Abstract
An Arn+ (n = 1–6000) gas cluster ion source has been utilized to map the chemical distribution of lipids in a mouse brain tissue section. We also show that the signal from high mass species can be further enhanced by doping a small amount of CH4 into the Ar cluster to enhance the ionization of several biologically important molecules. Coupled with secondary ion mass spectrometry instrumentation which utilizes a continuous Ar cluster ion projectile, maximum spatial resolution and maximum mass resolution can be achieved at the same time. With this arrangement, it is possible to achieve chemically resolved molecular ion images at the 4-µm resolution level. The focused Arn+/[Arx(CH4)y]+ beams (4–10 µm) have been applied to the study of untreated mouse brain tissue. A high signal level of molecular ions and salt adducts, mainly from various phosphocholine lipids, has been seen and directly used to map the chemical distribution. The signal intensity obtained using the pure Ar cluster source, the CH4-doped cluster source and C60 is also presented.
Keywords: molecular ion imaging, SIMS, mouse brain section, Ar cluster ion beam
Introduction
Secondary ion mass spectrometry (SIMS) generally exhibits improved spatial resolution compared to other imaging mass spectrometry techniques, such as matrix-assisted laser desorption/ionization (MALDI) and desorption electrospray ionization (DESI). However, the limited mass range and low ionization probability limit the study of biological materials. Various methods have been explored to enhance secondary ion yields. One approach is to develop new primary ion beams. For polyatomic primary beams such as and that lower the level of chemical damage induced by the primary ion beam bombardment compared with atomic primary ions,[1] the secondary ion yields have been reported to be enhanced by one to two orders of magnitude.[2] This development has moved time-of-flight-SIMS analysis out of the static regime enabling depth profiling and 3D imaging.[3] The recent development of a gas cluster ion beam (GCIB) such as Arn+ (n = ~10 000) enables delivery of larger secondary molecular ions almost ‘fragment-free’ when compared with .[4,5] Another new candidate, , delivers more than 10 times signal enhancement from a variety of biomolecules through the protonation process in the impact zone when compared with .[6] However, Ar-GCIBs are mainly used as a sputtering source to depth profile inorganic and organic materials[7] since they are difficult to focus. There are several other methods that have been reported to enhance the characteristic ions of organic materials, including matrix-enhanced SIMS,[8] metal-assisted SIMS,[9] nanoparticle-enhanced SIMS,[10] oxygen flooding-enhanced SIMS[11] and water vapor-enhanced SIMS[12] to name a few. These approaches have not found widespread use, however, due to issues of sample preparation, reproducibility and suitability for molecular depth profiling.
Here, we show that a 20-keV Arn+ (n = 1–6000) GCIB is capable of yielding rich chemical information about lipids in the mass range between m/z 650 and 1000 from various biological samples. In addition, we have found that by mixing less than 5% CH4 into the Ar cluster, the secondary ion yield of [M+H]+ from various biomaterials can be enhanced by 2–10 times relative to the pure Ar clusters.[13] In addition, the design of these sources has been optimized to the point where they are able to produce a continuous beam with a 4–10-µm spot size. The samples can be analyzed directly without matrix which is of course required for MALDI imaging. We report here the pure and mixed Ar cluster imaging on a mouse brain section, to reveal the unique lipid distribution of various phosphocholine (PC) lipids and cholesterol, and to investigate the signal enhancement on the real biosamples using a mixed Ar cluster. The results demonstrate that these projectiles can be used as an imaging ion source in addition to a sputtering source for SIMS. The developed methodology has promise to locate multiple lipid molecular ions in complex biological systems without sample pretreatment.
Experimental
Tissue sample and SIMS analysis
Mouse brain sections were kindly provided by Vanderbilt University. A 10-µm-thick mouse brain slice was mounted onto indium tin oxide-coated glass. SIMS analyses were performed directly on these samples at room temperature using the J105 3D Chemical Imager from Ionoptika, which has been detailed elsewhere.[14] The system is equipped with a 40-keV and a 20-keV gas cluster ion source (GCIB). This GCIB is fed with UHP Ar and generates Arn+ (n = 1–6000) at 18 bar of back pressure. Mixed cluster beams [Arx(CH4)y]+ were generated by filling a volume of Ar and 1.1% CH4 in a stainless steel mixing chamber inserted into the primary gas line. The gas tanks were connected to the bottom of the chamber for mixing before being introduced into the ion source. The cluster size distribution was checked by pulsing the gun and measuring the flight time spectrum of the projectile ions between the pulser and the target surface via the ion induced secondary electron emission signal. The resulting flight time spectrum can be converted into a cluster mass distribution by means of the known flight path (42.9 cm) and kinetic energy (20 keV) of the projectiles. The pure and mixed Ar cluster beams were focused to ~10 µm to raster across a very large area on the tissue sample in a tiled image mode. Each tile had an area of 1000 × 1000 µm2 with 128 × 128 pixels; the total ion dose was 3 × 1012 ions/cm2 per tile. The same dose of primary ions was used for both the and the mixed Ar cluster ion experiments.
Results and discussion
Tissue imaging using pure cluster beam
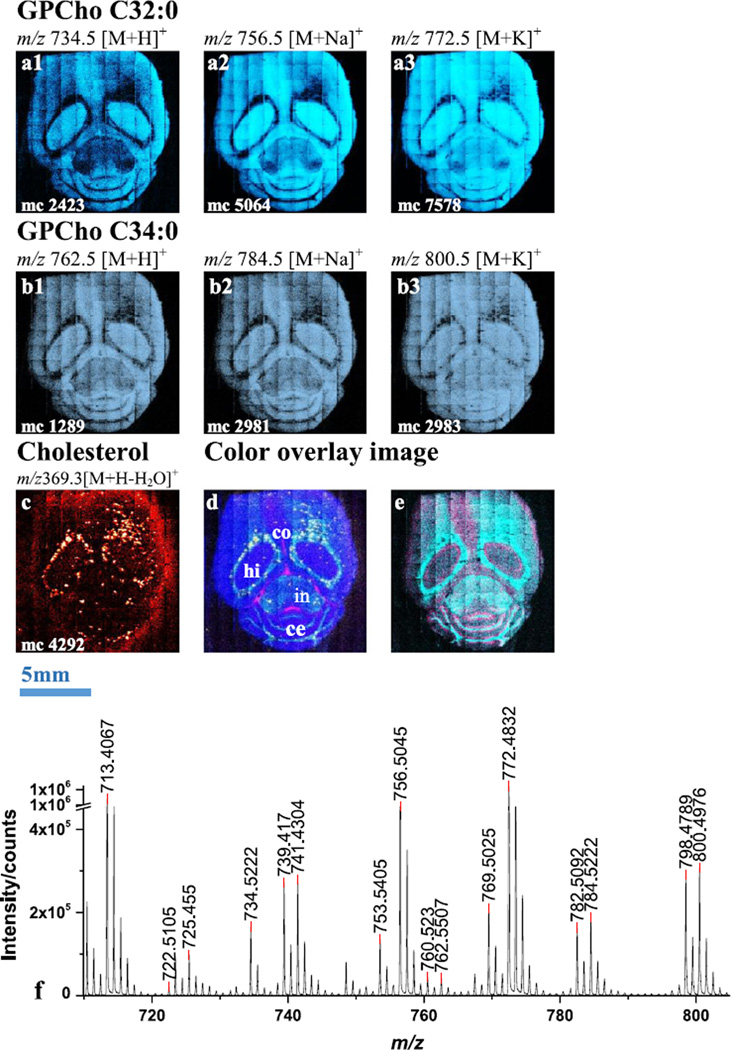
An cluster beam was used to analyze a horizontal section of mouse brain in positive ion mode. The tiled images of selected lipid molecular ions and the mass spectrum in the range of m/z 720~810 are shown in Fig. 1. The spectra [Fig. 1 (f)] demonstrate that high mass resolution (m/Δm ~7000) in the high mass range is retained which benefits the assignment of lipid species. The intensity ratio of molecular ion to fragment, represented by comparing m/z 734.5 with m/z 184.1 and m/z 772.5 with m/z 184.1, is about 1:31 and 1:4, which is significantly improved when compared with a projectile at 1: 64 and 1:15.[15] Because of the higher signal level, the molecular ions can be used directly to map the distributions on the tissue. Without any pre-treatment of the sample, the lipids are detected both as molecular ions and as their sodiated and potassiated adducts. Since the sample is unwashed, the natural chemical gradients are kept unchanged. Hence, the selected molecular ions and their salt adducts share the same distribution. As in Fig. 1 (a1, a2, a3), single ion images of [M+H]+, [M+Na]+ and [M+K]+ at m/z 734.5, 756.5 and 772.5 from GPCho C32:0 are abundant in the hippocampus and inner cerebellum; while detected ions from GPCho C34:0 at m/z 762.5, 784.5 and 800.5 in Fig. 1 (b1, b2, b3) have a similar distribution to GPCho C32:0. The ion image of cholesterol at m/z 369.3 shows the expected elevated intensity in the white matter, e.g. around the hippocampus and cerebellum as shown in Fig. 1 (c). The finding correlates well with other recent MALDI[16] and DESI[17] imaging experiments. The correlation of different lipids can be better demonstrated in a colored overlay image shown in Fig. 1 (d), where GPCho C32:0 in blue is concentrated in the hippocampus and inner cerebellum, and GPCho C34:3 in turquoise is localized around the hippocampus, internal capsule and the groove of the cerebellum. Cholesterol in yellow is spotted around the hippocampus internal capsule and the grooves of the cerebellum. Glucosyl ceramide (d18:1/16:0) in hot pink has a moustache-like pattern within the corpus callosum. There are other ions in the 500–700 m/z range that exhibit a unique distribution pattern such as m/z 534.3. By overlapping this ion with GPCho C36:3 or C36:1, it is found that these two type of ions are concentrated around the hippocampus and cerebellum and closely next to each other as shown in Fig. 1 (e).
Figure 1.
Selected ion images and color overlay images of mouse brain section in positive SIMS mode using . a1, a2, a3), Single ion images of [M+H]+, [M+Na]+ and [M+K]+ at m/z 734.5, 756.5 and 772.5 from GPCho C32:0, respectively. b1, b2, b3), Single ion images of [M+H]+, [M+Na]+ and [M+K]+ at m/z 762.5, 784.5 and 800.5 from GPCho C34:0, respectively. c), Single ion image of cholesterol at m/z 369.3. d), Color overlay image of GPCho C32:0 at m/z 734.5 in blue, GPCho C34:3 at m/z 822.6 in turquoise, cholesterol at m/z 369.3 in yellow and glucosyl ceramide (d18:1/16:0) at m/z 722.5 in hot pink. With the major brain structures indicated (co-corpus callosum, ce-cerebellum, hi-hippocampus, in-internal capsule). e), Color overlay image of DAG ion at m/z 534.3 in hot pink and GPCho C36:3 at m/z 822.6 in turquoise. f), Positive SIMS spectrum collected from the tissue in mass range between m/z 720 and 810.
Tissue imaging using a mixed cluster ion beam
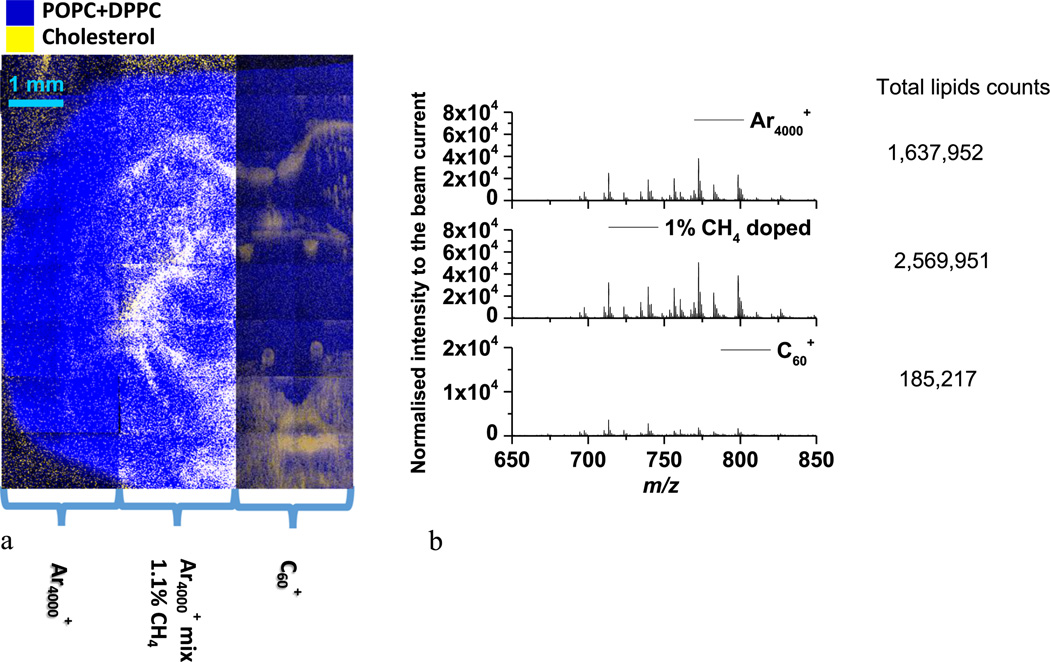
A coronal mouse brain section was imaged using , 1.1% methane doped , and in order to compare the relative response of each projectile. The section is symmetric which facilitates the comparison of the lipid signal in different parts of the tissue. As shown in Fig. 2, a part of the tissue section was divided into three panels and analyzed using the different projectiles. The color overlay image shows the distribution of GPCho C34:1 and GPCho C32:0 in blue and cholesterol in yellow. The PC lipids in the mass range of m/z 650–850 and cholesterol are anti-localized in the mouse brain tissue section.[18,19] For a comparison of signal levels, the lipid signal in the mass range of m/z 650–850 was summed from six areas of 19 × 19 pixels in each panel and normalized to the primary ion dose. The results shows that the total lipid signal for the 1.1% CH4 doped cluster delivers a 2× increase in signal compared with pure Ar4000+ cluster, the enhancement is a factor of 13 when compared to as shown in Fig. 2 (b).
Figure 2.
Color overlay image of mouse brain section using different cluster beams. a) The three panels on the tissue section are subjected to the analyses using , 1.1% methane-doped and . The color overlay image shows the distribution of GPCho C34:1 and GPCho C32:0 in blue and cholesterol in yellow. b) The normalized spectra and total lipids counts from the three panels on the tissue section in mass range of m/z 650–850.
Conclusion and outlook
SIMS imaging with a GCIB is optimized for the ionization of analytes such as lipids, metabolites and pharmaceuticals. The unique lipid distribution in the mass range of m/z 650–830 has been obtained for the untreated horizontal section of mouse brain using pure Ar clusters. The mixed clusters continue to yield a higher signal level of the high mass lipid species compared with the pure Ar cluster and on biological tissue. In the future, these developments open the opportunity of sharpening the probe size of the cluster beam to explore a smaller regime of biological systems, e.g. mammalian cells, to address the question of localization of endogenous lipids with exogenous drug molecules.
Acknowledgements
This project was supported by grants from the National Center for Research Resources (5P41RR031461), National Institutes of Health (2R01 473 EB002016-1) and Novartis. In addition, infrastructure support from the Division of Chemical Sciences at the Department of Energy grant number DE-FG02-06ERER15803 is acknowledged. Technical assistance in acquiring the images was provided by Paul Blenkinsopp of Ionoptika. We appreciate the efforts of the Caprioli group at Vanderbilt University for supplying tissue sections.
References
- 1.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. Anal. Chem. 2003;75:1754. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 2.Mahoney CM. Mass Spectrom. Rev. 2010;29:247. doi: 10.1002/mas.20233. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Anal. Chem. 2007;79:2199. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 4.Sheraz née Rabbani S, Barber A, Fletcher JS, Lockyer NP, Vickerman JC. Anal. Chem. 2013;85:5654. doi: 10.1021/ac4013732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoyagi S, Fletcher J, Sheraz S, Kawashima T, Berrueta Razo I, Henderson A, Lockyer N, Vickerman J. Anal. Bioanal. Chem. 2013;405:6621. doi: 10.1007/s00216-013-7139-z. [DOI] [PubMed] [Google Scholar]
- 6.Sheraz S, Barber A, Fletcher JS, Lockyer NP, Vickerman JC. Anal. Chem. 2013;85:5654. doi: 10.1021/ac4013732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cumpson PJ, Portoles JF, Barlow AJ, Sano N, Birch M. Surf. Interface Anal. 2013;45:1859. [Google Scholar]
- 8.Fitzgerald JJD, Kunnath P, Walker AV. Anal. Chem. 2010;82:4413. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- 9.Heile A, Lipinsky D, Wehbe N, Delcorte A, Bertrand P, Felten A, Houssiau L, Pireaux JJ, De Mondt R, Van Vaeck L, Arlinghaus HF. Appl. Surf. Sci. 2008;255:941. [Google Scholar]
- 10.Kim Y-P, Oh E, Hong M-Y, Lee D, Han M-K, Shon HK, Moon DW, Kim H-S, Lee TG. Anal. Chem. 2006;78:1913. doi: 10.1021/ac051500j. [DOI] [PubMed] [Google Scholar]
- 11.Liao H-Y, Lin K-Y, Kao W-L, Chang H-Y, Huang C-C, Shyue J-J. Anal. Chem. 2013;85:3781. doi: 10.1021/ac400214t. [DOI] [PubMed] [Google Scholar]
- 12.Mouhib T, Delcorte A, Poleunis C, Bertrand P. J. Am. Soc. Mass Spectrom. 2010;21:2005. doi: 10.1016/j.jasms.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Wucher A, Tian H, Winograd N. Rapid Commun. Mass Spectrom. 2014;28:396. doi: 10.1002/rcm.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Rapid Commun. Mass Spectrom. 2011;25:925. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 15.Rabbani S, Barber AM, Fletcher JS, Lockyer NP, Vickerman JC. Anal. Chem. 2011;83:3793. doi: 10.1021/ac200288v. [DOI] [PubMed] [Google Scholar]
- 16.Dufresne M, Thomas A, Breault-Turcot J, Masson JF, Chaurand P. Anal. Chem. 2013;85:3318. doi: 10.1021/ac3037415. [DOI] [PubMed] [Google Scholar]
- 17.Wu CP, Ifa DR, Manicke NE, Cooks RG. Anal. Chem. 2009;81:7618. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bich C, Touboul D, Brunelle A. Mass Spectrom. Rev. 2013 doi: 10.1002/mas.21399. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 19.Weaver EM, Hummon AB. Adv. Drug Deliv. Rev. 2013;65:1039. doi: 10.1016/j.addr.2013.03.006. [DOI] [PubMed] [Google Scholar]