Abstract
Peter Wildy first observed genetic recombination between strains of HSV in 1955. At the time, knowledge of DNA repair mechanisms was limited, and it has only been in the last decade that particular DNA damage response (DDR) pathways have been examined in the context of viral infections. One of the first reports addressing the interaction between a cellular DDR protein and HSV-1 was the observation by Lees-Miller et al. that DNA-dependent protein kinase catalytic subunit levels were depleted in an ICP0-dependent manner during Herpes simplex virus 1 infection. Since then, there have been numerous reports describing the interactions between HSV infection and cellular DDR pathways. Due to space limitations, this review will focus predominantly on the most recent observations regarding how HSV navigates a potentially hostile environment to replicate its genome.
Keywords: C-NHEJ, classic nonhomologous end-joining, DDR, DNA damage response, Herpes simplex virus 1, homologous recombination, HR, HSV-1, intrinsic antiviral defense, microhomology-mediated end joining, MMEJ, single-strand annealing, SSA, virally encoded recombinase, virus–host interactions
HSV-1 is a ubiquitous human pathogen responsible for significant disease during acute infection. In addition, HSV-1 causes latent infections in sensory neurons for the life of the host with the potential for reactivation and recurrent disease. It is becoming increasingly clear that viruses like HSV have evolved complex interactions with their hosts. Because viruses rely on host cellular machinery during infection, they have evolved to usurp cellular processes. On the other hand, cells have intracellular antiviral defenses designed to fight viral infections. Thus, although HSV-1 may utilize some components of the DNA damage response machinery to replicate its genome, other components are antiviral, and HSV-1 has developed mechanisms to avoid antiviral restriction. The complex interaction between HSV-1 and the cell reflects an evolutionary tug of war in which cells have evolved antiviral mechanisms that are, in turn, counteracted by viral strategies that promote lytic infection. This review will focus on recent examples that demonstrate the intricate interactions between HSV-1 and host cell DNA damage response pathways.
The earliest stages of HSV-1 infection
HSV-1 has a large double-stranded linear DNA genome (152 kb), and viral DNA synthesis takes place in the infected cell nucleus in large globular domains called replication compartments. Replication compartments serve to concentrate and partition viral and cellular proteins that are required for productive infection. At the earliest stages of infection, however, cellular proteins are recruited to the vicinity of viral genomes in an attempt to thwart the infection. For instance, PML and other ND10 proteins form virus-induced PML-nuclear bodies (viPML-NB) that are associated with repression of vial gene expression (reviewed in [8]). ViPML-NBs are subsequently disrupted by the E3 ubiquitin ligase activity of ICP0 [9]. As described below, other cellular proteins, many of which also exert antiviral effects, are recruited to viral genomes including cellular histones as well as components of the DNA damage response pathways. HSV has evolved to counteract antiviral mechanisms primarily through the action of ICP0. Some components of the DNA damage response (DDR) may also be beneficial to viral infection, and in this review, we will discuss how HSV navigates this complex cellular environment to create conditions that are conducive to productive viral infection.
HSV-1 DNA replication is closely associated with recombination
The virus encodes seven essential replication proteins: the origin-binding protein (UL9), the single-strand DNA-binding protein (SSB; ICP8), the heterotrimeric helicase/primase (UL5/8/52), the polymerase (UL30) and the polymerase processivity factor (UL42). Replication occurs in a biphasic manner, beginning with an UL9-dependent phase and later switching to a mechanism that does not require UL9 [10,11]. The HSV genome contains three origins of replication: two copies of oriS and one oriL (reviewed in [12]). Current models suggest that together ICP8 and UL9 trigger the melting of one of these origins followed by recruitment of the helicase/primase complex and the HSV polymerase to carry out unwinding and elongation, respectively [12].
HSV-1 DNA replication produces concatemers, which are required for the generation of progeny; however, the mechanism by which they are formed is unclear. It has long been recognized that HSV-1 genomes undergo a high degree of recombination [1,13–19]. Although it has been proposed that the viral genome circularizes and rolling circle replication leads to the formation of concatemers, several lines of evidence suggest that HSV DNA replication is more complex. Controversy still remains over whether the incoming viral genome circularizes prior to replication [20,21]. HSV-1 replication proteins are able to catalyze rolling circle replication in vitro [22–25], but it has not been shown conclusively that rolling circle replication occurs during infection. Simple rolling circle replication does not explain the observation that genomic inversions occur as soon as viral DNA synthesis can be detected [15,26,27]. In addition, replication of the HSV-1 genome produces X and Y branched structures that can be visualized by electron microscopy and 2D gel electrophoresis [27,28]. These structures are reminiscent of recombination intermediates and suggest a more complex mode of replication. We have suggested that the HSV replication machinery promotes a unique form of DNA replication that utilizes a recombination-dependent mechanism to produce concatemers, which are required for packaging infectious virus [3,29].
The notion that HSV replication machinery promotes recombination-dependent replication is supported by experiments using HSV-1 as a helper virus to facilitate replication of other viruses and amplicons. For instance, replication of SV40 DNA by the six-core HSV-encoded replication factors and SV40 large T antigen produces concatemers composed of X-shaped DNA structures that may represent recombination intermediates [30]. Since SV40 replication normally produces two circular daughter molecules, it is noteworthy that the presence of HSV replication proteins can alter the mode of replication to generate complex concatemeric DNA [31]. In addition, adeno-associated virus (AAV) propagated using HSV as a helper virus produces high molecular weight forms of DNA that are not observed when adenovirus is used as a helper [32]. Thus, in the context of an HSV-1 infection, recombination may play a role in the generation of high molecular weight AAV concatemers that have a complex structure. Taken together, these data are consistent with the notion that the HSV replication machinery is inherently recombinogenic, giving rise to complex concatemeric DNA.
In addition to the core HSV replication machinery, we have identified a virus-encoded two-subunit recombinase that is reminiscent of the well-studied RedExo/β system of phage lambda [33,34]. The lambda RedExo/β recombinase has been shown to perform strand annealing reactions in vitro. [33,34]. In addition, RedExo/β and related recombinases from other bacteriophages have been shown to promote in vivo recombination-mediated genetic engineering using short homologies – ‘recombineering’ in bacteria (reviewed in [29]). The HSV recombinase comprises UL12, a 5′–3′ exonuclease, and ICP8, which in addition to its role as a single-strand DNA-binding protein (SSB) can also function as a single-strand DNA annealing protein (SSAP). UL12 and RedExo share conserved sequence elements, and both proteins interact with their partner SSAPs, ICP8 and Red-β, respectively (reviewed in [29]). The precise role of the UL12/ICP8 complex during infection remains unclear. We initially proposed that UL12 might be responsible for processing replication intermediates into a form suitable for encapsidation [35]; however, recent work has suggested that the viral recombinase may be involved at an earlier step of infection during DNA synthesis to influence the mode of replication itself [36]. Thus, UL12 may stimulate a pathway of recombination-dependent replication required to produce concatemers that can be packaged into infectious virus.
A role for cellular DDR proteins in viral DNA replication has been suggested based on the observation that several cellular factors involved in homologous recombination (HR) including MRE11, RAD50, NBS1 and RAD51 are recruited to viral prereplicative sites and replication compartments [3,37–41]. In addition, both ICP8 and UL12 have been shown to interact with many DDR proteins [39,42–44]; however, attempts to identify the precise roles played by these proteins in HSV DNA replication have not been straightforward. For instance, although HSV may take advantage of cellular components to promote viral DNA replication, many DDR pathways promote antiviral mechanisms such as silencing and the induction of innate immune signaling. Because many components of cellular DDR pathways have complex and overlapping roles, it has been difficult to tease apart the precise functions of cellular DDR pathways during infection. Furthermore, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), ATM and the MRN complex also participate in other cellular processes and may play roles in HSV replication that are distinct from their roles in DDR pathways.
Cellular DNA damage response pathways
In order to maintain its genetic integrity, the cell encodes a variety of mechanisms collectively termed the DDR. The pathway by which cellular DNA is repaired depends on multiple factors, including cell type and cell cycle, as well as the nature and severity of the DNA lesion. Certain pathways are only activated during S phase at replication forks. Some types of repair require a template strand, while others do not, and some types of damage can be repaired by several mechanisms. The cell can utilize either direct chemical reversal of damage or excision repair to replace damaged or mismatched bases in DNA. Excision repair requires a template strand and the removal of stretches of nucleotides or bases. This mechanism encompasses base excision repair (BER), nucleotide excision repair and mismatch repair (MMR). For double-strand breaks, the cell employs recombination-mediated repair. When DNA damage cannot be repaired by any of these strategies, the cell may utilize damage tolerance mechanisms, like translesion synthesis, postreplication gap filling or replication fork regression. These mechanisms do not repair the damaged DNA, per se, but allow replication to proceed, often resulting in mutations. If damage is too great, the biological response tips from repair/tolerance toward cell cycle arrest and apoptosis.
MMR proteins are required for efficient HSV-1 replication
MMR overview
The MMR pathway is a highly conserved mechanism that is responsible for detecting and repairing mismatched bases, as well as insertion-deletions loops (IDLs) that arise during DNA replication. Single-base mismatches and 1–2 base IDLs are recognized and bound by the MSH2/MSH6 heterodimer, while larger IDLs are bound by the MSH2/MSH3 heterodimer. The MLH1/PMS2 heterodimer is then recruited to help organize other MMR proteins, such as EXOI and RPA, onto mismatched DNA to facilitate resection and repair.
MMR & HSV
Both MSH2 and MLH1 are required for efficient replication of HSV-1 in normal human cells and are localized to viral replication compartments [43]. In addition, interactions have been reported between ICP8 and UL12 and MMR proteins MSH2, MSH3, and MSH6 [39,43]. Interestingly, however, these proteins may have functions in HSV infection that are distinct from their canonical roles in recognizing mismatched DNA. MLH1 is recruited to viral genomes at the earliest stages of viral infection, and depletion of MLH1 in the context of viral infection inhibits immediate early gene expression [43]. On the other hand, MSH2, which is generally thought to function before MLH1, is apparently recruited after MLH1 and may play a later role in HSV infection [43]. These results suggest that although both MLH1 and MSH2 are required for efficient HSV infection, MLH1 may play a role in viral infection that is independent of MSH2 and may be distinct from the MMR pathway. MLH1 was also shown to be a component of PML-NBs, although unlike other components of these nuclear bodies, it is not degraded by ICP0 [43]. It will be of considerable interest to further explore the precise roles of both MLH1 and MSH2 during HSV infection.
HSV-1 influences pathway choice for double-strand break repair
DSBR overview
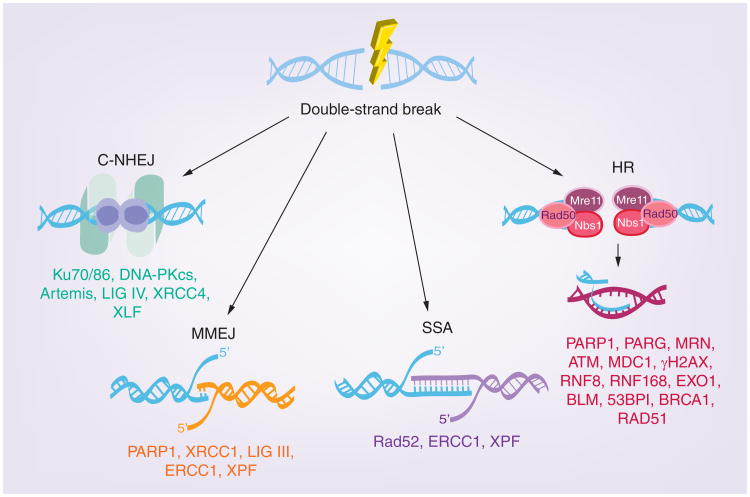
Probably the most well studied type of DNA repair is double-strand break repair (DSBR), which encompasses a variety of recombination-mediated pathways with distinct but overlapping functions. These mechanisms include: HR, single strand annealing (SSA), classic nonhomologous end joining (C-NHEJ), and microhomology-mediated end joining (MMEJ; Figure 1). The three pathways shown on the right side of Figure 1 (HR, SSA and MMEJ) require some degree of homology; whereas, C-NHEJ does not. The homology-driven pathways involve resection of DNA followed by annealing to a complementary strand. HR is generally more accurate than the other DSBR pathways, since HR employs strand invasion into a homologous chromosome or sister chromatid. SSA and MMEJ, on the other hand, are more error prone, often resulting in deletions or genomic translocations. Another type of homology driven repair, synthesis-dependent strand annealing (SDSA) occurs during DNA replication in response to stalled replication forks. C-NHEJ does not require homology and can directly fuse unrelated DNA molecules (reviewed in [45]). Despite the potential for generating errors, C-NHEJ is the preferred mechanism of repair in higher eukaryotes and can occur during G1, S and G2 phases of the cell cycle.
Figure 1. DNA double-strand break repair pathways.
In response to double-strand breaks, DNA can be repaired by one of four major pathways: C-NHEJ, MMEJ, SSA and HR. HR, SSA and MMEJ are homology-directed forms of repair; whereas, C-NHEJ does not require homology. Double-strand breaks arising at replication forks can also be repaired by synthesis-dependent strand annealing and break-induced repair mechanisms via ATR-CHK1 signaling (not shown in figure).
C-NHEJ: Classic nonhomologous end joining; HR: Homologous recombination;
MMEJ: Microhomology-mediated end joining; SSA: Single strand annealing.
DSBR & HSV
As shown in Figure 1, several cellular DDR mechanisms are available for recombination/repair during HSV-1 infection. In order to determine whether HSV utilizes one or more of these pathways during infection, chromosomally integrated GFP correction assays were used to measure the frequency of DSBR by C-NHEJ, MMEJ, HR and SSA in infected cells [36]. Using these assays, we have shown that HSV infection stimulates SSA; however, HR, C-NHEJ and MMEJ were inhibited [36]. These results suggest that HSV has evolved to utilize SSA, and in the following sections we will explore possible reasons and mechanisms for the apparent inhibition of the other three pathways in infected cells.
HSV-1 inhibits classic nonhomologous end joining
C-NHEJ overview
As stated above, classic nonhomologous end joining (C-NHEJ) involves the direct fusion of nonhomologous dsDNA ends. This process involves at least three steps: recognition of DSB, end trimming/processing of nonligatable termini and ligation. C-NHEJ is promoted by the Ku70/86 heterodimer, which recognizes DSBs and binds to dsDNA ends. Ku70/86 recruits the DNA-PKcs, which aligns DNA ends. End processing occurs if the DNA ends are not easily ligatable, containing either nonhomologous regions or unusual structures such as DNA hairpins. Cellular enzymes thought to play a role in the end-processing step include DNA-PKcs, Artemis, PNKP, TdT and polymerases (pols) λ and μ (reviewed in [46]). The ligation step of C-NHEJ requires XRCC4, which interacts with both LIG IV and DNA, and is thought to be necessary for LIG IV recruitment to the DSB. XRCC4 also interacts with XLF; however, the precise role of XLF in NHEJ is not yet known [47].
C-NHEJ & HSV
The HSV genome contains nicks and gaps that are randomly located and present on both strands. Because the HSV-1 genome has dsDNA ends in addition to nicks and gaps, it is tempting to speculate that one or more of the DDR pathways might be activated by the genome as soon as it enters the nucleus. In fact, cells transfected with viral DNA exhibit RPA32 S4/S8 phosphorylation, a mark specific for DNA-PK activation [48]; however, DNA-PK is not activated in HSV-infected cells. It has been recognized since 1996 that DNA-PKcs activity is attenuated in HSV-infected cells in an ICP0-dependent manner (Figure 2) [2,48,49], leading to the suggestion that components of the C-NHEJ pathway are antiviral. In fact, HSV-1 replication is more efficient in cells lacking the catalytic subunit of DNA-PKcs [49], and in Ku-deficient murine embryonic fibroblasts, viral yields are increased by almost 50-fold [39]. In addition, we have recently demonstrated that ICP0 is required to relieve suppression of HSV-1 DNA infectivity caused by DNA-PKcs [48]. Possible reasons for the antiviral properties of C-NHEJ will be discussed below.
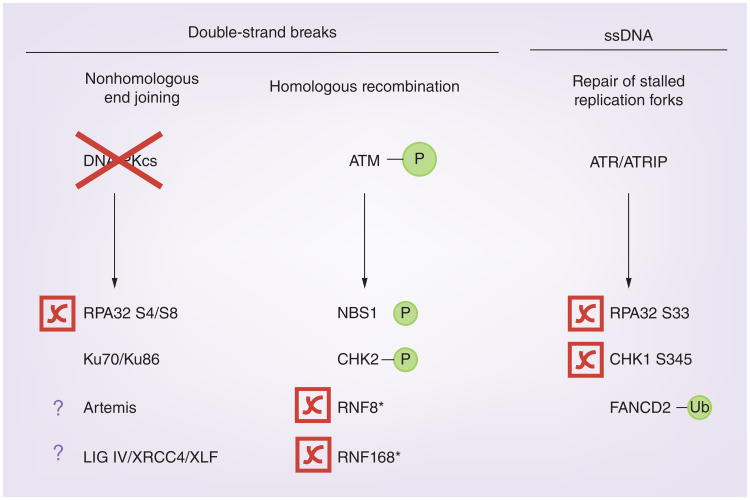
Figure 2. HSV-1 manipulates components of the cellular DNA damage response.
During infection, HSV-1 inhibits phosphorylation of RPA32 S4/S8 by degrading or inhibiting DNA-PKcs in an ICP0-dependent fashion [2,40,48]. Although ATM, NBS1 and CHK2 are phosphorylated during infection, the HR pathway is inhibited due to ICP0-dependent degradation of RNF8 and RNF168 [37,38,40,59]. ATR/ATRIP phosphorylation of CHK1 S345 and RPA32 S33 is inhibited; however, FANCD2 is activated by ubiquitination during HSV-1 infection [44,62,94]. Asterisk (*) indicates proteins that are degraded by ICP0.
DNA-PKcs: DNA-dependent protein kinase catalytic subunit; HR: Homologous recombination.
Homologous recombination components play positive & negative roles in HSV-1 infection
HR overview
HR is mediated by the PI3 kinase-like kinase, ATM, which is activated when the MRN complex (MRE11, RAD50 and NBS1) senses a double-strand break. ATM has numerous substrates, including the histone variant, H2AX, the phosphorylated form of which is called γH2AX [50]. The γH2AX signal can spread as far as one to two megabases from the initial site of damage in an ATM- and MDC1-dependent manner [51]. This extensive phosphorylation is responsible for the appearance of damage foci observed by immunofluorescence microscopy [52]. Additional downstream effectors are then recruited to damage foci in a sequential fashion following ubiquitination of H2A-type histones by RNF8 and RNF168 (reviewed in [53]). RNF8/RNF168-dependent ubiquitin conjugation is required to recruit and stabilize downstream repair proteins, such as BRCA1, 53BP1 and RAD51 ([54–56]; and reviewed in [53]). MRE11 and CtIP facilitate the initial end resection step, after which EXO1 and BLM carry out extensive resection [57,58]. Following end resection, ssDNA is coated by RPA, which is important for activation of the ATR pathway. RAD51 filaments assemble on RPA-coated DNA and facilitate strand invasion, resulting in a D-loop structure. During HR, this process results in Holliday junctions, which must be resolved to produce repaired, linear dsDNA. Strand invasion can also proceed via another mechanism, SDSA, which does not produce Holliday junctions (described below). Once the DSB has been repaired, DDR proteins dissociate from the DNA resulting in the resolution of damaged foci.
Manipulation of HR during HSV-1 infection
Although chromosomal integration assays suggested that HR is suppressed during HSV-1 infection [36], several components of the cellular HR machinery are required for efficient virus production. Virus production is deficient in cells lacking ATM, MRN (MRE11, NBS1, RAD50) and WRN [37,39,42]. In addition, we and others have demonstrated that HSV-1 induces ATM activation, as evidenced by the recruitment and phosphorylation of ATM, MDC1, NBS1 and CHK2 [37,38,40]. Lilley et al. demonstrated that damage foci containing γH2AX and MDC1 are still able to form in IR-treated HSV-infected cells [37]; however, BRCA1 and 53BP1 are not recruited to damage foci because RNF8 and RNF168 are degraded by ICP0 (Figure 2) [59]. Thus, although ATM is activated in HSV-infected cells, HR itself is inhibited, consistent with the observation that HR is inhibited in HSV-infected cells using the chromosomal reporter assay [36]. Interestingly, since HR proteins upstream of RNF8 and RNF168 are recruited to replication compartments and are required for efficient replication, it is possible that some of these components play positive roles during infection that are distinct from HR [37,39,42].
ATR-CHK1 signaling is disrupted in HSV-infected cells
ATR-CHK1 pathways
In uninfected cells, the checkpoint kinase, ATR, is activated in response to stretches of ssDNA adjacent to dsDNA, like those found at stalled replication forks. ATR is also activated by substrates produced during ATM-mediated end resection. Thus, ATM activation generally results in activation of the ATR pathway [60,61]. RPA is recruited to stretches of ssDNA, and recruits ATR and ATRIP. ATR signaling also requires the recruitment of the 9–1–1 (RAD9-RAD1-HUS1) checkpoint clamp, which in turn recruits the ATR-activator, TopBP1, which results in phosphorylation of CHK1 on S317 and S345 and RPA32 on S33.
ATR-CHK1 inhibition during HSV-1 infection
In uninfected cells, activation of ATM would be expected to result in the activation of ATR. Interestingly, ATR-CHK1 signaling is disrupted during HSV-1 infection even though HSV-1 DNA replication activates ATM signaling (Figure 2) [37,38,40,62]. ATR is not activated in HSV-infected cells, even if these cells are treated with hydroxyurea (HU), which causes replication fork stalling [62,63]. We have recently identified the mechanism by which HSV-1 inhibits ATR signaling: four replication proteins (ICP8 and the three components of the helicase/primase complex) can bind to substrates that contain ssDNA adjacent to dsDNA, which are similar to substrates recognized by RPA and ATR. Thus, these four viral proteins prevent the loading of the 9–1–1 complex and the subsequent recruitment of TopBP1, effectively disabling ATR signaling. It is still not clear why HSV has evolved to prevent ATR signaling. It is known that in uninfected cells, ATR signaling can stabilize stalled forks and prevent fork collapse in order to prevent the formation of DSB [64]. Although DSB formation would be deleterious for cells, it may be beneficial during HSV replication, perhaps by stimulating recombination. This is supported by the observation that artificial activation of ATR results in reduced recombination between coinfecting viruses [65]. Although HR is inhibited in infected cells, other mechanisms of recombination such as SSA may be stimulated under these conditions (discussed below). Despite the observation that ATR signaling is prevented in HSV-infected cells, ATR and several proteins in this pathway are recruited to viral replication compartments and are essential for efficient virus production [62,65]. These results suggest that ATR pathway proteins play positive roles in HSV infection that are distinct from ATR signaling. Alternatively, it may be beneficial for HSV-1 to recruit these proteins to viral genomes as a way to inhibit cellular DNA replication.
The Fanconi anemia pathway plays a positive role in HSV infection
FA pathway overview
In uninfected cells, the Fanconi anemia (FA) pathway is activated by ATR during S-phase in response to replication stress caused by stalled forks and interstrand cross-links (ICLs) [66,67]. The FA pathway has been shown to promote replication restart by coordinating homology-mediated repair (HR and SSA) and translesion synthesis [66,68,69], modulating MMR [70,71] and suppressing C-NHEJ [72]. The FA pathway is composed of at least 15 proteins, which are divided into three functional groups: the core complex, the ID complex and downstream effectors. Activation of the FA pathway requires monoubiquitination of the ID complex (FANCI and FANCD2) by the FA core complex (FANCA, B, C, E, F, G, L and M), which together with FAAP24 and FAAP100 form a multisubunit E3 ubiquitin ligase, and the E2-conjugating enzyme, UBE2T. FA activation is also dependent on phosphorylation of FANCI and FANCD2 by ATR [73,74].
The FA pathway & HSV-1 infection
Recently, the Mohr lab demonstrated that FA proteins are necessary for efficient HSV-1 replication and transcription and suggested that these proteins act as regulators of DNA repair pathway choice during infection [44]. HSV-1 potently activates the FA pathway, by monoubiquitination of FANCI-D2, which seems to require HSV pol and DNA replication (Figure 2) [44]. In addition, they demonstrated that FANCI interacts with ICP8, pol, UL42, UL12 and dUTPase and that FANCD2 interacts with the helicase subunit, UL5 [44]. Furthermore, they showed that HSV-1 replication was restricted in FA-deficient cells, and that this restriction was partially eliminated by treatment with the DNA-PKcs inhibitor, NU7441 [44]. These results suggest that the FA pathway may play a role in restricting DNA-PKcs activity during HSV-1 infection. Previous reports demonstrate that FA pathway proteins stimulate SSA. This is consistent with the notion that these proteins direct repair toward the SSA pathway while inhibiting C-NHEJ in HSV-infected cells.
SSA is stimulated in HSV-1 infected cells
SSA overview
SSA is a form of homology-mediated repair, although it is more error prone than HR, and can cause deletions and chromosomal translocations. SSA is initiated when a double-strand break occurs between two repeated sequences oriented in the same direction. Homologous regions of single-stranded DNA are exposed through extensive end resection by a 5′–3′ exonuclease. Annealing is facilitated by RAD52, which is an SSAP [75,76]. Following annealing, it is believed that nonhomologous 3′ overhangs are cleaved by ERCC1/XPF (reviewed in [77]). Green fluorescent protein (GFP) reporter assays have been used to identify the cellular components required for SSA, and interestingly, most appear to overlap with other DDR pathways [75,76,78]. For instance, the single-strand annealing protein Rad52 is also involved in assembly of Rad51 filaments during HR, and ERCC1/XPF are also implicated in nucleotide excision repair and MMEJ (reviewed in [77]).
HSV-1 stimulates SSA
GFP correction assays have demonstrated that HSV stimulates SSA, raising the question of whether SSA is carried out by viral or cellular proteins (or both). As mentioned above, HSV encodes a two-subunit recombinase (UL12 and ICP8). The alkaline nuclease (UL12) component of the HSV-1 recombinase is necessary and sufficient to stimulate SSA [36]. Stimulation of SSA was abrogated in the nuclease-dead mutant, UL12 D340E, suggesting that UL12 nuclease activity may be required for end-resection prior to SSA [36]. Direct involvement of ICP8 in this process has been difficult to demonstrate because ICP8 is also essential for DNA synthesis. Additional experimentation will be required to assess the possible involvement of cellular SSA proteins such as RAD52 and the ERCC1/XPF complex.
HSV-1 may also utilize MMEJ/SDSA to repair DSBs during replication
MMEJ & SDSA overview
Perhaps the least-understood DSBR mechanism is MMEJ. Like SSA, this process requires resection to expose regions of homology and annealing of homologous regions. Unlike SSA, however, MMEJ is thought to require a small amount of homology (5–25nt) for end joining to occur. Although MMEJ is not well defined, several proteins are thought to participate in this process, including: PARP1, XRCC1, ERCC1/XPF, LIG III and possibly the MRN complex [79–84]. Interestingly, MMEJ has been shown to have an important role in the repair of DSBs at collapsed replication forks [85] and may play a role in SDSA (reviewed in [86]).
A model for A-NHEJ/SDSA during HSV-1 infection
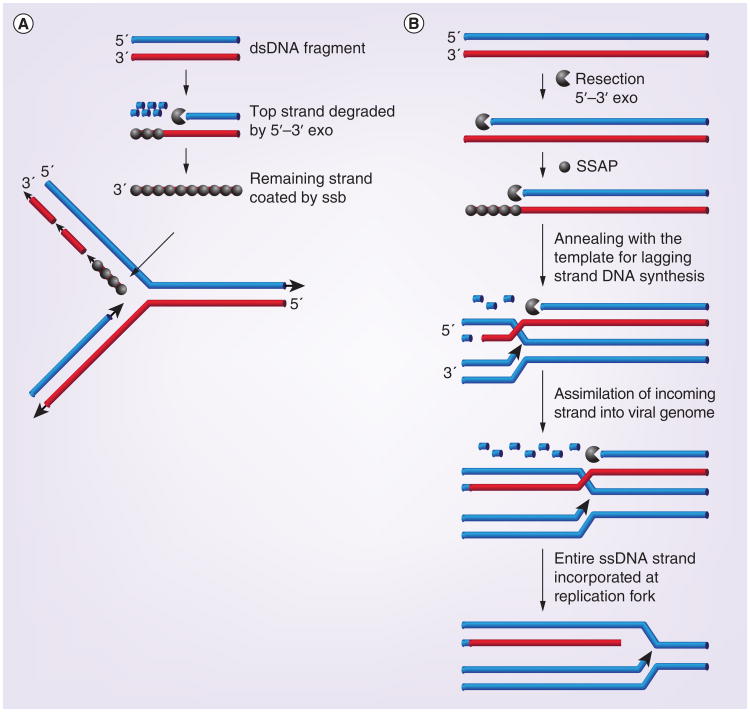
As stated above, it is clear that recombination is tightly linked to DNA replication during HSV-1 infection. Thus, in addition to classic SSA, it is possible that the HSV-1 may use an SDSA mechanism (reviewed in [29]). SDSA utilizes strand exchange, in which the 5′ end of dsDNA break is resected, and the 3′ end is annealed to a homologous region at a single-strand region of a growing replication fork (reviewed in [86]). Interestingly, the recombineering machinery employed by the lambda RedExo/β and other bacteriophage-encoded recombination systems such as RecE/T are also dependent on DNA synthesis (reviewed in [29]). Recombineering can be performed with either single- or double-stranded oligonucleotides and is used to efficiently incorporate mutations into bacterial genomes. Single-strand oligonucleotides are coated by an SSAP such as lambda Redß or RecT and inserted at the DNA replication fork. When double-stranded oligonucleotides are used, the exonuclease degrades one entire strand. In the model shown in Figure 3A, the remaining strand is coated with the SSAP and incorporated on the lagging strand template (reviewed in [29]). A more elaborate situation has been proposed to occur during replication of viral genomes (Figure 3B) [87]. According to this scenario, a DSB on one viral genome is concurrently resected and annealed at the lagging strand template. This model was originally proposed by Kuzminov ([88]; and reviewed [29]). Additional experiments will be required to determine whether HSV utilizes this type of synthesis-dependent annealing reaction during replication; however, it is known that the HSV-1 recombinase, UL12/ICP8, is capable of performing strand exchange in vitro and that recombination is linked to DNA synthesis [34].
Figure 3. Possible synthesis-dependent strand annealing strategies for HSV-1 recombination-dependent replication.
(A) Step-wise model for strand annealing. One entire strand of a dsDNA molecule is resected by exonuclease and the remaining strand is coated with SSAP prior to incorporation at the lagging strand of a replication fork. (B) Concerted model for strand annealing. Resection of one strand of a long dsDNA molecule occurs concomitantly with strand annealing, resulting in incorporation of the entire strand into the lagging strand at a replication fork. This model was originally suggested by Kuzminov [88].
Adapted with permission from [29].
PARP/PARG may play positive & negative roles during HSV-1 infection
PARP overview
PARP proteins utilize NAD+ to catalyze the covalent attachment of poly-ADP ribose (PAR) chains to proteins. Although there are 17 members of the PARP family, PARP-1 is responsible for nearly all PARylation that takes place in the cell, and only PARP-2 has been shown to complement a PARP-1 mutant [89]. Interestingly, PARP-1 plays both pro- and anti-recombinogenic roles and regulates many DDR pathways. In addition to its role in MMEJ, PARP-1 also plays a crucial role in recovery from replication fork stalling through stimulation of HR. PARP-1 senses and is recruited to nicks, ssDNA breaks and dsDNA breaks and PARylates itself and other proteins. The PAR post-translational modification is thought to control the activity and function of several DDR proteins such as MRE11, NBS1 and DNA-PKcs. Thus, PARP-1 plays an important role in chromatin remodeling and the recruitment and regulation of cellular DDR proteins. On the other hand, excess DNA damage can result in overactivation of PARP and lead to cell death. The enzyme, PARG,, catalyzes the removal of PAR chains and is required to prevent cell death and promote replication restart via HR [90,91]. In the absence of PARG, PAR chains are thought to accumulate on DNA, preventing RPA binding and as a result, RPA32 phosphorylation by DNA-PKcs is inhibited [91]. Thus, the PARP/PARG balance plays an important role in modulating the DNA damage response.
PARP/PARG & HSV
A study to examine metabolic changes in HSV-infected cells revealed that PARP is activated during infection. Vastag et al. reported that NAD+ levels are decreased during HSV-1 infection [92], and PARylation carried out by activated PARP1/2 during HSV-1 infection was found to be responsible for the observed NAD+ depletion [93]. Since PARP1/2 sense nicks and ssDNA breaks, it is possible that nicks and gaps in the viral genome are responsible for PARP activation. Whether nicks and gaps in viral genomes are repaired during the early stages of infection by DDR machinery is not known; however, PARP binding and the subsequent recruitment of repair proteins may represent an attempt to fill in gaps. Interestingly, PARG is degraded by ICP0 during HSV-1 infection [93]. Thus, it is possible that although PARP is activated by infection, the downstream DNA repair processes facilitated by PARG are prevented by ICP0. This may be a mechanism by which HSV counteracts the antiviral activity of some DDR pathways such as C-NHEJ. Although ICP0 degrades DNA-PKcs in some cells, the activity of DNA-PKcs is inhibited even in Vero cells in which DNA-PKcs are not degraded [40,48,94]. As mentioned above, in the absence of PARG, DNA-PKcs activity is inhibited. The degradation of PARG by ICP0 may thus contribute to the inhibition of DNA-PKcs, implying that HSV has evolved more than one mechanism to inhibit DNA-PKcs and C-NHEJ. We are intrigued by the possibility that gap filling facilitated by PARP/PARG and circularization by C-NHEJ are antiviral and contribute to genome silencing. The ability of ICP0 to degrade both DNA-PKcs and PARG may be a means by which HSV prevents circularization, consistent with the demonstration by Jackson and Deluca that in the presence of ICP0, circularization of the viral genome is prevented [20]. Further experimentation will be required to test this model and address the controversy over whether circularization occurs during lytic infection.
Some DDR proteins function as DNA sensors in intrinsic & innate immune responses
DDR & antiviral defense
When HSV-1 infects a cell, the viral genome is released from the capsid into the host cell nucleus, which may be an intrinsically hostile environment for invading pathogens. Cellular defense strategies include three inter-related arms: intrinsic antiviral mechanisms, innate immune signaling and the adaptive immune responses. We are intrigued by the observation that DNA damage-sensing proteins are able to sense ‘foreign’ DNA and trigger various types of antiviral responses. It is tempting to speculate that these responses are part of a larger network of antiviral defense mechanisms and that some DDR pathways may have evolved initially to counteract environmental pathogens such as viruses.
Intrinsic antiviral mechanisms
Intrinsic antiviral proteins are cellular factors that are constitutively expressed and poised to inhibit infection immediately following viral entry [95]. The first recognized intrinsic factors target retroviruses, and the number of retroviral defense factors has now grown to include Fv-1, TRIM5α, APOBEC3G and SAMHD1. More recently, it has been recognized that nuclear factors such as PML (TRIM19) exert antiviral effects that target herpesviruses, especially HSV and HCMV [96,97]. As described above, incoming viral genomes recruit PML into viPML-NBs that have been associated with repression of viral gene expression [96]. Other proteins that are recruited to viral genomes at the earliest stages of infection may also be part of the intrinsic antiviral network. For example, DNA-PKcs may modulate transcription of viral genes through several mechanisms. It has been reported to modulate RNAP II activity and inhibit the ability of RNAP II to bypass DSBs [98,99]. Alternatively, if DNA-PKcs and Ku proteins are recruited to viral DNA ends, they might be expected to activate C-NHEJ resulting in the circularization of viral genomes and the subsequent promotion of chromatinization and epigenetic silencing [2,100]. Activation of RNF8 and RNF168 and the ubiquitination of H2A has also been suggested to cause genome silencing [59]. In addition, IFI16 is a DNA sensor involved in innate IRF-3-mediated signaling as well as intrinsic antiviral responses. IFI16 has also been shown to sense microbial DNA and promote epigenetic silencing of DNA lacking or loosely associated with chromatin, such as HSV-1 genomes [101]. Thus, the ability of various cellular components, including DDR proteins, to silence viral gene expression appears to be a common mechanism by which cells have evolved to counteract viral infections. The ability of ICP0 to degrade PML, DNA-PKcs, RNF8/168 and IFI16 demonstrates that HSV has evolved to evade intrinsically antiviral silencing mechanisms.
DDR proteins may act as DNA sensors that trigger innate immune responses
In addition to intrinsic defenses such as repression of viral gene expression, cells have elaborate signaling mechanisms to trigger innate and adaptive immune responses to viral infection. Viral nucleic acids are the predominant pathogen-associated molecular patterns produced during viral infection and are recognized by pattern recognition receptors. It has been suggested that the cell distinguishes between viral and endogenous nucleic acids based in part on their cellular compartmentalization and chemical differences in the DNA itself. For example, DNA that is present in a compartment other than the nucleus, such as in endosomes or in the cytoplasm, may be identified as foreign. Cytosolic sensors of viral/microbial DNA include: DNA-dependent activator of IFN (DAI), RNA polymerase III, PHYIN family proteins (such as IFI16 and AIM2), DExD/H-box helicases (like RIG-I), DNA-PKcs, cGAS and STING (reviewed in [102]). On the other hand, in the nucleus the cell relies on chemical differences in DNA to distinguish between foreign and endogenous DNA. For example, unmethylated CpG DNA is chemically distinct from cellular DNA and will flag viral DNA as a pathogen-associated molecular pattern [103].
In addition to the possible roles in intrinsic antiviral mechanisms described above, it appears that DNA-PKcs may also stimulate innate immune signaling. DNA-PKcs has been reported to sense and respond to DNA and to induce transcription of Cxcl10, IL-6 and IFN-β as part of the IRF-3 innate immune response [104]. In that report, Ferguson et al. also demonstrated that HSV-1 infection stimulates IL-6 transcription in MEFs and that this stimulation is partially relieved in DNA-PKcs knockout cells (Prkdc-/- MEFs). It is thus possible that DNA-PK exerts its antiviral effects by several different mechanisms including initiating an innate signaling cascade. Recent studies have demonstrated that IFI16 also may play roles in both the intrinsic and innate antiviral responses [105–107]. HSV-1 triggers IRF-3 induction and activation of the inflammasome in human fibroblasts in an IFI16-dependent manner [106,108]. ICP0 degrades IFI16 as part of the immune-evasion strategy of HSV-1 [106].
The future outlook of HSV-1 & cellular DDR research
Can drugs that target DDR be used as antiviral treatments?
HSV-1 is a ubiquitous pathogen that infects approximately 95% of the human population and causes oral and genital lesions; and ocular infections, like herpes keratitis, that can cause visual impairment. In immunocompromised individuals, HSV infection is far more serious, causing severe illness and even mortality. Resistance to frontline herpes antiviral drugs, such as acyclovir, occurs at low frequency in immunocompetent individuals and is more common in immunocompromised patients [109,110]. Thus, there is a significant need for additional therapeutic strategies to supplement traditional nucleoside analog drug treatments and limit drug resistance caused by overuse.
An exciting potential for novel therapeutics is emerging as a result of our growing understanding of the role viral and cellular recombination/repair proteins play in HSV-1 replication. Since many viral and cellular proteins associated with HSV-1 recombination/repair are essential for viral growth, they can be considered as potential targets for new therapies, for example, Yan et al. recently demonstrated that the HIV integrase inhibitor, XZ45, was capable of blocking α, β and γ herpesvirus replication [111]. In the presence of XZ45, recombination between two mutant HSV-1 viruses was significantly inhibited perhaps by targeting ICP8 [111]. Cellular DDR proteins are also possible candidates for treatment of acute HSV-1 infection. For example, the ATM inhibitor, KU-55933 has recently been shown to suppress HSV-1 replication and cytopathic effects in herpes keratitis [112]. The observation that several DDR proteins are important antiviral modulators may also be exploited in the future for novel antiviral strategies. In addition, the finding that the viral recombinase is important in pathway choice needed for productive infection suggests that UL12 or ICP8 may also be exploited for antiviral therapy.
Conclusion & future perspective
In this review, we have described several examples of DDR pathways that are manipulated by HSV infection. DNA sensors such as DNA-PKcs and IFI16 appear to play overlapping roles in intrinsic and innate antiviral responses; however, it is likely that researchers have only scratched the surface regarding the identification of cellular proteins that can trigger antiviral responses. We predict that additional cellular factors that exert antiviral effects by sensing foreign DNA will be identified in the future.
We have also explored how HSV has evolved an unusual mechanism by which to replicate its genome. If filling in nicks and gaps and circularization are antiviral as we have suggested, it is possible that recombination-dependent replication pathways using SSA and SDSA provide a mechanism that evades these antiviral mechanisms and produces DNA concatemers that can be packaged into infectious virus. It is our hope that, within the next 5–10 years, we will gain a better understanding of the mechanism of HSV-1 replication and how it has evolved to evade cellular antiviral strategies. We are struck by the possibility that linear DNA viruses from bacteria, protozoa, yeast, mammals and insects that replicate through concatemeric DNA may all encode two subunit recombinases similar to UL12 and ICP8 [113,114]. The evolutionary conservation between the recombinases from these different DNA viruses suggests that they have evolved replication strategies that are distinct from cellular replication mechanisms and utilize an unusual form of recombination-dependent DNA replication. In the case of HSV, which has coevolved with its mammalian host, there has also been evolutionary pressure to evade intrinsic and innate antiviral mechanisms.
Executive Summary.
HSV-1 DNA replication is closely associated with recombination.
Mismatch repair proteins are required for efficient HSV-1 replication.
- HSV-1 influences pathway choice for double-strand break repair:
- –HSV-1 inhibits classic nonhomologous end joining;
- –Homologous recombination components play positive and negative roles in HSV-1 infection;
- –ATR-Chk1 signaling is disrupted in HSV-infected cells;
- –The Fanconi anemia pathway plays a positive role in HSV infection;
- –Single-strand annealing is stimulated in HSV-1 infected cells;
- –HSV-1 may utilize microhomology-mediated end joining/synthesis-dependent strand annealing during replication;
- –PARP/PARG may play positive and negative roles during HSV-1 infection;
Some DDR proteins function as DNA sensors in intrinsic and innate immune responses.
Future outlook: viral recombination may be a potential target for the development of second-line antiviral drugs.
Acknowledgments
The authors would like to thank the members of their laboratory for scholarly discussion and suggestions on the manuscript. They also thank KN Mohni for helpful comments and J Sawitzke for help with the design of Figure 3.
Financial & competing interests disclosure: This project was supported by National Institutes of Health grants AI069136 and AI021747, awarded to SK Weller. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Wildy P. Recombination with herpes simplex virus. J Gen Microbiol. 1955;13(2):346–360. doi: 10.1099/00221287-13-2-346. [DOI] [PubMed] [Google Scholar]
- 2.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70(11):7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55(8):451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 4.Everett RD. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol. 2006;8(3):365–374. doi: 10.1111/j.1462-5822.2005.00677.x. [DOI] [PubMed] [Google Scholar]
- 5.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 7.Weller SK. Herpes simplex virus reorganizes the cellular DNA repair and protein quality control machinery. PLoS Pathog. 2010;6(11):e1001105. doi: 10.1371/journal.ppat.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20(49):7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 9.Everett RD, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73(1):417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blümel J, Matz B. Thermosensitive UL9 gene function is required for early stages of herpes simplex virus type 1 DNA synthesis. J Gen Virol. 1995;76(Pt 12):3119–3124. doi: 10.1099/0022-1317-76-12-3119. [DOI] [PubMed] [Google Scholar]
- 11.Schildgen O, Graper S, Blumel J, Matz B. Genome replication and progeny virion production of herpes simplex virus type 1 mutants with temperature-sensitive lesions in the origin-binding protein. J Virol. 2005;79(11):7273–7278. doi: 10.1128/JVI.79.11.7273-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weller SK, Coen DM. Herpes simplex viruses:mechanisms of DNA replication. Cold Spring Harb Perspect Biol. 2012;4(9):a013011. doi: 10.1101/cshperspect.a013011. • Reviews current models for HSV-1 replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delius H, Clements JB. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- 14.Hayward GS, Jacob RJ, Wadsworth SC, Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci USA. 1975;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202(2):530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 16.Brown SM, Ritchie DA, Subak-Sharpe JH. Genetic studies with herpes simplex virus type 1 The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 17.Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- 18.Dutch RE, Bianchi V, Lehman IR. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J Virol. 1995;69(5):3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu X, Wang H, Zhang X. High-frequency intermolecular homologous recombination during herpes simplex virus-mediated plasmid DNA replication. J Virol. 2002;76(12):5866–5874. doi: 10.1128/JVI.76.12.5866-5874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson SA, DeLuca NA. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc Natl Acad Sci USA. 2003;100(13):7871–7876. doi: 10.1073/pnas.1230643100. • Demonstrates that circularization of the HSV-1 genome does not occur in cells infected with wild-type virus; however, circularization can be detected in cells infected with an ICP0 mutant virus. This result suggests that ICP0 inhibits circularization of the HSV-1 genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strang BL, Stow ND. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J Virol. 2005;79(19):12487–12494. doi: 10.1128/JVI.79.19.12487-12494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkenberg M, Lehman IR, Elias P. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc Natl Acad Sci USA. 2000;97(8):3896–3900. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves-Woodward KL, Gottlieb J, Challberg MD, Weller SK. Biochemical analyses of mutations in the HSV-1 helicase-primase that alter ATP hydrolysis, DNA unwinding, and coupling between hydrolysis and unwinding. J Biol Chem. 1997;272(7):4623–4630. doi: 10.1074/jbc.272.7.4623. [DOI] [PubMed] [Google Scholar]
- 24.Skaliter R, Lehman IR. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc Natl Acad Sci USA. 1994;91(22):10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skaliter R, Makhov AM, Griffth JD, Lehman IR. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected human cells. J Virol. 1996;70(2):1132–1136. doi: 10.1128/jvi.70.2.1132-1136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamberti C, Weller SK. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226(2):403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 27.Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70(5):3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weller SK, Sawitzke JA. Recombination promoted by DNA viruses: phage lambda to herpes simplex virus. Annu Rev Microbiol. 2014;68:237–258. doi: 10.1146/annurev-micro-091313-103424. •• Explores recombination-dependent replication employed by HSV-1 and the phage λ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blumel J, Graper S, Matz B. Structure of simian virus 40 DNA replicated by herpes simplex virus type 1. Virology. 2000;276(2):445–454. doi: 10.1006/viro.2000.0574. [DOI] [PubMed] [Google Scholar]
- 31.Matz B. Herpes simplex virus infection generates large tandemly reiterated simian virus 40 DNA molecules in a transformed hamster cell line. J Virol. 1987;61(5):1427–1434. doi: 10.1128/jvi.61.5.1427-1434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolas A, Alazard-Dany N, Biollay C, et al. Identification of rep-associated factors in herpes simplex virus type 1-induced adeno-associated virus type 2 replication compartments. J Virol. 2010;84(17):8871–8887. doi: 10.1128/JVI.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs invasion in phage lambda recombination. Genetics. 1997;147(3):961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuven NB, Willcox S, Griffith JD, Weller SK. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J Mol Biol. 2004;342(1):57–71. doi: 10.1016/j.jmb.2004.07.012. • Demonstrates that UL12 and ICP8 form a two-subunit recombinase that can perform strand exchange in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein JN, Weller SK. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244(2):442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- 36.Schumacher AJ, Mohni KN, Kan Y, Hendrickson EA, Stark JM, Weller SK. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012;8(8):e1002862. doi: 10.1371/journal.ppat.1002862. •• Demonstrates that HSV-1 stimulates single-strand annealing and inhibits homologous recombination and nonhomologous end joining. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci USA. 2005;102(16):5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirata N, Kudoh A, Daikoku T, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280(34):30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 39.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol. 2004;78(11):5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78(9):4783–4796. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory DA, Bachenheimer SL. Characterization of mre11 loss following HSV-1 infection. Virology. 2008;373(1):124–136. doi: 10.1016/j.virol.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balasubramanian N, Bai P, Buchek G, Korza G, Weller SK. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J Virol. 2010;84(24):12504–12514. doi: 10.1128/JVI.01506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohni KN, Mastrocola AS, Bai P, Weller SK, Heinen CD. DNA mismatch repair proteins are required for efficient herpes simplex virus 1 replication. J Virol. 2011;85(23):12241–12253. doi: 10.1128/JVI.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karttunen H, Savas JN, McKinney C, et al. Co-opting the Fanconi anemia genomic stability pathway enables herpesvirus DNA synthesis and productive growth. Mol Cell. 2014;55(1):111–122. doi: 10.1016/j.molcel.2014.05.020. •• Demonstrates the novel discovery that HSV-1 interacts with and utilizes components of the Fanconi anemia pathway for efficient replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dueva R, Iliakis G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl Cancer Res. 2013;2:163–177. [Google Scholar]
- 46.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011;711(1–2):73–86. doi: 10.1016/j.mrfmmm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124(2):301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 48.Smith S, Reuven N, Mohni KN, Schumacher AJ, Weller SK. Structure of the herpes simplex virus 1 genome: manipulation of nicks and gaps can abrogate infectivity and alter the cellular DNA damage response. J Virol. 2014;88(17):10146–10156. doi: 10.1128/JVI.01723-14. •• Shows that the viral genome can activate DNA-dependent protein kinase catalytic subunit in the absence of viral proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73(1):650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 51.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Fernandez-Capetillo O, Celeste A, Nussenzweig A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle. 2003;2(5):426–427. [PubMed] [Google Scholar]
- 53.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49(5):795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Huen MS, Grant R, Manke I, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mailand N, Bekker-Jensen S, Faustrup H, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 56.Stewart GS, Panier S, Townsend K, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 57.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85(4):509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 58.Bolderson E, Tomimatsu N, Richard DJ, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucl Acids Res. 2010;38(6):1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lilley CE, Chaurushiya MS, Boutell C, et al. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29(5):943–955. doi: 10.1038/emboj.2009.400. • Identifies RNF8 and RNF168 as targets for degradation by ICP0 during HSV-1 infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks Nat. Cell Biol. 2006;8(1):37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 61.Sartori AA, Lukas C, Coates J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohni KN, Livingston CM, Cortez D, Weller SK. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J Virol. 2010;84(23):12152–12164. doi: 10.1128/JVI.01643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gordin M, Olshevsky U, Rosenkranz HS, Becker Y. Studies on herpes simplex virus DNA: denaturation properties. Virology. 1973;55(1):280–284. doi: 10.1016/s0042-6822(73)81031-1. [DOI] [PubMed] [Google Scholar]
- 64.Peasland A, Wang LZ, Rowling E, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105(3):372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohni KN, Dee AR, Smith S, Schumacher AJ, Weller SK. Efficient herpes simplex virus 1 replication requires cellular ATR pathway proteins. J Virol. 2013;87(1):531–542. doi: 10.1128/JVI.02504-12. •• Identifies certain components of the ATR pathway that contribute to efficient HSV-1 replication. Also identifies the mechanism by which HSV-1 inhibits ATR-CHK1 signaling during infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shigechi T, Tomida J, Sato K, et al. ATR-ATRIP kinase complex triggers activation of the Fanconi anemia DNA repair pathway. Cancer Res. 2012;72(5):1149–1156. doi: 10.1158/0008-5472.CAN-11-2904. [DOI] [PubMed] [Google Scholar]
- 67.Tomida J, Itaya A, Shigechi T, et al. A novel interplay between the Fanconi anemia core complex and ATR-ATRIP kinase during DNA cross-link repair. Nucl Acids Res. 2013;41(14):6930–6941. doi: 10.1093/nar/gkt467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakanishi K, Yang YG, Pierce AJ, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102(4):1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhi G, Wilson JB, Chen X, et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009;69(22):8775–8783. doi: 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng M, Xie J, Ucher A, Stavnezer J, Cantor SB. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 2014;33(15):1698–1712. doi: 10.15252/embj.201387530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adamo A, Collis SJ, Adelman CA, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39(1):25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 73.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18(16):1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26(18):7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA. 2002;99(21):13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24(21):9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amsterdam) 2013;12(8):620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucl Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 81.Cheng Q, Barboule N, Frit P, et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucl Acids Res. 2011;39(22):9605–9619. doi: 10.1093/nar/gkr656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16(8):819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 83.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16(8):814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem. 2009;284(44):30565–30573. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Truong LN, Li Y, Shi LZ, et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci USA. 2013;110(19):7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ottaviani D, LeCain M, Sheer D. The role of microhomology in genomic structural variation. Trends Genet. 2014;30(3):85–94. doi: 10.1016/j.tig.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 87.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in lambda red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63(4):751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amsterdam) 2004;3(8–9):1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 90.Min W, Cortes U, Herceg Z, Tong WM, Wang ZQ. Deletion of the nuclear isoform of poly(ADP-ribose) glycohydrolase (PARG) reveals its function in DNA repair, genomic stability and tumorigenesis. Carcinogenesis. 2010;31(12):2058–2065. doi: 10.1093/carcin/bgq205. [DOI] [PubMed] [Google Scholar]
- 91.Illuzzi G, Fouquerel E, Ame JC, et al. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucl Acids Res. 2014;42(12):7776–7792. doi: 10.1093/nar/gku505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7(7):e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grady SL, Hwang J, Vastag L, Rabinowitz JD, Shenk T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J Virol. 2012;86(15):8259–8268. doi: 10.1128/JVI.00495-12. • Identifies PARG as a target for degradation by ICP0 during HSV-1 infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mohni KN, Smith S, Dee AR, Schumacher AJ, Weller SK. Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling. PLoS Pathog. 2013;9(10):e1003652. doi: 10.1371/journal.ppat.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5(11):1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 96.Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94(Pt 3):465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 97.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80(16):8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maldonado E, Shiekhattar R, Sheldon M, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381(6577):86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 99.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19(3):276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 100.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 101.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci USA. 2013;110(47):E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Orzalli MH, Knipe DM. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. •• Excellent review of the mechanisms by which mammalian cells sense foreign DNAs to trigger antimicrobial responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Unterholzner L. The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218(11):1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 104.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. • Identifies a novel role for DNA-dependent protein kinase catalytic subunit as a DNA sensor for the innate immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Unterholzner L, Keating SE, Baran M, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109(44):E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Z, Yamauchi Y, Kamakura M, et al. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J Virol. 2012;86(1):492–503. doi: 10.1128/JVI.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87(9):5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bacon TH, Levin MJ, Leary JJ, Sarisky RT, Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin Microbiol Rev. 2003;16(1):114–128. doi: 10.1128/CMR.16.1.114-128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Velzen M, Missotten T, van Loenen FB, et al. Acyclovir-resistant herpes simplex virus type 1 in intra-ocular fluid samples of herpetic uveitis patients. J Clin Virol. 2013;57(3):215–221. doi: 10.1016/j.jcv.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 111.Yan Z, Bryant KF, Gregory SM, et al. HIV integrase inhibitors block replication of alpha-, beta-, and gammaherpesviruses. MBio. 2014;5(4):e01318–e01314. doi: 10.1128/mBio.01318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alekseev O, Donovan K, Azizkhan-Clifford J. Inhibition of ataxia telangiectasia mutated (ATM) kinase suppresses herpes simplex virus type 1 (HSV-1) keratitis. Invest Ophthalmol Vis Sci. 2014;55(2):706–715. doi: 10.1167/iovs.13-13461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Myers RS, Rudd KE. Mining DNA sequences for molecular enzymology: the Red alpha superfamily defines a set of recombination nucleases. In: Ahmed F, editor. Proceedings of the 1998 Miami Nature Biotechnology Winter Symposium. Oxford; United Kingdom: 1998. pp. 49–50. [Google Scholar]
- 114.Lo Piano A, Martinez-Jimenez MI, Zecchi L, Ayora S. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011;160(1–2):1–14. doi: 10.1016/j.virusres.2011.06.009. [DOI] [PubMed] [Google Scholar]