cancer. 2014;134:306–318. doi: 10.1002/ijc.28371. [DOI] [PubMed] [Google Scholar]
- 187.Li H, Bian C, Liao L. et al. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126:565–575. doi: 10.1007/s10549-010-0954-4. [DOI] [PubMed] [Google Scholar]
- 188.Leung CM, Chen TW, Li SC. et al. MicroRNA expression profiles in human breast cancer cells after multifraction and single-dose radiation treatment. Oncol Rep. 2014;31:2147–2156. doi: 10.3892/or.2014.3089. [DOI] [PubMed] [Google Scholar]
- 189.Perez-Madrigal D, Finegan KG, Paramo B, Tournier C. The extracellular-regulated protein kinase 5 (ERK5) promotes cell proliferation through the down-regulation of inhibitors of cyclin dependent protein kinases (CDKs) Cell Signal. 2012;24:2360–2368. doi: 10.1016/j.cellsig.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 190.Zhang G, Zhou H, Xiao H. et al. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59:98–107. doi: 10.1007/s10620-013-2858-8. [DOI] [PubMed] [Google Scholar]
- 191.Li L, Shi JY, Zhu GQ, Shi B. MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem. 2012;113:1235–1244. doi: 10.1002/jcb.23457. [DOI] [PubMed] [Google Scholar]
- 192.Kondo N, Toyama T, Sugiura H. et al. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 193.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 194.Liu H, Cao YD, Ye WX, Sun YY. Effect of microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in MDA-MB-231 cells. Tumori. 2010;96:751–755. doi: 10.1177/030089161009600518. [DOI] [PubMed] [Google Scholar]
- 195.Li Y, Hong F, Yu Z. Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res. 2013;41:596–602. doi: 10.1177/0300060513485856. [DOI] [PubMed] [Google Scholar]
- 196.Elliman SJ, Howley BV, Mehta DS. et al. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis. 2014;3:e113. doi: 10.1038/oncsis.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shimono Y, Zabala M, Cho RW. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ma L, Young J, Prabhala H. et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sochor M, Basova P, Pesta M. et al. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236–1243. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 201.Gong J, Luk F, Jaiswal R, Bebawy M. Microparticles Mediate the Intercellular Regulation of microRNA-503 and Proline-Rich Tyrosine Kinase 2 to Alter the Migration and Invasion Capacity of Breast Cancer Cells. Front Oncol. 2014;4:220. doi: 10.3389/fonc.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Blenkiron C, Goldstein LD, Thorne NP. et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lowery AJ, Miller N, Devaney A. et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Foekens JA, Sieuwerts AM, Smid M. et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gasparini P, Cascione L, Fassan M. et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rothe F, Ignatiadis M, Chaboteaux C. et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6:e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Weber JA, Baxter DH, Zhang S. et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Roth C, Rack B, Muller V. et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Heneghan HM, Miller N, Lowery AJ. et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 210.Wu Q, Lu Z, Li H. et al. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hu Z, Dong J, Wang LE. et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33:828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 212.Lyng MB, Laenkholm AV, Sokilde R. et al. Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant tamoxifen mono-therapy: a DBCG study. PLoS One. 2012;7:e36170. doi: 10.1371/journal.pone.0036170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rodriguez-Gonzalez FG, Sieuwerts AM, Smid M. et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;127:43–51. doi: 10.1007/s10549-010-0940-x. [DOI] [PubMed] [Google Scholar]
- 214.Maillot G, Lacroix-Triki M, Pierredon S. et al. Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- 215.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Valabrega G, Montemurro F, Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann Oncol. 2007;18:977–984. doi: 10.1093/annonc/mdl475. [DOI] [PubMed] [Google Scholar]
- 217.Ichikawa T, Sato F, Terasawa K. et al. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS One. 2012;7:e31422. doi: 10.1371/journal.pone.0031422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Fassan M, Pizzi M, Realdon S. et al. The HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesis. Hum Pathol. 2013;44:1804–1810. doi: 10.1016/j.humpath.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 219.Scott GK, Goga A, Bhaumik D. et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 220.Dangi-Garimella S, Yun J, Eves EM. et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Le XF, Almeida MI, Mao W. et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One. 2012;7:e41170. doi: 10.1371/journal.pone.0041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jung EJ, Santarpia L, Kim J. et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sun Y, Wang M, Lin G. et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Joosse SA, Muller V, Steinbach B. et al. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br J Cancer. 2014;111:909–917. doi: 10.1038/bjc.2014.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kleivi Sahlberg K, Bottai G, Naume B. et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21:1207–1214. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 226.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 227.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 229.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 230.Krutzfeldt J, Rajewsky N, Braich R. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 231.Saito Y, Hashimoto Y, Arai M. et al. Chemistry, properties, and in vitro and in vivo applications of 2'-O-methoxyethyl-4'-thioRNA, a novel hybrid type of chemically modified RNA. Chembiochem. 2014;15:2535–2540. doi: 10.1002/cbic.201402398. [DOI] [PubMed] [Google Scholar]
- 232.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yang X, Cheng Y, Li P. et al. A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 2015;36:383–391. doi: 10.1007/s13277-014-2617-2. [DOI] [PubMed] [Google Scholar]
- 235.Huang S, Chen Y, Wu W. et al. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 237.Rosi NL, Giljohann DA, Thaxton CS. et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 238.Elbakry A, Zaky A, Liebl R. et al. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 239.Lee JS, Green JJ, Love KT. et al. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ekin A, Karatas OF, Culha M, Ozen M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J Gene Med. 2014;16:331–335. doi: 10.1002/jgm.2810. [DOI] [PubMed] [Google Scholar]
- 241.Yang G, Wu D, Zhu J. et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep. 2013;30:877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]
- 242.Gong C, Yao Y, Wang Y. et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Allen KE, Weiss GJ. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther. 2010;9:3126–3136. doi: 10.1158/1535-7163.MCT-10-0397. [DOI] [PubMed] [Google Scholar]
- 244.Bai H, Xu R, Cao Z. et al. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011;585:402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 245.Park EY, Chang E, Lee EJ. et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 246.Yin J, Zheng G, Jia X. et al. A Bmi1-miRNAs cross-talk modulates chemotherapy response to 5-fluorouracil in breast cancer cells. PLoS One. 2013;8:e73268. doi: 10.1371/journal.pone.0073268. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 247.Xin H, Li X, Yang B. et al. Blood-based multiple-microRNA assay displays a better diagnostic performance than single-microRNA assay in the diagnosis of breast tumor. Tumour Biol. 2014;35:12635–12643. doi: 10.1007/s13277-014-2587-4. [DOI] [PubMed] [Google Scholar]