Abstract
Cytosolic isocitrate dehydrogenase 1 (IDH1) with an R132H mutation in brain tumors loses its enzymatic activity for catalyzing isocitrate to α-ketoglutarate (α-KG) and acquires new activity whereby it converts α-KG to 2-hydroxyglutarate. The IDH1 mutation induces down-regulation of tricarboxylic acid cycle intermediates and up-regulation of lipid metabolism. Sterol regulatory element-binding proteins (SREBPs) regulate not only the synthesis of cholesterol and fatty acids but also acyclin-dependent kinase inhibitor p21 that halts the cell cycle at G1. Here we show that SREBPs were up-regulated in U87 human glioblastoma cells transfected with an IDH1R132H-expression plasmid. Small interfering ribonucleic acid (siRNA) for SREBP1 specifically decreased p21 messenger RNA (mRNA) levels independent of the p53 pathway. In IDH1R132H-expressing U87 cells, phosphorylation of Retinoblastoma (Rb) protein also decreased. We propose that metabolic changes induced by the IDH1 mutation enhance p21 expression via SREBP1 and inhibit phosphorylation of Rb, which slows progressionof the cell cycle and may be associated with non-aggressive features of gliomas with an IDH1 mutation.
Keywords: isocitrate dehydrogenase 1 (IDH1) mutation, sterol regulatory element-binding proteins (SREBP), p21, lipid metabolism, tricarboxylic acid (TCA) cycle
Introduction
Malignant gliomas are the most frequent and lethal cancers originating in the central nervous system. The most biologically aggressive subtype is glioblastoma (World Health Organization Grade IV). The current standard therapy for glioblastoma is surgical resection followed by adjuvant radiation therapy and administration of an alkylating agent temozolomide, and produces a median survival of only 14.6 months.21)
Genomic analysis of gliomas has identified mutations in the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) genes.16,25) IDH1 and IDH2 convert isocitrate to α-ketoglutarate (α-KG) producing nicotinamide adenine dinucleotide phosphate. A mutation affecting codon 132 of the IDH1 gene, located on chromosome 2q22, has been found in 12% of glioblastomas,16) resulting in an Arg to His substitution. Notably, IDH mutations are detected in 95% of secondary gliomas (diffuse astrocytoma, oligodendroglioma, oligoastrocytoma, anaplastic astrocytoma, anaplastic oligodendroglioma, and anaplastic oligoastrocytoma.25) The presence of the IDH1 mutation has been shown to be associated with a significantly better prognosis (IDH1 mutation versus wild IDH1; 31 months versus 15 months).
IDH1R132H shows decreased enzymatic activity for isocitrate, leading to lower α-KG production. However, it has an altered enzymatic activity and uses α-KG as a substrate to synthesize 2-hydroxyglutarate (2-HG).6,9,10,26,27) It has been purposed that 2-HG can competitively inhibit α-keto acid transaminase.18) In addition, levels of tricarboxylic acid (TCA) cycle intermediates were down-regulated and lipid metabolism was up-regulated in mutant IDH1-expressing cells. The IDH mutation has also been related to the methylation status of CpG sites, in particular hypermethylation of the methyl guanine methyl transferase (MGMT) promoter.3,19) Although the mechanism for a favorable prognosis in IDHR132H gliomas has not been fully elucidated, these altered metabolism and methylation statuses are thought to be associated with nonaggressive features.
The sterol regulatory element-binding protein (SREBP) family of transcription factors controls lipid metabolism. SREBP1 consists of SREBP1a and SREBP1c.7) SREBP1a and SREBP1c are alternate transcripts from a single gene that differ in the first exon.20) SREBP1c controls the expression of enzymes involved in the synthesis of fatty acids and triglycerides in lipogenic organs. Meanwhile, SREBP1a is highly expressed in cells, which enhances a wide range of enzymes for the synthesis of fatty acids, cholesterol, and phospholipids. SREBP2 regulates cholesterol synthesis. Interestingly, cyclin-dependent kinase inhibitor p21, which halts the cell cycle at the G1 stage, is regulated by SREBP. SREBP1a activates the p21 promoter as strongly as p53, a tumor suppressor positively regulating p21. Increased expression of SREBP1a activates p21 expression, resulting in cell growth arrest.12)
In the present study, we explore the association between the IDH1 mutation and p21 activation via SREBP and propose a mechanism for a nonaggressive profile in gliomas bearing the IDH1 mutation.
Materials and Methods
I. Plasmids
cDNA of human IDH1 (NM_005896) was purchased from OriGene Technologies (Rockville, Maryland, USA). Site-directed mutagenesis was performed using the KOD-Plus-Mutagenesis Kit (Toyobo, Tokyo) to change G395 to A in IDH1, resulting in an R132H mutation. Wild-type and IDH1R132H cDNAs were excised by EcoRI and EcoRV. The resulting cDNA fragments were cloned into the EcoRI–SmaI sites of pIRES2-EGFP (Addgene, Cambridge, Massachusetts, USA) (Fig. 1A).
Fig. 1.
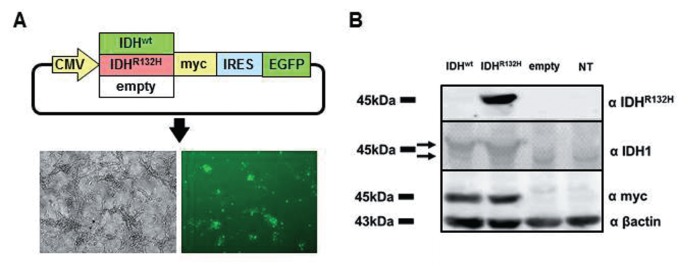
A: Construction of plasmids and transfection. An R132H mutation in IDH1 was made by converting guanine at position 395 to adenine with the KOD-Plus-Mutagenesis Kit (Toyobo). IDH1wt or IDH1R132H cDNA was inserted downstream of a CMV promoter in pIRES2-EGFP (Addgene). U87 human glioblastoma cells were transfected with Lipofectamine 2000 (Invitrogen). Transfection efficiency is about 40%. B: Western analysis. U87 cells were transfected with IDH1wt, IDH1R132H, or an empty-vector plasmid. Expressions of IDH1R132H and IDH1wt were confirmed by western blotting of cell lysates obtained 24 h after transfection. IDH1R132H was specifically recognized with an anti-IDH1R132H antibody. The upper arrow indicates the expression of IDH1 from the transfected plasmid and the lower one indicates endogenous IDH1 protein. IDH1: isocitrate dehydrogenase 1, NT: no transfection.
II. Cell culture, transfection
U87 cells, a cell line established from glioblastoma, harbor the normal IDH1 gene and the wild-type p53 gene.23) U87 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, California, USA) with 10% fetal bovine serum. Plasmid transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfection efficiency is reproducibly about 40%, as estimated by counting EGFP-positive cells, and did not differ among plasmids transfected (Fig. 1A).
For siRNA experiments, U87 cells were transfected in the same way. siRNA oligonucleotides were synthesized by Sigma-Aldrich (Tokyo). The sequences of siRNA targeting SREBP1 were as follows:
5′-rCrGrGrArGrArArGrCUrGrCrCUrAUrArATT-3′ (sense) and
5′-UUrGrAUrArGrGrCrArGrCUUrCUUrCUUr-CUrCrCrGTT-3′ (antisense).
The sequences of siRNA targeting SREBP2 were as follows:
5′-GUrGUrGrG3rAUUrGUrGUrCrCUrGrArGrCr-GUTT-3′ (sense) and
5′-ArCrGrCUrCrArGrGrArCrArAUrCrArCrArCTT-3′ (antisense).
III. RNA micro array, quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA from transfected U87 cells was purified using the RNeasy Mini kit (Qiagen, Tokyo) according to the manufacturer's instructions. RNA microarray analysis was done by Filgen, Inc (Nagoya, Aichi). Total RNA was converted to cDNA with the SuperScript VILO cDNA synthesis kit (Invitrogen). Ten nanograms of cDNA was used as a template for quantitative PCR with gene-specific primers (Table 1) and SYBR green (Qiagen) by 40 cycles of denaturing at 95°C for 15 sec and annealing and extension at 60–62°C for 30 sec. These PCRs were repeated six times. The data were analyzed by the Student's t-test.
Table 1.
Primer sequences and specific annealing temperatures used for quantitative PCR
| Gene | Primer sequence | Annealing temperature (°C) |
|---|---|---|
| SREBP1 | 5′-AGGACAGCCTGGCTACCACA-3′ | 60 |
| 5′-AGAAGCAGGTCACACAGGAACA-3′ | ||
| SREBP2 | 5′-CAAGGCCCTGGAAGTGACA-3′ | 60 |
| 5′-AGGAACTCTGCTGCCCATCTG-3′ | ||
| SREBP1a | 5′-CTGCTGACCGACATCGAAGAC-3′ | 62 |
| 5′-GATGCTCAGTGGCACTGACTCTTC-3′ | ||
| SREBP1c | 5′-CGGAGCCATGGATTGCACTTTC-3′ | 62 |
| 5′-GATGCTCAGTGGCACTGACTCTCC-3′ | ||
| p53 | 5′-AGAGCTGAATGAGGCCTTGGAA-3′ | 60 |
| 5′-GAGTCAGGCCCTTCTGTCTTGAAC-3′ | ||
| p21 | 5′-AAGACCATGTGGACCTGTCACTGT-3′ | 60 |
| 5′-GAAGATCAGCCGGCGTTTG-3′ | ||
| MDM2 | 5′-TGGGCAGCTTGAAGCAGTTG-3′ | 60 |
| 5′-CAGGCTGCCATGTGACCTAAGA-3′ | ||
| ACLY | 5′-ATGCAGCAGCCAAGATGTTCA-3′ | 60 |
| 5′-CACTCGCATGTCTGGGTTGTTTA-3′ | ||
| ACO2 | 5′-TTTGACAAGTGGGATGGCAAG-3′ | 60 |
| 5′-CAATGAGCAGGTTGTTGGAGATG-3′ | ||
| IDH2 | 5′-GAGTGGAGCCATGACCAAGGA-3′ | 60 |
| 5′-TGCTCTTGATGGTGTCGAGGA-3′ | ||
| IDH3A | 5′-TTTACGCGAATGTCCGACCA-3′ | 60 |
| 5′-TGATACTCTGCACGACTCCCATCAAC-3′ | ||
| MDH2 | 5′-TCTGAGCCACATCGAGAGACCAA-3′ | 60 |
| 5′-GACTCCAGCCGGAATAACTACCAC-3′ | ||
| OGDH | 5′-TGTCAATTCGATTCAAAGCTGGAG-3′ | 60 |
| 5′-AAGGGTTGGCCACCAAGGA-3′ |
SREBP1a and SREBP1c primers were synthesized by Sigma-Aldrich. The other primers were obtained from Takara Bio. ACLY: ATP citrate lyase, ACO2: mitochondrial aconitase 2, IDH: isocitrate dehydrogenase, MDH2: malate dehydrogenase 2, MDM2: murine double minute 2, OGDH: oxoglutarate dehydrogenase, PCR: polymerase chain reaction, SREBP: sterol regulatory element-binding protein.
IV. Western blotting
Cells were lysed 24 h after transfection with a lysis buffer (1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% TritonX-100, 0.01 M Tris-HCl [pH 8.0], and 0.14 M NaCl) and protease inhibitors were incubated for 10 min on ice. Equal amounts of protein extracts were separated by SDS-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane. Membranes were blocked for 1h in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat milk and were then probed with anti-IDH1 (N-20) (sc-49996; Santa Cruz Biotechnology, Dallas, Texas, USA), anti-IDH1R132H (DIA H09; Dianova, Warburgstr, Hamburg, Germany), anti-myc (R950-25; Invitrogen), anti-Retinoblastoma (Rb) (4H1) (#9309; Cell Signaling Technologies, Danvers, Massachusetts, USA), anti-phospho-Rb (Ser795) (#9301; Cell Signaling Technologies), or anti-β actin (C4) (sc-47778; Santa Cruz Biotechnology) antibodies. Membranes were then incubated with anti-mouse or anti-rabbit immunoglobulin G labeled with horseradish peroxidase. Chemiluminescent signals were detected on an imaging analyzer.
V. ELISA
ELISA kits were used for the measurement of p21 (#7167; Cell Signaling Technologies) and p53 protein (CY-7049; CycLex, Ina, Nagano).
Results
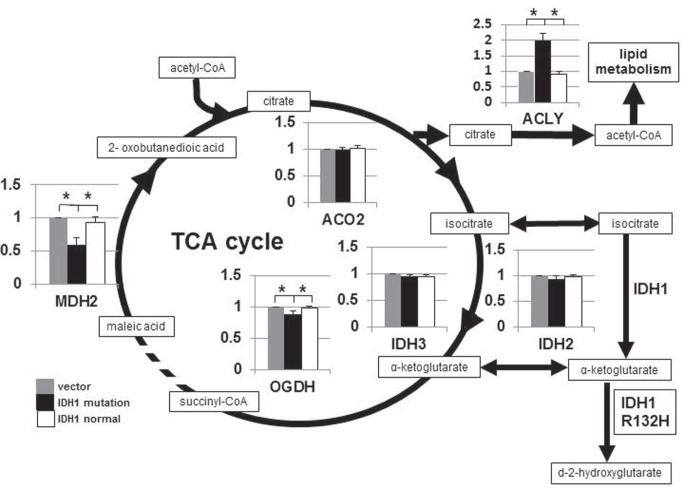
I. U87 glioblastoma cells with IDH1R132H altered the TCA cycle and lipid metabolism
A recent report showed that 2-HG increased specifically in IDH1-mutated gliomas and affected TCA cycle metabolism.18) To examine the effect of the IDH1R132H mutation on glioma cells, we constructed a plasmid expressing normal IDH1 or IDH1R132H with a myc-tag (Fig. 1) and transfected them into U87 human glioblastoma cells. The result of qRT-PCR and RNA microarrays of IDHR132H-transfected U87 cells relative to IDHWT-transfected cells is summarized in Fig. 2 and Table 2. The expression levels of enzymes involved in the TCA cycle downstream of α-KG appeared lower in IDH1R132H-transfected cells. In particular, an approximately 40% reduction in malate dehydrogenase 2 (MDH2) expression was detected relative to IDHWT-transfected cells. However, the other TCA cycle enzymes upstream of α-KG were not affected by IDH1 mutation, probably due to relative depletion of α-KG in IDH1R132H-transfected cells by competitive inhibition by 2-HG.18) Interestingly, the level of ATP citrate lyase (ACLY) that converts citrate to acetyl-CoA in the cytoplasm significantly increased in IDH1R132H-transfected cells, as revealed by RNA microarrays and qRT-PCR. Acetyl-CoA is then utilized for lipid synthesis as well. The other enzymes involved in lipid metabolism were up-regulated (Table 2). It is suggested that acetyl-CoA is preferentially metabolized for lipid synthesis rather than in the TCA cycle in IDHR132H U87 cells.
Fig. 2.
Expression of IDH1R132H alters metabolism in the TCA cycle in U87 human glioblastoma cells. The mRNA levels of enzymes involved in the TCA cycle were examined by quantitative reverse transcription-PCR. Notably, MDH2 mRNA levels were reduced in IDH1R132-transfected cells while ACLY mRNA levels increased relative to IDH1wt-transfected cells. ACLY: ATP citrate lyase, ACO2: mitochondrial aconitase 2, IDH1: isocitrate dehydrogenase 1, MDH2: malate dehydrogenase 2, OGDH: oxoglutarate dehydrogenase, PCR: polymerase chain reaction, TCA: tricarboxylic acid. *p < 0.05.
Table 2.
RNA microarray analysis of genes involved in TCA cycle and lipid metabolism
| TCA cycle |
|||
|---|---|---|---|
| Genes | Accession number | Mutation/normal* | |
| ACO2 | Aconitase 2, mitochondrial | NM_001098 | 1.01 |
| IDH2 | Isocitrate dehydrogenase 2 (NADP+) | NM_002168 | 1.14 |
| IDH3 | Isocitrate dehydrogenase 3 (NAD+) | NM_004135 | 0.98 |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase | NM_001003941 | 0.87 |
| DLD | Dihydrolipoamide dehydrogenase | NM_000108 | 0.93 |
| SUCLG | Succinate-CoA ligase | NM_001177599 | 0.92 |
| SDH | Succinate dehydrogenase | NM_003000 | 0.94 |
| MDH2 | Malate dehydrogenase 2 | NM_005918 | 0.84 |
| ACLY | ATP citrate lyase | NM_001096 | 1.46 |
| Lipid metabolism | |||
| SREBF1 | Sterol regulatory element-binding transcription factor 1 | NM_001005291 | 1.51 |
| SREBF2 | Sterol regulatory element-binding transcription factor 2 | NM_004599 | 1.35 |
| SCAP | SREBF chaperone | NM_012235 | 1.57 |
| FASN | Fatty acid synthase | NM_004104 | 2.06 |
| ACACA | Acetyl-CoA carboxylase alpha | NM_198834 | 1.43 |
| ACACB | Acetyl-CoA carboxylase beta | NM_001093 | 2.51 |
| HMGCR | 3-Hydroxy-3-methylglutaryl-CoA reductase | NM_000859 | 2.03 |
*The ratio of the amount of mRNA in IDH1-mutation-expressing cells to IDH1-normal cells. IDH1: isocitrate dehydrogenase 1, mRNA: messenger ribonucleic acid, TCA: tricarboxylic acid.
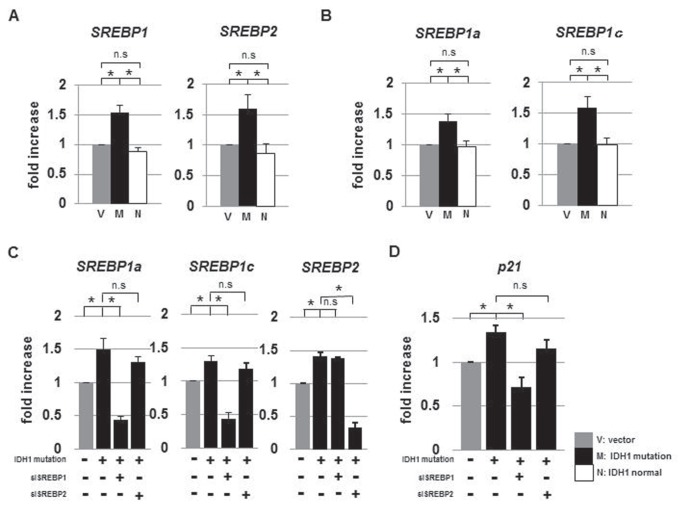
II. Up-regulation of p21 depends on SREBP1a
Increases in the expression levels of SREBP1 and -2 prompted us to examine p21, a cyclin-dependent kinase inhibitor. p21, which halts the cell cycle at G1, is regulated by SREBP.12) qRT-PCR analysis showed that IDH1R132H-transfected cells up-regulated SREBP1a, 1c, and 2 (Fig. 3A, B). In particular, SREBP1a has been shown to activate p21 expression and leads to cell growth arrest.12) In order to investigate the relationship between p21 and SREBP1 in U87 cells, we performed siRNA experiments. siRNA for SREBP1 specifically knocked down SREBP1a and 1c and siRNA for SREBP2 suppressed its target mRNA (Fig. 3C). Knock-down of SREBP1, but not SREBP2, specifically induced a decrease in p21 expression in IDHR132H-transfected cells (Fig. 3D). These results indicated that the R132H mutation in IDH1 induced the expression of SREBPs and that SREBP1 enhanced the expression of p21 in IDH1R132H-transfected cells.
Fig. 3.
The SREBP family of transcription factors were up-regulated in IDH1R132H-transfected U87 cells. Quantitative reverse transcription-PCR analysis of SREBP1 and 2 (A), and SREBP1a and SREBP1c (B) mRNA levels increased in IDH1R132H-U87 cells. C: siRNA for SREBP1 and 2 used in the present study specifically suppressed its target mRNA. D: p21 mRNA levels that increased in IDH1R132H-U87 cells were suppressed by knock-down of SREBP1, not SREBP2. IDH1: isocitrate dehydrogenase 1, M: IDH1R132H transfection, N: IDH1wt transfection, n.s: not significant, PCR: polymerase chain reaction, SREBP: sterol regulatory element-binding protein, V: empty vector. *p < 0.05.
III. IDHR132H up-regulates p21 independent of p53
p53, a tumor suppressor, regulates p21 expression and is negatively regulated by murine double minute 2 (MDM2).5) To examine the effect of IDH1R132H on the p53–p21 signaling pathway, we measured p21, p53, and MDM2 mRNA levels in IDH1wt- and IDH1R132H-transfected cells by qRT-PCR and assayed p21 and p53 by ELISA (Fig. 4, Table 3). p21 was up-regulated in IDH1R132H-transfected cells. However, p53 and MDM2 mRNA levels did not differ among the three groups (Fig. 4). These results suggested that the up-regulation of p21 protein was mediated by direct activation by SREBP, but not by p53.
Fig. 4.
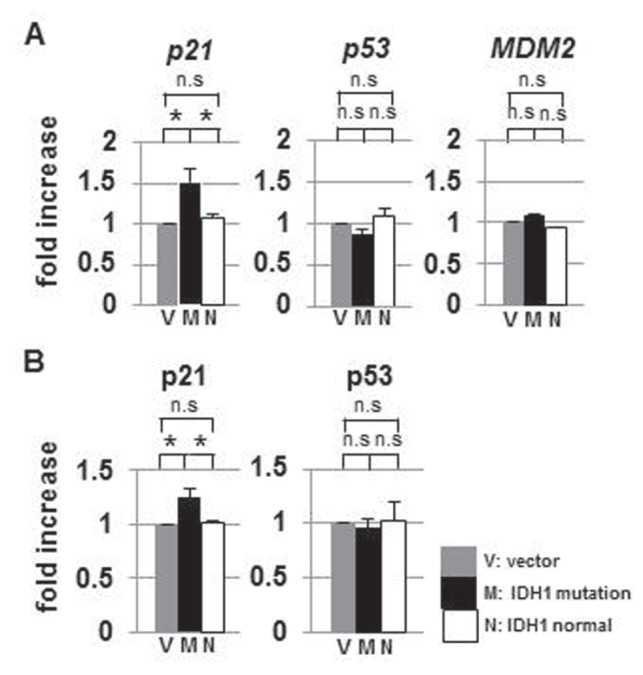
p21 was specifically up-regulated in IDH1R132H-expressing U87 cells. A: Quantitative reverse transcription-PCR analysis of p21, p53, and MDM2 mRNA levels in IDH1wt- and IDH1R132H-transfectedU87 cells. p21 mRNA was significantly increased in IDH1R132H cells. However, p53 and its negative regulator MDM2 mRNA levels did not show significant changes, relative to IDHwt-transfected cells. B: ELISA of p21 and p53. p21 significantly increased in IDH1R132H-expressing U87 cells. IDH: isocitrate dehydrogenase, M: IDH1R132H transfection, MDM2: murine double minute 2, N: IDH1wt transfection, n.s: not significant, PCR: polymerase chain reaction, V: empty vector. *p < 0.05.
Table 3.
Representative genes associated with cell cycle progression in IDH1-mutation-expressing cells
| Genes | Accession number | Mutation/normal* |
|---|---|---|
| MDM2 | NM_002392 | 1.03 |
| p15 | NM_004936 | 1.47 |
| p16 | NM_000077 | 1.37 |
| p18 | NM_001262 | 1.03 |
| p19 | NM_001800 | 1.35 |
| p21 | NM_000389 | 1.50 |
| p27 | NM_004064 | 1.22 |
| p45 | NM_005983 | 2.03 |
| CDK2 | NM_001798 | 0.95 |
| CDK4 | NM_000075 | 0.85 |
| CDK6 | NM_00114530 | 1.07 |
| CDC6 | NM_001254 | 1.16 |
| Cyclin A1 | NM_001111045 | 0.98 |
| Cyclin A2 | NM_001237 | 1.13 |
| Cyclin B1 | NM_031966 | 0.80 |
| Cyclin B2 | NM_004701 | 0.75 |
| Cyclin B3 | NM_033031 | 1.19 |
| Cyclin D3 | NM_001136017 | 0.85 |
| Cyclin E1 | NM_001238 | 0.98 |
| Cyclin E2 | NM_057749 | 0.78 |
| E2F1 | NM_005225 | 1.12 |
| E2F3 | NM_001949 | 1.17 |
| E2F4 | NM_001950 | 1.00 |
| E2F5 | NM_001083588 | 0.92 |
| E2F6 | NM_198256 | 0.82 |
| E2F8 | NM_024680 | 1.28 |
*The ratio of the amount of mRNA in IDH1-mutation-expressing cells relative to IDH1-normal cells. CDC6: cell division cycle 6, CDK: cyclin-dependent kinase, IDH1: isocitrate dehydrogenase 1, MDM2: murine double minute 2, mRNA: messenger ribonucleic acid.
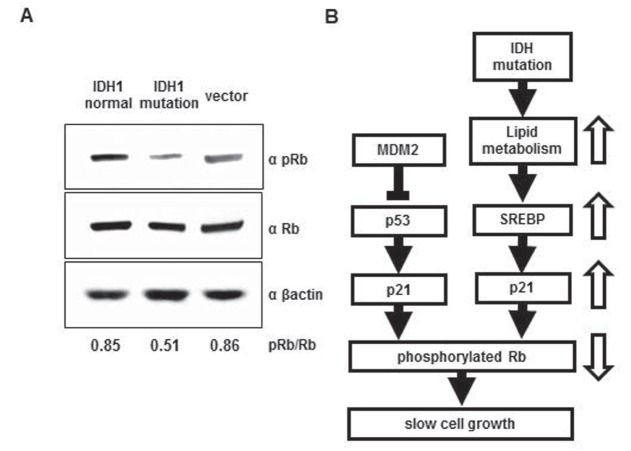
p21 protein inhibits the phosphorylation of Rb protein, one of the tumor suppressors. Nonphosphorylated Rb protein binds E2F transcription factor and prevents E2F from activating gene expression. Phosphorylated Rb fails to bind E2F and permits cells entering from the G1 phase to the S phase in the cell cycle.4) Western blot analysis of Rb protein indicated that the level of the phosphorylated form, relative to total Rb protein, decreased in IDH1R132H-transfected cells (Fig. 5A), suggesting slowing in progression of the cell cycle in IDH1-mutated U87 cells (Fig. 5B).
Fig. 5.
A: Rb protein in IDH1R132H-U87 cells. Cells were lysed 24 h after transfection with IDH1wt, IDH1R132H, and a vector plasmid. Western analysis of pRb and total Rb shows that pRb decreased in IDHR132H-transfected cells. For comparison, the intensity of each band was measured and the ratio of pRb to Rb (pRb/Rb) was calculated as shown at the bottom. B: A proposed signaling cascade leading to nonaggressive cancer associated with the IDH1 mutation. IDH1: isocitrate dehydrogenase 1, pRb: phosphorylated Rb, Rb: retinoblastoma.
Discussion
Since gliomas invasively grow into normal brain tissue and is thus difficult to surgically resect completely, the therapeutic outcome of gliomas was extremely poor. Although improved surgical techniques in combination with an alkylating agent temozolomide have made the median survival time of glioblastomas longer (21.4 months),15) the outcomes of glioblastomas are still poor relative to other carcinomas. However, gliomas bearing the IDH1 mutation are associated with better outcomes.16,24)
The mutated IDH1 produces 2-HG from α-KG in the cytoplasm.6) 2-HG competitively inhibits α-keto acid transaminase, thereby suppressing the TCA cycle.18) The present analyses confirmed that the TCA cycle was down-regulated in IDH1R132H-cells (Fig. 2), which has been proposed as the Warburg effect.11,13,22,24,25) Furthermore, it has been said that this down-regulation is associated with a selective advantage for cancer cells because nutrients are converted to building blocks such as lipids to be used for proliferation rather than being oxidized in the TCA cycle.18) The present study showed that down-regulation of the TCA cycle and up-regulation of ACLY, which converts citrate in the cytoplasm into acetyl-CoA, resulted in enhanced lipid metabolism (Fig. 2, Table 2). The R132H mutation in IDH1 induced shunting of carbons from glycolysis into de novo synthesis of lipid rather than into the TCA cycle. The enhanced lipid metabolism by the mutated IDH1can be relevant to increased SREBP expression.
The transcription factor to regulate lipid synthesis from acetyl-CoA in the cytoplasm is SREBP. Our data clearly showed that IDH1R132H induced increases in the mRNA levels of all SREBP family transcripts, 1a, 1c, and 2 (Fig. 4). SREBP1a and 2 have been shown to enhance p21 promoter activity,12) which was also confirmed in U87glioblastoma cells by qRT-PCR and siRNA knock-down experiments (Fig. 3C, D). Another pathway that regulates p21 is the p53-MDM2 cascade. As shown in Fig. 4, p53 and its mRNA levels in IDH1R132H-transfected cells did not differ from IDHwt-transfected cells, which supported that p21 was up-regulated via the SREBP pathway independent of the p53 pathway (Fig. 4). In addition, it has been reported that glycolysis is enhanced in glioma with the IDH1 mutation,18) and that glycolysis suppresses p53.14) This line of evidence supports p53 not playing a role in p21 activation in IDH1R132H U87 cells. Recently, IDH1R132H has been reported to be associated with SREBP1a activation and cellular proliferation.28) However, the precise mechanism how IDH1R132H induces SREBP1a activation was not revealed. Although IDH1R132H is associated with slow tumor progression, it is controversial whether IDH1R132H mutation induces or suppresses cell growth in cultured glioma cells. Another study reported that stably IDH1R132H expressing U87 cells decreased cellular proliferation.2)
In an attempt to demonstrate the direct association between the IDH1R132H and the retardation of cell growth, we analyzed the cell cycle profile of the transfected U87 cells. Unfortunately, we failed to get reproducible data, probably due to a subtle difference between IDH1wt- and IDH1R132H-cells (data not shown). We next measured the proliferation rate of IDH1wt- and IDH1R132H-transfected cells. Although the difference was not statistically significant, the U87 cells transfected with IDH1R132H plasmid tended to slower growth (Fig. 6). Accumulation of subtle growth retardation after a number of cell division in IDH1R132H glioma may lead to smaller tumor burden. The results obtained in the present study is based on the experiments with the U87 glioblastoma cell line, one of the widely employed in cultured brain tumor cells. However, it is desirable to examine other brain tumor cell lines and patients' glioblastomas in order to confirm the present results.
Fig. 6.
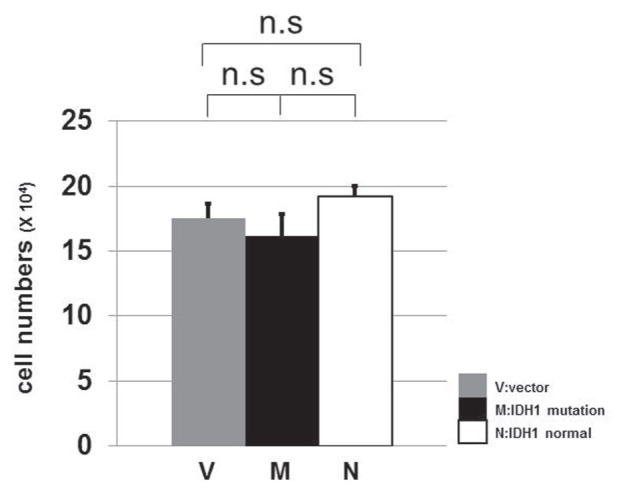
The comparison of growth of U87 cells after transfection. 1 × 104 cells of U87 were transfected with IDH1R132H (M), IDH1wt (N), or a vector plasmid (V). Three days after transfection, we counted the number of cells. The experiment was performed in triplicate. The U87 cells transfected with IDH1 mutation plasmid were less proliferative compared to those transfected with IDH1 normal or a vector plasmid although the difference was not statistically significant. IDH1: isocitrate dehydrogenase 1, n.s: not significant.
Several reports implicate doxidative stress1,6,8,17,25) or methylation of the MGMT promoter portion in gliomas with the IDH1 mutation3,19) important for a nonaggressive profile. We propose that suppression of the TCA cycle and subsequent enhancements in lipid metabolism induce up-regulation of the SREBP family, which results in the increased activity of p21 and decrease in phosphorylation of Rb protein (Fig. 5B). The R132H mutation in IDH1 appears to give rise to diverse metabolic changes, such as increased oxidative stress, inhibition of the TCA cycle, and enhanced lipid metabolism. The sum of all these alterations may make tumor cells nonaggressive. More detailed analysis of the metabolic changes induced by the IDH1 mutation will help us understand the mechanism of the low-grade malignant profile of an IDH1R132H glioma.
Acknowledgments
Satsuki Miyata received a Research Award to JMU graduate students. Metabolome analysis was supported by Human Metabolome Technologies, Inc.
References
- 1). Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, Van Noorden CJ: The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 119: 487– 494, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2). Bralten LB, Kloosterhof NK, Balvers R, Sacchetti A, Lapre L, Lamfers M, Leenstra S, de Jonge H, Kros JM, Jansen EE, Struys EA, Jakobs C, Salomons GS, Diks SH, Peppelenbosch M, Kremer A, Hoogenraad CC, Smitt PA, French PJ: IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol 69: 455– 463, 2011. [DOI] [PubMed] [Google Scholar]
- 3). Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, Kelsey KT, Wiencke JK: DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst 103: 143– 153, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4). Cmielová J, Rezáčová M: p21Cip1/Waf1 protein and its function based on a subcellular localization. J Cell Biochem 112: 3502– 3506, 2011. [DOI] [PubMed] [Google Scholar]
- 5). Colman MS, Afshari CA, Barrett JC: Regulation of p53 stability and activity in response to genotoxic stress. Mutat Res 462: 179– 188, 2000. [DOI] [PubMed] [Google Scholar]
- 6). Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739– 744, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7). Edwards PA, Tabor D, Kast HR, Venkateswaran A: Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta 1529: 103– 113, 2000. [DOI] [PubMed] [Google Scholar]
- 8). Fu Y, Huang R, Du J, Yang R, An N, Liang A: Glioma-derived mutations in IDH: from mechanism to potential therapy. Biochem Biophys Res Commun 397: 127– 130, 2010. [DOI] [PubMed] [Google Scholar]
- 9). Fu Y, Huang R, Zheng Y, Zhang Z, Liang A: Glioma-derived mutations in isocitrate dehydrogenase 2 beneficial to traditional chemotherapy. Biochem Biophys Res Commun 410: 218– 223, 2011. [DOI] [PubMed] [Google Scholar]
- 10). Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW: Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339– 344, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11). Hsu PP, Sabatini DM: Cancer cell metabolism: Warburg and beyond. Cell 134: 703– 707, 2008. [DOI] [PubMed] [Google Scholar]
- 12). Inoue N, Shimano H, Nakakuki M, Matsuzaka T, Nakagawa Y, Yamamoto T, Sato R, Takahashi A, Sone H, Yahagi N, Suzuki H, Toyoshima H, Yamada N: Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol Cell Biol 25: 8938– 8947, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13). Kondoh H: Cellular life span and the Warburg effect. Exp Cell Res 314: 1923– 1928, 2008. [DOI] [PubMed] [Google Scholar]
- 14). Mason EF, Zhao Y, Goraksha-Hicks P, Coloff JL, Gannon H, Jones SN, Rathmell JC: Aerobic glycolysis suppresses p53 activity to provide selective protection from apoptosis upon loss of growth signals or inhibition of BCR-Abl. Cancer Res 70: 8066– 8076, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15). Muragaki Y, Maruyama T, Iseki H, Tanaka M, Shinohara C, Takakura K, Tsuboi K, Yamamoto T, Matsumura A, Matsutani M, Karasawa K, Shimada K, Yamaguchi N, Nakazato Y, Sato K, Uemae Y, Ohno T, Okada Y, Hori T: Phase I/IIa trial of autologous formalin-fixed tumor vaccine concomitant with fractionated radiotherapy for newly diagnosed glioblastoma. Clinical article. J Neurosurg 115: 248– 255, 2011. [DOI] [PubMed] [Google Scholar]
- 16). Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW: An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807– 1812, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17). Reitman ZJ, Yan H: Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 102: 932– 941, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18). Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H: Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 108: 3270– 3275, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19). Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY: Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27: 4150– 4154, 2009. [DOI] [PubMed] [Google Scholar]
- 20). Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL: Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest 99: 846– 854, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21). Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987– 996, 2005. [DOI] [PubMed] [Google Scholar]
- 22). Vander Heiden MG, Cantley LC, Thompson CB: Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029– 1033, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23). Van Meir EG, Kikuchi T, Tada M, Li H, Diserens AC, Wojcik BE, Huang HJ, Friedmann T, de Tribolet N, Cavenee WK: Analysis of the p53 gene and its expression in human glioblastoma cells. Cancer Res 54: 649– 652, 1994. [PubMed] [Google Scholar]
- 24). Yan H, Bigner DD, Velculescu V, Parsons DW: Mutant metabolic enzymes are at the origin of gliomas. Cancer Res 69: 9157– 919, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25). Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD: IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765– 773, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26). Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB: Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene 31: 2491– 2498, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27). Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225– 234, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28). Zhu J, Cui G, Chen M, Xu Q, Wang X, Zhou D, Lv S, Fu L, Wang Z, Zuo J: Expression of R132H mutational IDH1 in human U87 glioblastoma cells affects the SREBP1a pathway and induces cellular proliferation. J Mol Neurosci 50: 165– 171, 2013. [DOI] [PubMed] [Google Scholar]