Myosin 1b functions as an effector of EphB2/ephrinB signaling and controls cell morphology and cell repulsion.
Abstract
Eph receptors and their membrane-tethered ligands, the ephrins, have important functions in embryo morphogenesis and in adult tissue homeostasis. Eph/ephrin signaling is essential for cell segregation and cell repulsion. This process is accompanied by morphological changes and actin remodeling that drives cell segregation and tissue patterning. The actin cortex must be mechanically coupled to the plasma membrane to orchestrate the cell morphology changes. Here, we demonstrate that myosin 1b that can mechanically link the membrane to the actin cytoskeleton interacts with EphB2 receptors via its tail and is tyrosine phosphorylated on its tail in an EphB2-dependent manner. Myosin 1b regulates the redistribution of myosin II in actomyosin fibers and the formation of filopodia at the interface of ephrinB1 and EphB2 cells, which are two processes mediated by EphB2 signaling that contribute to cell repulsion. Together, our results provide the first evidence that a myosin 1 functions as an effector of EphB2/ephrinB signaling, controls cell morphology, and thereby cell repulsion.
Introduction
EphB receptors (erythropoietin-producing hepatoma-amplified sequence) are a large family of transmembrane tyrosine kinase receptors that interact with ephrinB ligands—also transmembrane proteins—triggering a cell signaling cascade (Klein, 2012). Eph/ephrin signaling contributes to the establishment of the precise organization of tissues during embryonic development and maintains tissue patterning and controls tissue homeostasis in the adult (Batlle et al., 2002; Rohani et al., 2011; Batlle and Wilkinson, 2012). Experimental evidence suggests that the establishment and maintenance of cell segregation by Eph/ephrin signaling involved different mechanisms including contact repulsion restricting cell migration (Xu et al., 1999; Marston et al., 2003; Zimmer et al., 2003; Poliakov et al., 2008; Astin et al., 2010; Rohani et al., 2011). Restricted cell migration mediated by the activation of EphB receptors involves significant changes in cell morphology including cell contraction and formation of cell protrusions as well as remodeling of the actin cytoskeleton (Marston et al., 2003; Zimmer et al., 2003; Moeller et al., 2006; Groeger and Nobes, 2007; Kayser et al., 2008). Tyrosine phosphorylation of EphB downstream effectors modulates the remodeling of the actin network (Irie and Yamaguchi, 2002; Evans et al., 2007; Tolias et al., 2007; Mohamed et al., 2012). However, the plasma membrane and the cortical actin network need to be mechanically coupled to bring about these morphological changes and cell repulsion (Raucher et al., 2000; Sheetz, 2001).
With their ability to generate mechanical force and bind actin filaments as well as cellular membranes, the widely expressed class 1 myosins link the cytoskeleton to membranes (McConnell and Tyska, 2010; Tyska and Nambiar, 2010). Myosins 1 are single headed members of the myosin super family. They are involved in membrane remodeling and regulation of actin dynamics. They have been implicated in various aspects of membrane trafficking along the endocytic and exocytic pathways, in the regulation of membrane tension, and in the formation or stability of membrane protrusions at the plasma membrane such as microvilli of enterocytes (Raposo et al., 1999; Salas-Cortes et al., 2005; Nambiar et al., 2009, 2010). We have recently shown that one of these vertebrate myosins 1, myosin 1b (Myo1b) elongates membrane tubules originating from sorting endosomes and the trans-Golgi network along actin bundles (Salas-Cortes et al., 2005; Almeida et al., 2011; Yamada et al., 2014). Myo1b is also associated with the plasma membrane and it has been suggested that this motor protein controls directed cell migration during zebrafish embryo development (Raposo et al., 1999; Tang and Ostap, 2001; Diz-Muñoz et al., 2010). Similar to other myosins 1, Myo1b interacts with cell membrane acidic phospholipids via its highly basic C-terminal tail domain. Myo1b’s association with membrane protrusions in HeLa cells required the interaction of its pleckstrin homology domain with phosphatidylinositol 4,5-bisphosphate (Komaba and Coluccio, 2010). However, the various roles of this motor protein from organelle to the plasma membrane suggest that it may possess additional mechanisms for membrane targeting (Mazerik and Tyska, 2012). In this study, we identify a new Myo1b membrane-binding partner: the transmembrane EphB2 receptors. By using live-cell imaging and biochemical analysis, we demonstrate that Myo1b is a downstream effector of EphB–ephrinB signaling controlling cell repulsion by regulating the redistribution of myosin II in actomyosin fibers and the formation of filopodia at the interface of ephrinB1 and EphB2 cells.
Results
EphB2 interacts via the tail region of Myo1b
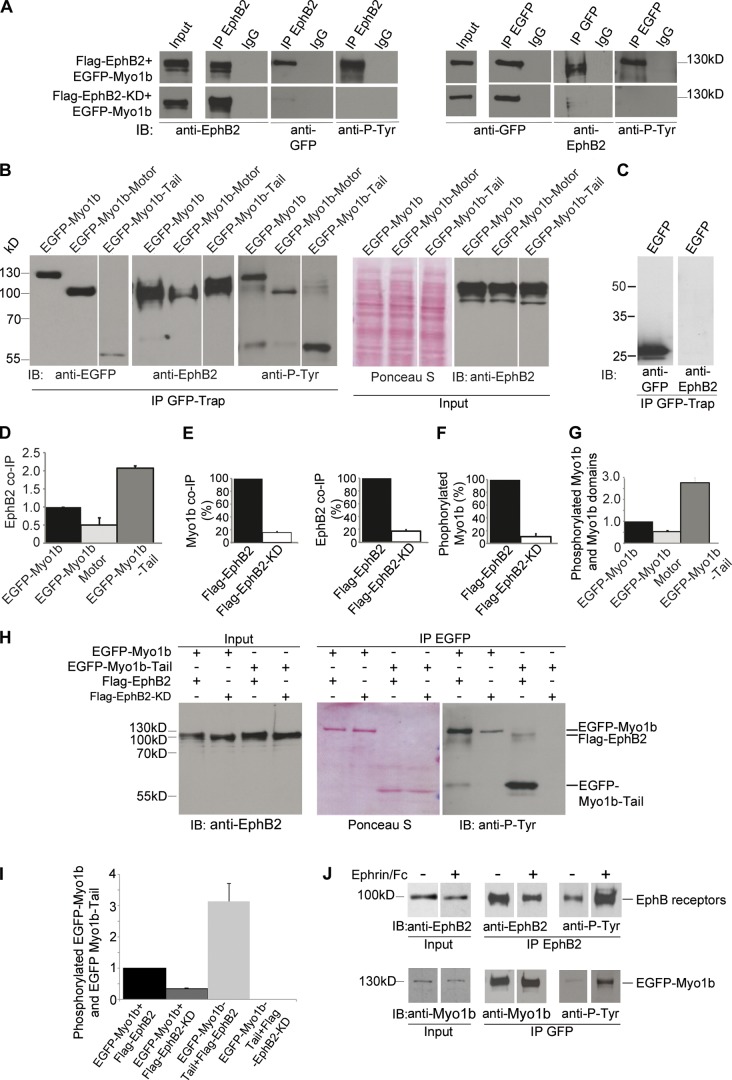
We first observed that Myo1b coimmunoprecipitated (coIP) with the EphB2 transmembrane receptors and conversely EphB2 coIP with Myo1b when the two proteins were coexpressed in Hek293T cells (Fig. 1 A). Myo1b is composed of an N-terminal motor domain, a light chain binding domain (LCBD) with one or more IQ (IQxxxRGxxxR) motifs (a helical sequence of ∼23 amino acids with a core consensus sequence) depending on the splicing isoform that binds calmodulin in a calcium-dependent manner, and a tail region with a highly basic C-terminal tail homology 1 domain that binds membranes (McConnell and Tyska, 2010). We compared the ability of the motor and tail domains to bind EphB2 using EGFP-tagged versions of Myo1b. Although the expression level of EGFP-Myo1b-Tail was lower than EGFP-Myo1b and EGFP-Myo1b motor, EGFP-Myo1b-Tail pulled down more than twofold of Flag-EphB2 compared with EGFP-Myo1b (Fig. 1, B and D), whereas no Flag-EphB2 was pulled down with EGFP tag alone (Fig. 1 C). Furthermore, EGFP-Myo1b-motor pulled down only 50% of Flag-EphB2 compared with EGFP-Myo1b (Fig. 1, B and D), suggesting that EphB2 binds Myo1b-Tail preferentially. YFP-EphB2 extracted from a cell lysate and immobilized on agarose beads pulled down the soluble recombinant GST-Myo1b-Tail but not the GST alone, confirming that Myo1b interacts with EphB2 via its Tail domain (Fig. S1 A).
Figure 1.
Myo1b-Tail interacts with EphB2 receptors and is phosphorylated depending on EphB2 kinase activity. (A) Flag-EphB2, Flag-EphB2-KD, or EGFP-Myo1b immunoprecipitated from Hek293T cell lysate (Input) using anti-EphB2 or anti-GFP antibodies or normal IgG were analyzed by SDS-PAGE and immunoblotted (IB) with anti-EphB2, anti-GFP, and anti-phosphorylated tyrosine (anti-P-Tyr) antibodies. 70% of the pull-down was loaded to detect the coIPs, whereas 20% was loaded to detect the immunoprecipitations and EGFP-Myo1b tyrosine phosphorylation. The antibodies did not detect any material when the immunoprecipitations were performed with normal IgG and tyrosine phosphorylation of Flag-EphB2 that coIP with Myo1b was hardly detectable in these conditions. (B) The different EGFP-tagged Myo1b recombinant domains pulled down with GFP-Trap from cells lysates (Input) also expressing Flag-EphB2 were analyzed by SDS-PAGE and immunoblotted with anti-GFP, anti-EphB2, or anti-phosphorylated tyrosine antibodies. Cell lysates contained a similar amount of total proteins as detected by Ponceau S and EphB2 receptors as detected with anti-EphB2 antibodies. (C) EphB2 does not coIP with EGFP when coexpressed with Flag-EphB2 in Hek293T cells. (D) The amount of Flag-EphB2 pulled down with Myo1b recombinant domains was quantified and normalized to the amount of Myo1b recombinant domains expressed. Data are shown as the mean of three experiments. Error bars represent ± SEM. (E) EGFP-Myo1b that coIP with Flag-EphB2 or Flag-EphB2-KD and Flag-EphB2 or Flag-EphB2-KD that coIP with EGFP-Myo1b were quantified and normalized to the amount of EGFP-Myo1b and Flag-EphB2 or Flag-EphB2-KD expressed in lysates. EGFP-Myo1b that coIPs with Flag-EphB2-KD is expressed as a percentage of EGFP-Myo1b that coIPs with Flag-EphB2 and Flag-EphB2-KD that coIPs with EGFP-Myo1b is expressed as percentage of Flag-EphB2 that coIPs with EGFP-Myo1b. Data are shown as the mean of two experiments. Error bars represent ± SEM. (F) Phosphorylated EGFP-Myo1b that coIP with Flag-EphB2 or Flag-EphB2-KD was quantified and normalized to the amount of EGFP-Myo1b expressed in the lysates and expressed as a percentage of phosphorylated EGFP-Myo1b that coIP with Flag-EphB2. Data are shown as the mean of two experiments. Error bars represent ± SEM. (G) Phosphorylated EGFP-Myo1b domains in cell also expressing Flag-EphB2 were quantified and normalized to the amount of phosphorylated EGFP-Myo1b. Data are shown as the mean of three experiments. Error bars represent ± SEM. (H) EGFP-Myo1b or EGFP-Myo1b-Tail were pulled down by GFP-Trap beads from Hek293T cells expressing also Flag-EphB2 or Flag-EphB2-KD. Flag-EphB2 and Flag-EphB2-KD were expressed at similar levels as judged by immunoblotting of the cell lysates (Input) with anti-EphB2 antibodies. Similar amounts of EGFP-Myo1b or EGFP-Myo1b-Tail were immunoprecipitated from cells expressing Flag-EphB2 and Flag-EphB2-KD as judged by Ponceau S. 20% of Flag-EphB2 and Flag-EphB2-KD that coIP with EGFP-Myo1b or EGFP-Myo1b-Tail were analyzed by SDS-PAGE and immunoblotted with anti-phosphotyrosine antibodies. (I) Phosphorylated EGFP-Myo1b and EGFP-Myo1b-Tail were quantified, normalized to the expression of EGFP-Myo1b or EGFP-Myo1b-Tail, and expressed as a percentage of phosphorylated EGFP-Myo1b. Data are shown as the mean of two experiments. Error bars represent ± SEM. (J) EphB2 or EGFP-Myo1b were immunoprecipitated from LS174T cell lysates (Input) expressing or not EGFP-Myo1b and treated or not with ephrinB1-Fc for 10 min with anti-EphB2 antibodies or GFP-Trap before being analyzed by SDS-PAGE and immunoblotting with anti-EphB2 or anti-Myo1b and anti-phosphotyrosine antibodies.
EphB2 kinase activity is required for Myo1b–EphB2 interaction and induced Myo1b phosphorylation
The autophosphorylation of the EphB2 receptors that depend on their own kinase activity results in a conformational change, allowing some of the effectors to bind to the juxtamembrane domain (Wybenga-Groot et al., 2001). We thus, investigated whether the Myo1b–EphB2 interaction requires EphB2 kinase activity. Only 16% of EGFP-Myo1b that coIP with Flag-EphB2 coIP with Flag-EphB2 mutated for a single amino acid that inhibits its kinase activity (Genander et al., 2009; Flag-EphB2-KD). Similarly 17% of Flag-EphB2-KD coIP with EGFP-Myo1b compared with Flag-EphB2 (Fig. 1, A and E). Treatment with the tyrosine kinase inhibitor genistein also impaired Myo1b–EphB2 interaction (Fig. S1 B) as demonstrated by 29% of Flag-EphB2 that coIP with EGFP-Myo1b coIP after genistein treatment (Fig. S1 C). Thus, the kinase activity of EphB2 is required for Myo1b–EphB2 interaction.
Activated EphB2 may also trigger Myo1b phosphorylation. Indeed, EGFP-Myo1b appeared to be tyrosine phosphorylated in cells expressing EphB2 but this phosphorylation decreased by 89% in cells expressing Flag-EphB2-KD and by 76% in cells treated with genistein (Fig. 1, A and F; and Fig. S1, B and C). In addition, EGFP-Myo1b-Tail was highly phosphorylated compared with the motor domain or full-length Myo1b in cells expressing Flag-EphB2 (Fig. 1, B and G). In contrast to what was observed with full-length Myo1b we could not detect phosphorylation in Myo1b-Tail when expressed with Flag-EphB2-KD, suggesting that Myo1b-Tail phosphorylation relies only on EphB2 kinase activity (Fig. 1, H and I). We confirmed that the level of Myo1b phosphorylation depends on the stimulation of the EphB receptors by analyzing Myo1b phosphorylation when endogenous EphB receptors were stimulated in the LS174T cells (Batlle et al., 2002). Tyrosine phosphorylation of EGFP-Myo1b increased with EphB phosphorylation in the LS174T cells stimulated with clustered ephrinB1-Fc (Fig. 1 J).
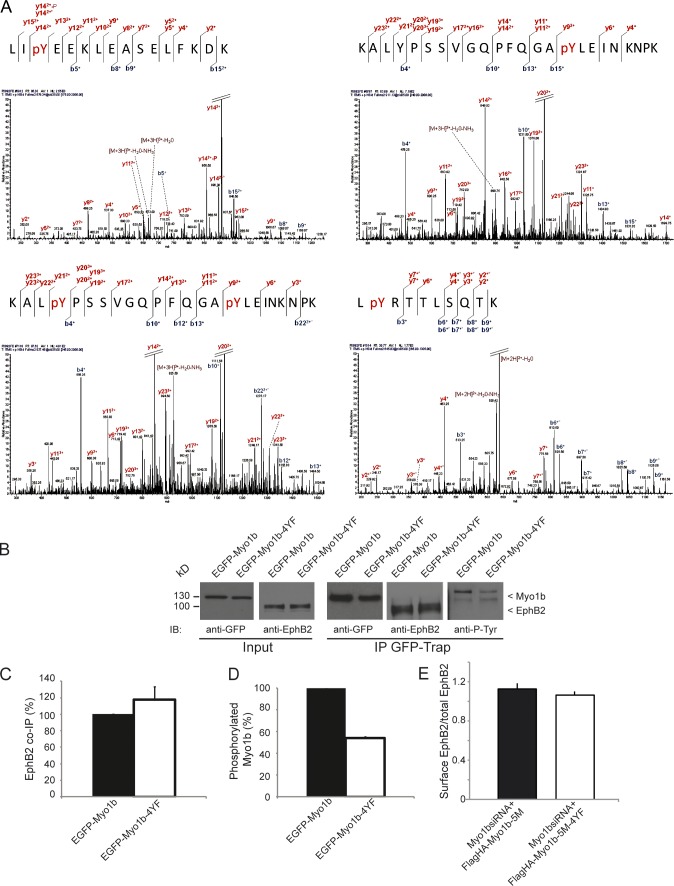
Next we identified the phosphorylated tyrosines of Myo1b when EGFP-Myo1b-Tail was coexpressed with Flag-EphB2 by mass spectroscopy. Myo1b-Tail was mainly phosphorylated on four tyrosine residues: Y909, Y926, Y938, and Y1049 (Fig. 2 A). We generated an EGFP-Myo1b mutant where we replaced these four residues with four phenylalanines (EGFP-Myo1b-4YF). Myo1b phosphorylation mutant still coIP with Flag-EphB2 and did not affect the delivery of EphB2 to the plasma membrane (Fig. 2, B, C, and E). However, phosphorylation of Myo1b mutant was decreased by 46% compared with EGFP-Myo1b when expressed with Flag-EphB2 (Fig. 2 D). The remaining phosphorylation of Myo1b mutant may reflect additional phosphorylation on its motor domain (see Fig. 1 B) and/or the appearance of new cryptic sites for tyrosine phosphorylation caused by insertion of the four mutations.
Figure 2.
Myo1b-4YF is less phosphorylated than Myo1b but coIP with EphB2 and does not alter EphB2 delivery to the plasma membrane. (A) Representative tandem mass spectra (simultaneous fragmentation of neutral loss product and precursor) for identification of EGFP-Myo1b-Tail phosphorylation sites after its immunoprecipitation with GFP-Trap from Hek293T cells also expressing Flag-EphB2. Liquid chromatography tandem mass spectrometry is shown for EGFP-Myo1b-Tail peptides, with the position of the phosphate group monophosphorylated LIpY909EEKLEASELFKDK (679.34(3+) m/z), KALYPSSVGQPFQGApY938LEINKNPK (911.13(3+) m/z), LpY1049RTTLSQTK (645.83(2+) m/z), and diphosphorylated KALpY926PSSVGQPFQGApY938LEINKNPK (937.45(3+) m/z). The fragmentation spectra shown are Lys-C–derived peptides from EGFP-Myo1b-Tail. The corresponding peptide sequences and observed ions obtained from the phosphopeptides are shown above the spectra. Tandem mass spectrum are labeled to show singly, doubly, and triply charged b and y ions, as well as ions corresponding to neutral losses of phosphoric acid (P), water (circles), and NH3 (asterisks); M, parent ion mass. (B) EGFP-Myo1b and EGFP-Myo1b-4YF were pulled down with GFP-Trap from Hek293T cell lysates (Input) also expressing Flag-EphB2 and analyzed by SDS-PAGE and immunoblotting with anti-GFP, anti-EphB2, and anti-phospho-tyrosine (anti-P-Tyr) antibodies. Note the decrease of phosphorylation of EGFP-Myo1b-4YF compared with EGFP-Myo1b. (C) The amount of EphB2 that coIPs with EGFP-Myo1b or EGFP-Myo1b-4YF was quantified, normalized to the amount of the recombinant proteins pulled down, and expressed as a percentage of the amount that coIP with EGFP-Myo1b. Data are shown as the mean of three experiments. Error bars represent ± SEM. (D) The amount of phosphorylated EGFP-Myo1b and EGFP-Myo1b-4YF was quantified, normalized to the amount of the recombinant proteins pulled down, and expressed as a percentage of phosphorylated EGFP-Myo1b. Data are shown as the mean of three experiments. Error bars represent ± SEM. (E) YFP-EphB2-HCT cells transfected with Myo1b siRNAs and plasmid encoding Flag-HA-Myo1b-5M or Flag-HA-Myo1b-5M-4YF were incubated with clustered ephrinB1-Fc. The ratio of fluorescence detected at the cell surface for bound ephrinB1 over the fluorescence detected for YFP-EphB2 corresponding to the total amount of receptors was calculated for both experimental conditions and expressed in arbitrary units. Data are shown as the mean of two experiments (n = 84 for cells transfected with Myo1bsiRNA and Flag-HA-Myo1b-5M and n = 79 for cells transfected with Myo1bsiRNA and Flag-HA-Myo1b-5M-4YF). Note that the difference is not significant.
Collectively, these observations suggest that conformational changes induced by the autophosphorylation of EphB2 are required for its interaction with Myo1b-Tail. Furthermore, EphB2 or a kinase that is activated by EphB2 kinase activity phosphorylates Myo1b-Tail on four tyrosine residues.
Myo1b regulates cell segregation that is mediated by EphB2–ephrinB1 signaling
Complementary expression of Eph receptors and ephrins has been implicated in boundary formation and segregation of different cell populations in many tissues during development and in adulthood (Rohani et al., 2011; Batlle and Wilkinson, 2012; Fagotto et al., 2013). Taking advantage of the capability of cells with reciprocal expression of EphB receptors and their ligand to segregate, we investigated the role of Myo1b in EphB2–ephrinB1 signaling (Mellitzer et al., 1999; Cortina et al., 2007; Poliakov et al., 2008). After transfection of plasmids, we isolated cellular pools expressing YFP-EphB2, Cherry-ephrinB1, or Cherry from the colorectal cancer cell line HCT116 and the human embryonic kidney cell line Hek293T that do not express endogenous EphB receptors and ephrinB ligands (Fig. S2, A, B, E, and F). We then verified that YFP-EphB2 and Cherry ephrinB1 were cell surface accessible and that YFP-EphB2 could be activated by its ligand in these cells (Fig. S2, C–F).
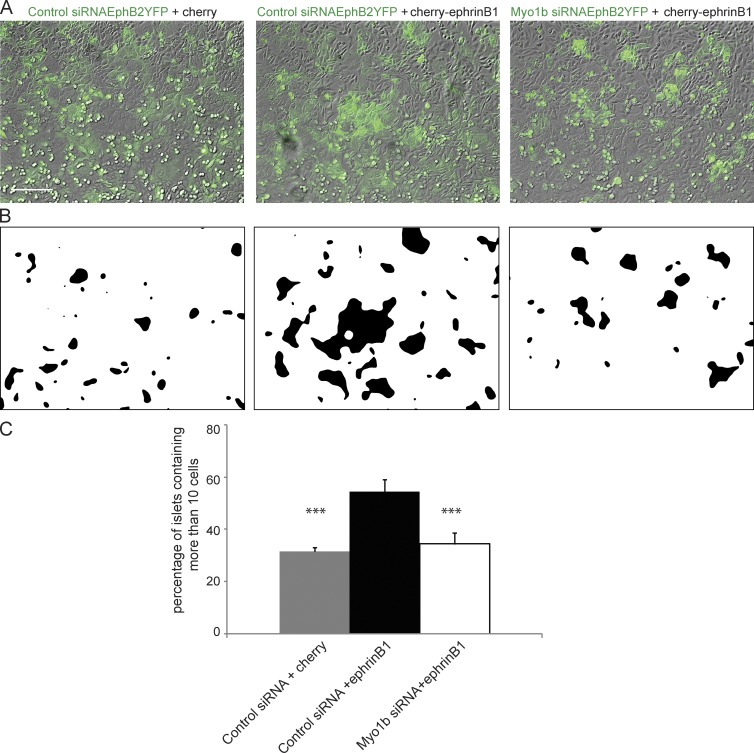
We then analyzed whether YFP-EphB2-HCT116 and Cherry-ephrinB1-HCT116 cells or Cherry-HCT116 cells segregated by counting the number of islets containing >10 YFP-EphB2-HCT116 cells after 48 h of co-culture. These islets represented >50% of the total number of islets observed when YFP-EphB2-HCT116 cells were cocultivated with Cherry-ephrinB1-HCT116 cells but only 30% when they were cocultivated with Cherry-HCT116 cells (Figs. 3 and S3), suggesting that YFP-EphB2 cells respond to ephrinB1 signaling generated by Cherry-ephrinB1 cells and segregate.
Figure 3.
Myo1b KD reduces the number of large islets formed by YFP-EphB2-HCT116 cells when they are cocultivated with Cherry-ephrinB1-HCT116 cells. (A and B) YFP-EphB2-HCT116 cells transfected with control or Myo1b siRNAs and cocultivated with Cherry-ephrinB1-HCT116 or Cherry-HCT116 cells were analyzed by phase-contrast and fluorescent microscopy. (A) The overlay of phase-contrast images (gray) and YFP images (green). (B) The same images as A after treatment as described in Fig. S3 to visualize the outlines of the islets formed by YFP-EphB2-HCT116 cells. Bar, 150 µm. (C) The number of cells per islets was quantified in the different experimental conditions based on the nuclear staining and as described in Fig. S3. Data are shown as the mean of three experiments. Error bars represent ± SEM. n = 191 and 346 islets for control siRNA-treated YFP-EphB2-HCT116 cells cocultivated with Cherry-HCT116 cells or with Cherry-ephrinB1-HCT116 cells, respectively. n = 198 islets for Myo1b siRNA-treated YFP-EphB2-HCT116 cells cocultivated with Cherry-ephrinB1-HCT116 cells. Paired Student’s t test was used to analyze the probabilities of these data. ***, P = 7.7 × 10−7 for control siRNA YFP-EphB2-HCT116 cells cocultivated with Cherry-HCT116 cells versus those cells cocultivated with Cherry-ephrinB1-HCT116. ***, P = 1.7 × 10−8 for Myo1b siRNA YFP-EphB2-HCT116 cells cocultivated with Cherry-ephrinB1-HCT116 cells versus control siRNA YFP-EphB2-HCT116 cells.
To determine whether Myo1b contributes to this EphB2–ephrinB1 cell segregation we knocked down its expression by transfecting Myo1b siRNA. Myo1b was barely detectable by immunoblotting after transfection with siRNA in the EphB2-HCT116 and EphB2-Hek293T cells (Fig. S4, A and C). Although Myo1b knockdown (KD) did not significantly affect the amount of EphB2 receptors at the surface of both cell types (Fig. S4, B and D), the percentage of islets with >10 cells after Myo1b KD was of the same range as that observed when YFP-EphB2-HCT116 cells were cocultivated with Cherry-HCT116 cells (Fig. 3), indicating that Myo1b contributes to cell segregation mediated by EphB2/ephrin signaling.
EphB2-induced cell repulsion requires non-muscle myosin 2 (NMM2) and Myo1b
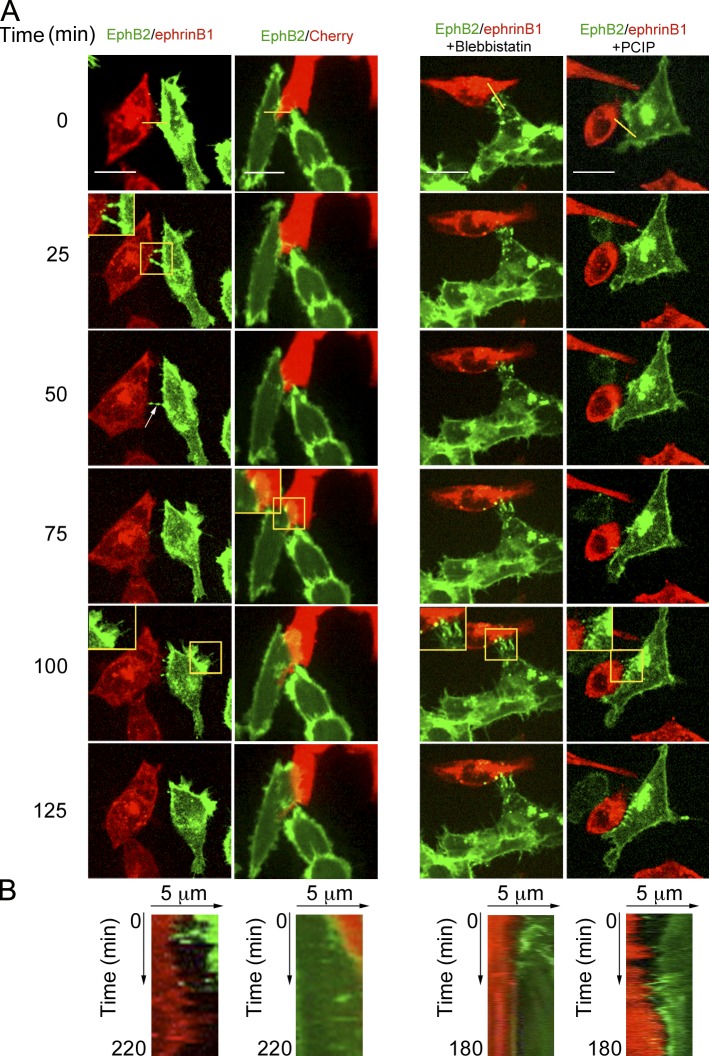
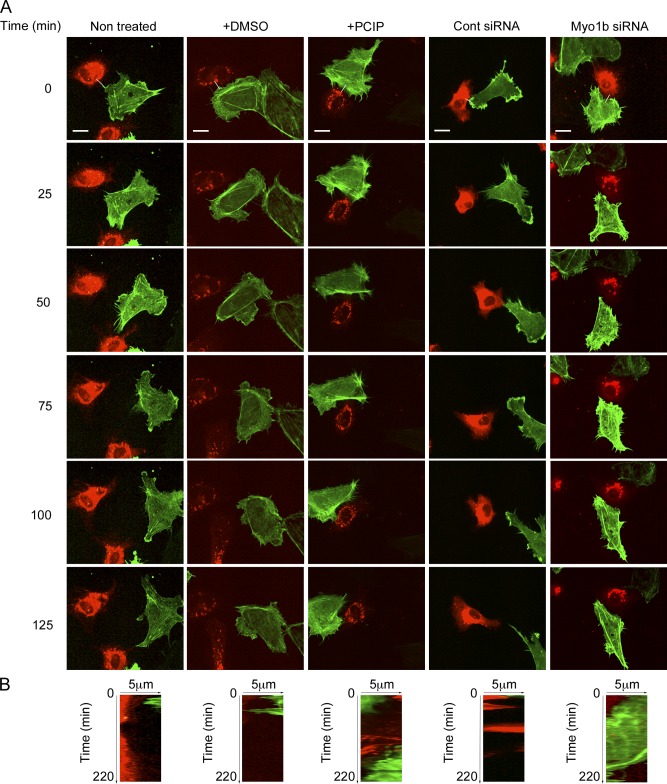
Because cell repulsion mediated by Eph/ephrin signaling has been proposed to be a possible mechanism for cell segregation (Batlle and Wilkinson, 2012), we next investigated by time-lapse confocal spinning microscopy whether Myo1b is required for cell repulsion. We observed repulsion between YFP-EphB2 and Cherry-ephrinB1 cells, whereas YFP-EphB2 cells overlap Cherry cells when they were cocultivated (Fig. 4 and Video 1). 44% of the YFP-EphB2 cells contacting Cherry-ephrinB1 cells repulsed (Table 1). Treatment with blebbistatin that inhibits NMM2 ATPase activity inhibits repulsion between YFP-EphB2 and Cherry-ephrinB1 cells compared with cells treated with the diluent (Table 1, Fig. 4, and Video 2). These observations confirm that EphB2 receptors expressed in HCT116 cells are ligand dependent activated and that NMM2 contributes to cell repulsion. In parallel we analyzed cell repulsion in primary human umbilical vein endothelial cells (HUVECs) that express endogenously EphB receptors and can be stimulated with ephrinB ligands (Groeger and Nobes, 2007). HUVECs expressing GFP-LifeAct to visualize actin filaments in the protrusions (Riedl et al., 2008) that contacted HUVEC-expressing Cherry ephrinB1 repulsed (Fig. 5 and Video 3). Repulsion in both HCT116 cells and HUVECs was inhibited by pentachloropseudilin (PCIP) that inhibits myosin 1 ATPase activity (Table 1, Figs. 4 and 5, and Videos 2 and 3; Martin et al., 2009; Chinthalapudi et al., 2011). Similarly to PCIP treatment, and although Myo1bKD was less efficient in HUVECs than in HCT116 cells Myo1b KD inhibited HUVEC and HCT116 cell repulsion (Table 1; Figs. 5 and 6; Fig. S4, A and J; and Videos 4 and 5). Thus, in addition to NMM2 motor activity Myo1b motor activity is necessary to achieve cell repulsion mediated by EphB2–ephrinB1 signaling.
Figure 4.
Myosin 1 and NMM2 control HCT116 cell repulsion. YFP-EphB2-HCT116 cells were cocultivated with Cherry-ephrinB1-HCT116 or Cherry-HCT116 cells and treated or not with blebbistatin or PCIP. Representative sequences of merged YFP and Cherry fluorescent focal planes at the base of the cells are shown (see also Videos 1 and 2). Bars, 15 µm. The yellow boxes mark the regions enlarged by 1.4 and shown in the insets. The yellow lines on the merged images at time 0 represent the region of the kymographs shown in B. (B) Kymographs at the interface of EphB2 and the ephrinB1- or Cherry-expressing cells from Videos 1and 2. EphB2 cells remain in contact with ephrinB1 cells treated with blebbistatin or PCIP or with nontreated Cherry cells.
Table 1. Number of repulsions per cell–cell contact.
| Cells | Treatments | Number of videos | Number of cell–cell contacts | Number of repulsions | Repulsions/cell–cell contacts |
|---|---|---|---|---|---|
| % | |||||
| HCT116 | No treatment | 22 | 36 | 16 | 44 |
| DMSO (0.3%) | 19 | 32 | 20 | 62 | |
| Blebbistatin | 10 | 25 | 0 | 0 | |
| PCIP | 12 | 35 | 1 | 3 | |
| Control siRNA (10 nM) | 21 | 47 | 20 | 42 | |
| Control siRNA (30 nM) | 20 | 44 | 22 | 50 | |
| Myo1b siRNA (10 nM) | 20 | 48 | 3 | 6 | |
| Fascin siRNA (30 nM) | 21 | 51 | 10 | 20 | |
| HUVECs | DMSO (0.1%) | 12 | 11 | 9 | 81 |
| PCIP | 17 | 23 | 5 | 22 | |
| Control siRNA (10 nM) | 24 | 19 | 16 | 84 | |
| Myo1b siRNA (10 nM) | 20 | 20 | 7 | 35 |
Figure 5.
Myosin 1 controls HUVEC repulsion. HUVECs expressing GFP-LifeAct transfected or not with control or Myo1b siRNA were cocultivated with HUVECs expressing Cherry-ephrinB1 and treated or not with PCIP or DMSO. Representative sequences of merged GFP and Cherry fluorescent focal planes at the base of the cells are shown (see also Videos 3 and 4). Bars, 15 µm. The white lines on the merged images at time 0 represent the region of the kymographs shown in B. (B) Kymographs at the interface of LifeAct and the ephrinB1 cells from Videos 3 and 4.
Figure 6.
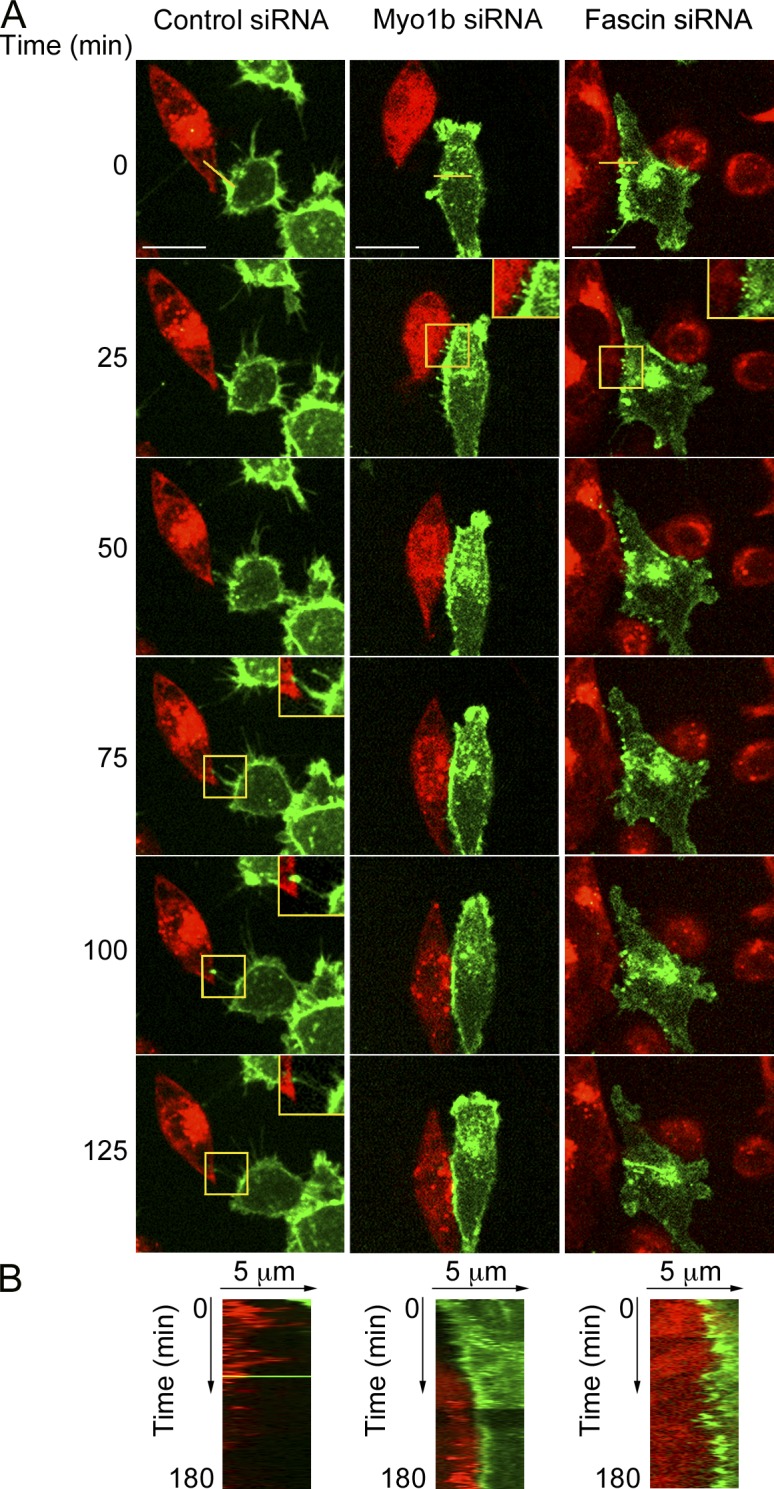
Myo1b and fascin are required for HCT116 cell repulsion. (A) YFP-EphB2-HCT116 cells transfected with control, Myo1b, or fascin siRNAs were cocultivated with Cherry-ephrinB1-HCT116. Representative sequences of merged YFP and Cherry fluorescent focal plane at the base of the cells illustrating the behavior of YFP-EphB2-HCT116 cells that contact Cherry-ephrinB1-HCT116 and correspond to Video 5 are shown. Bars, 15 µm. The yellow boxes mark the regions shown in the insets and enlarged by 1.4. The yellow lines on the merged images at time 0 represent the region of the kymographs shown in B. Filopodia with concentration of EphB2 at their tips are formed only in cells transfected with control siRNA. (B) Kymographs at the interface of EphB2 and the ephrinB1-expressing cells from Video 5. In the absence of Myo1b or fascin, EphB2 cells remain in contact with ephrinB1 cells.
Filopodia at the EphB2–ephrinB1 cell interface contribute to cell repulsion
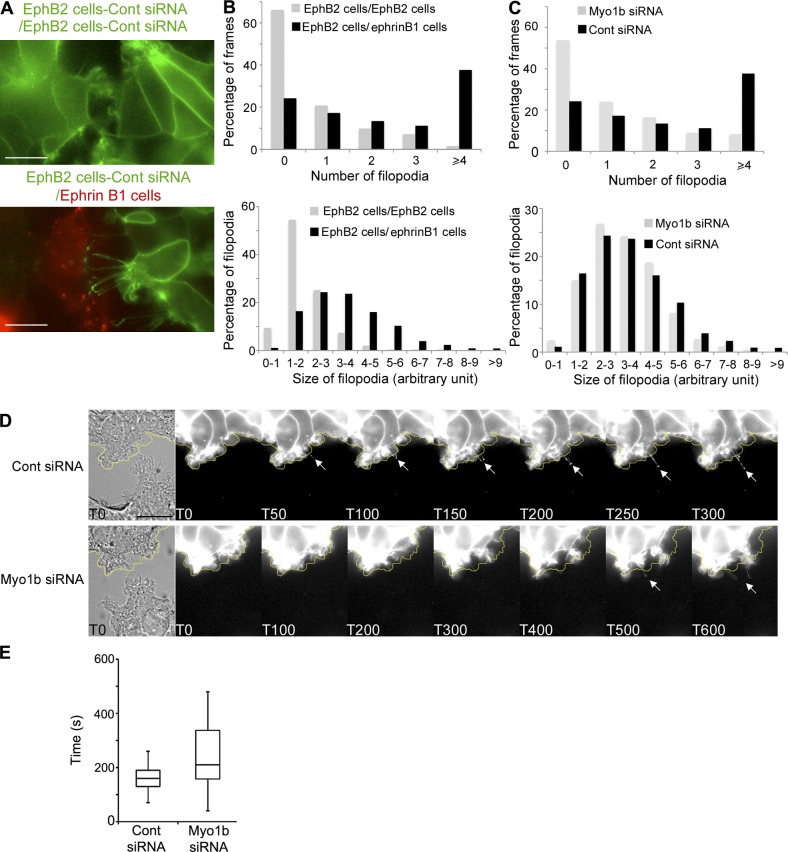
Previous studies indicate that EphB–ephrinB signaling induces important changes in cell morphology including formation of cell protrusions (Marston et al., 2003; Zimmer et al., 2003; Riedl et al., 2005; Moeller et al., 2006; Groeger and Nobes, 2007; Kayser et al., 2008; Astin et al., 2010). Similarly, we observed formation of different protrusions during cell repulsion. A new leading edge including lamellipodia and short filopodia was formed at the opposite of EphB2–ephrinB1 cell contact (Fig. 4 A, inset at 100 min; Fig. 6 A; and Videos 1 A and 5). In addition, long thin protrusions enriched in EphB2 appeared at EphB2–ephrinB1 cell–cell interface before the formation of the new leading edge (Fig. 4 A, inset at 25 min; and Video 1 A). The thin EphB2-enriched protrusions were formed in the presence of ephrinB1 cells but not in the presence of cells expressing only the Cherry tag, indicating that these protrusions are dependent on EphB2–ephrinB1 signaling (Fig. 4 and Video 1 A). Thin protrusions were also formed at the interface of GFP-LifeAct HUVECs and Cherry-ephrinB1-HUVEC before cell repulsion (Fig. S5 B and Video 3, A and B). To determine whether the thin protrusions at the EphB2–ephrinB1 cell interface contribute to cell repulsion, we knocked down fascin, which is one of the major constituents of filopodia. Fascin depletion in HCT116-YFP-EphB2 cells altered the morphology of these protrusions, indicating that they are filopodia (Fig. 6 and Video 5). Although fascin KD was less efficient than Myo1b KD it decreased by 60% the number of repulsions observed after cell–cell contact and compared with cells transfected with control siRNA (Fig. S4 G and Table 1). Because depletion of fascin did not affect the reorganization of NMM2 in fibers upon ephrinB1 stimulation (Fig. S4 I), this data suggests that EphB2-enriched filopodia, depending on fascin expression, contribute to cell repulsion independently on cell contraction and retraction fibers.
Myo1b is required to form filopodia at the EphB2–ephrinB1 cell interface
We observed that PCIP treatment or Myo1b depletion in EphB2-expressing HCT116 cells also altered EphB2-enriched filopodia at the EphB2–ephrinB1 cell interface (Fig. 4 A, inset at 100 min; Fig. 6 A, inset at 25 min; and Videos 2 and 5). However, the random migration of the two cell populations limits the number of contacts between EphB2 and ephrinB1cells and precluded a statistical analysis of the impact of Myo1b siRNA on the formation of EphB2-enriched filopodia (Table 1). To overcome this limitation and confirm the formation of filopodia at the EphB2 and ephrinB1 cell interface we cocultivated EphB2- and ephrinB1-Hek293T cells in Ibidi culture wells. After plating the two cell populations in two separated wells, the silicone barrier between the two wells was removed, allowing the two cell populations to migrate toward each other and thereby increasing the number of contacts between EphB2 and ephrinB1 cells observed per video. Similarly to the YFP-EphB2-HCT116 cells (Fig. 4), numerous and long filopodia displaying high concentrations of EphB2 at their tip were formed at the YFP-EphB2-Hek293T and Cherry-ephrinB1-Hek293T cell interface, whereas essentially lamellipodia were formed when YFP-EphB2 cells contacted other YFP-EphB2 cells (Fig. 7, A and B; and Video 6). The first contact between EphB2 and ephrinB1 cells was mediated by lamellipodia and the filopodia that were sometimes interconnected elongated out from these structures (Fig. 7, D and E; and Video 7). The number of filopodia formed when Myo1b was knocked down in the YFP-EphB2-Hek293T cells that touched Cherry-ephrinB1-Hek293T cells decreased considerably (Fig. 7 C). However, the remaining protrusions in Myo1b KD cells displayed similar size to those (Fig. 7 C) in control siRNA-transfected cells. Furthermore, the time observed between the first cell–cell contact via lamellipodia and the appearance of the first filopodia was more variable and significantly increased in the absence of Myo1b (Fig. 7, D and E; and Video 8), suggesting that Myo1b was required rather for the initiation of these filopodia and not for elongation.
Figure 7.
Myo1b is required to form filopodia at EphB2–ephrinB1 cell interface. (A) YFP-EphB2-Hek293T transfected with control siRNA cells were cultivated with Cherry-ephrinB1-Hek293T cells or themselves in Ibidi inserts and their behavior was monitored by time-lapse microscopy (Video 6). Representative frames for YFP-EphB2-Hek293T cells cultivated with other YFP-EphB2-Hek293T cells or with Cherry-ephrinB1-Hek293T cells are shown. Bars, 10 µm. (B) Distribution of the number of filopodia observed per frame normalized to the total number of frames analyzed (1 frame/5 min during 350 min) and distribution of the size of filopodia observed when YFP-EphB2-Hek293T cells transfected with control siRNA were facing themselves (11 videos) or Cherry-ephrinB1-Hek293T cells (11 videos). The number of frames with more than four filopodia and the length of the filopodia increase when YFP-EphB2-Hek293T cells contact ephrinB1-Hek293T cells. (C) Distribution of the number of filopodia observed per frame normalized to the total number of frames analyzed (1 frame/5 min during 350 min) and distribution of the size of filopodia observed in YFP-EphB2-Hek293T cells treated with control (11 videos) or Myo1b (12 videos) siRNAs and facing Cherry-ephrinB1-Hek293T cells. Myo1b KD increases the number of frames without filopodia but does not affect the size of the remaining filopodia. (D) YFP-EphB2-Hek293T cells treated with control or Myo1b siRNA were cultivated with Cherry-ephrinB1-Hek293T cells in Ibidi inserts and their behavior was monitored by time-lapse microscopy (Videos 7 and 8). Phase contrast of the first image and sequence of YFP fluorescent images illustrating the formation of filopodia after the first contact via lamellipodia are shown in the presence or absence of Myo1b. Arrows mark the elongating filopodia. Bar, 10 µm. (E) Time observed between the first contact formed by YFP-EphB2-Hek293T cells treated with control or Myo1b siRNA and Cherry-ephrinB1-Hek293T cells and the observation of the first filopodia was quantified and presented as box plots. Mann-Whitney test = 0.04.
Collectively, this live-cell imaging study suggests that Myo1b is required to initiate the formation of long, thin EphB2-enriched filopodia at the interface of ephrinB1 and EphB2 cells and Myo1b may thereby contribute to cell repulsion.
Myo1b motor activity and its phosphorylation controls NMM2 distribution and filopodia mediated by EphB2 signaling
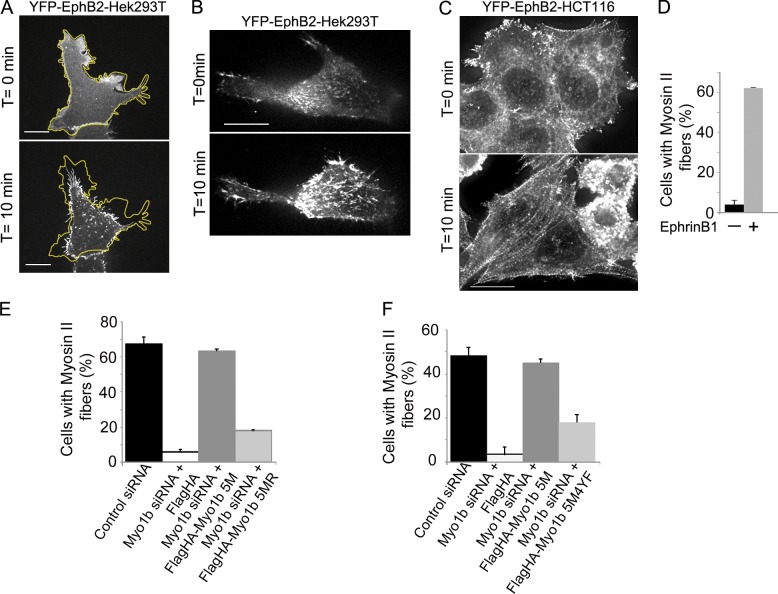
Myo1b can regulate filopodia involved in cell repulsion by regulating membrane tension and mechanically coupling the actin network involved in filopodia formation to the plasma membrane (Nambiar et al., 2009, 2010; Almeida et al., 2011). However, Myo1b may regulate membrane tension by also coupling cortical acto-NMM2 network to the plasma membrane (Diz-Muñoz et al., 2010) and thus contributes to cell repulsion by regulating NMM2 distribution. Taking advantage of clustered soluble recombinant ephrinB1-Fc that induces rapid morphological changes of EphB2 cells, we first analyzed whether Myo1b controls NMM2 distribution. YFP-EphB2-Hek293T cells contracted and formed protrusions when they were activated by clustered ephrinB1-Fc (Fig. 8 A and Video 9). Furthermore, NMM2 visualized by expressing the myosin regulatory light chain (MRLC)–RFP formed more fibers in YFP-EphB2-Hek293T cells treated by clustered ephrinB1-Fc than in nontreated cells (Fig. 8 B and Video 10). Similarly, 60% of YFP-EphB2-HCT116 cells displayed NMM2 fibers after ephrinB1-Fc treatment compared with 4% in nontreated cells (Fig. 8, C and D). Depletion of Myo1b in the YFP-EphB2-HCT116 cells decreased the number of cells showing alignments of NMM2 after EphB2 stimulations by clustered ephrinB1-Fc (Fig. 8, E and F) although it did not alter the level of serine-phosphorylated MRLC upon EphB2 stimulation (Fig. S4, E and F). The number of cells forming NMM2 fibers was rescued by expressing FlagHA-Myo1b-5M that was resistant to Myo1b siRNA (Fig. 8, E and F), confirming the specificity of our Myo1b siRNA for endogenous Myo1b (Almeida et al., 2011). We used our ability to rescue Myo1b KD with FlagHA-Myo1b-5M to determine whether the motor activity of Myo1b- and/or EphB2-dependent phosphorylation of Myo1b were required to form NMM2 fibers. We have previously designed a Myo1b rigor mutant by introducing the mutation N160A in the ATPase pocket of FlagHA-Myo1b-5M (FlagHA-Myo1b-5MR) and characterized it in vivo as in vitro (Almeida et al., 2011). Myo1b rigor failed to rescue NMM2 distribution after EphB2 stimulation of Myo1b KD YFP-EphB2-HCT116 cells (Fig. 8 E). Myo1b phosphorylation mutant expressed in cells depleted for the endogenous Myo1b was also unable to rescue the distribution of NMM2 (Fig. 8 F). Together these observations indicate that Myo1b motor activity and its EphB2-dependent phosphorylation control NMM2 distribution induced by EphB2–ephrinB1 signaling.
Figure 8.
NMM2 alignment induced by EphB2 stimulation requires Myo1b motor activity and tyrosine phosphorylation of its tail. (A) YFP-EphB2-Hek293T cells transfected with control siRNA were analyzed by live-cell imaging (Video 9). One frame before and one after 10 min of stimulation with clustered ephrinB1-Fc are shown. Bars, 5 µm. The yellow line outlines the cell periphery on the first frame before treatment. (B) YFP-EphB2-Hek293T cells, transfected with a plasmid encoding MRLC-RFP, were analyzed by live-cell imaging (Video 10) before and after stimulation with clustered ephrinB1-Fc. One frame before and one frame after 8 min of stimulation are shown. Bar, 5 µm. (C) YFP-EphB2-HCT116 cells were immunolabeled with anti-NMM2 antibodies and analyzed before or after 10 min of stimulation with clustered ephrinB1-Fc. Note the alignment of NMM2 after EphB2 stimulation. Bar, 6 µm. (D) YFP-EphB2-HCT116 cells transfected with Myo1b or control siRNAs were stimulated for 10 min or not with clustered ephrinB1-Fc and immunolabeled with anti-NMM2 antibodies, and the number of cells showing NMM2 alignment was quantified. Data are shown as the mean of three experiments (n = 1032 for control siRNA and 918 for Myo1b siRNA). Error bars represent ± SEM. (E and F) YFP-EphB2-HCT116 cells transfected with control siRNAs, Myo1b siRNA plus the empty Flag-HA plasmid (E and F), Myo1bsiRNA+Flag-HA-Myo1b5M (E and F), Myo1bsiRNA+Flag-HA-Myo1b5MR (E), or Myo1bsiRNA+Flag-HA-Myo1b5M4YF (F) were immunolabeled with anti-NMM2 antibodies and the number of cells showing NMM2 alignment was quantified. Data are shown as the mean of three experiments (E: n = 82 for control siRNA, 92 for Myo1b siRNA+Flag-HA, 74 for Myo1b siRNA+Flag-HA-Myo1b5M, and 74 for Myo1b siRNA+Flag-HA-Myo1b5MR; F: n = 70 for control siRNA, 57 for Myo1b siRNA+Flag-HA, 78 for Myo1b siRNA+Flag-HA-Myo1b5M, and 70 for Myo1b siRNA+Flag-HA-Myo1b5M4YF). Error bars represent ± SEM.
We next analyzed the protrusions formed after stimulation of YFP-EphB2 cells by clustered ephrinB1-Fc. These protrusions showed EphB2, actin filaments, and Myo1b (Figs. 8 A and 9 A, insets) and their number increased by 50% in YFP-EphB2-Hek293T cells treated for 10 min with clustered ephrinB1-Fc and compared with nonstimulated cells (Fig. 9 B). Myo1b KD decreased the number of protrusions after treatment of YFP-EphB2-HCT116 cells by clustered ephrinB1-Fc (Fig. 9, C and D), and this number was rescued by expressing FlagHA-Myo1b-5M (Fig. 9, C and D). In contrast, expression of Myo1b rigor mutant or Myo1b phosphorylation mutant failed to rescue several protrusions (Fig. 9, C and D), indicating that Myo1b motor activity and its EphB2-dependent phosphorylation control the formation of protrusions induced by EphB2 that was stimulated by ephrinB1-Fc.
Figure 9.
Filopodia mediated by EphB2 signaling required Myo1b motor activity and tyrosine phosphorylation of its tail. (A) YFP-EphB2-HCT116 cells stimulated or not with clustered ephrinB1-Fc were fixed and labeled with phalloidin and anti-Myo1b antibodies. Note the increase of protrusions labeled for EphB2, F-actin, and Myo1b after stimulation with clustered ephrinB1-Fc (see insets at higher magnification). Bars, 10 µM. (B) Mean number of protrusions observed per cell before and after stimulation of YFP-EphB2-Hek293T cells stimulated 10 min after ephrinB1-Fc. n = 44. (C and D) Membrane protrusions formed after 10 min of stimulation with clustered ephrinB1-Fc were quantified in YFP-EphB2-HCT116 cells transfected with control siRNA, Myo1b siRNA plus the empty Flag-HA plasmid (C and D), Myo1bsiRNA+Flag-HA-Myo1b5M (C and D), Myo1bsiRNA+Flag-HA-Myo1b5MR (C), or Myo1bsiRNA+Flag-HA-Myo1b5M4YF (D). Data are shown as the mean of the number of filopodia counted in three experiments. Error bars represent ± SEM. (C) n = 53 for control siRNA, 63 for Myo1b siRNA+Flag-HA, 75 for Myo1b siRNA+FlagHA-Myo1b-5M, and 58 for Myo1b siRNA+FlagHA-Myo1b-5MR. The probabilities of these data were analyzed with Paired Student’s t test. ***, P = 3 × 10−15 for Myo1b siRNA+Flag-HA and 1.4 × 10−12 for Myo1bsiRNA+FlagHA-Myo1b-5MR versus control siRNA-treated cells. (D) n = 59 for control siRNA, 58 for Myo1bsiRNA+FlagHA, 64 for Myo1b siRNA+FlagHA-Myo1b-5M, and 60 for Myo1b siRNA+Flag-HA-Myo1b-5M-4YF. ***, P = 2 × 10−20 for Myo1b siRNA+FlagHA and 2.5 × 10−13 for Myo1b siRNA+Flag-HA-Myo1b-5M-4YF–treated cells versus control siRNA-treated cells. (E) Sequence of fluorescent images of YFP-EphB2-Hek293T cells stimulated with clustered ephrinB1-Fc that illustrate filopodia formation growing from the cell edge and retraction fibers left behind after the retraction of the cell edge. Bars, 1 µm. The cell edge is marked by a yellow dashed line. (F) The increase of the number of protrusions after 10 min of stimulation with ephrinB1-Fc of YFP-EphB2-Hek293T cells transfected with control, Myo1b, or fascin siRNAs or treated with DMSO or 100 µM CK666 for 30 min at 37°C has been calculated from the total number of protrusions counted before and after stimulation and expressed as a percentage of the increase of the number of protrusions calculated in cells transfected with control siRNA. Data are shown as the mean of the number of protrusions counted for 44, 40, 32, and 39 control siRNA–, Myo1b siRNA–, fascin siRNA–, and CK666-treated cells, respectively. Error bars represent ± SEM. The probabilities of these data were analyzed with Paired Student’s t test: ***, P < 0.001 for protrusions formed in Myo1b siRNA–treated cells versus control siRNA–treated cells and CK666-treated cells versus DMSO-treated cells. (G) Filopodia growing from the cell edges (Fig. 8 A) after 10 min of stimulation with ephrinB1-Fc of YFP-EphB2-Hek293T cells transfected with control, Myo1b, or fascin siRNAs or treated with DMSO or CK666 were quantified and expressed as a percentage of the filopodia counted in cells transfected with control siRNA. Data are shown as the mean of the number of protrusions counted for 44, 40, 32, and 39 control siRNA–, Myo1b siRNA–, fascin siRNA–, and CK666-treated cells, respectively. Error bars represent ± SEM. The probabilities of these data were analyzed with Paired Student’s t test: ***, P < 0.001 for filopodia formed in Myo1b or fascin siRNA–treated cells versus control siRNA–treated cells and CK666-treated cells versus DMSO-treated cells.
The protrusions induced by ephrinB1-Fc treatment may correspond to filopodia, similar to those observed at the EphB2–ephrinB1 cell interface. They may also correspond to retraction fibers. To differentiate between these possibilities we considered protrusions as potential filopodia when they elongated out of YFP-EphB2-Hek293T cells (Fig. 9 E and Video 9). To confirm that these protrusions were indeed filopodia, we probed whether their formation can be altered by fascin KD and CK666, an inhibitor of the Arp2/3 complex that controls the polymerization of dendritic actin network required to form filopodia (Yang and Svitkina, 2011). Fascin was barely detectable in Hek293T cell KD for fascin and did not affect the redistribution of NMMII after EphB2 stimulation (Fig. S4, H and I). The number of protrusions that elongated out of the cells decreased by 60% after fascin KD and 73% after CK666 treatment, indicating that a part of the protrusions induced by ephrinB1-Fc treatment are indeed filopodia (Fig. 9 G). We then analyzed whether Myo1b regulates the protrusions and/or the filopodia induced by clustered ephrinB1-Fc treatment. Myo1b KD slightly decreased the total number of protrusions and up to 50% of the filopodia after ephrinB1 treatment (Fig. 9, F and G). Thus Myo1b controls both retraction fibers and filopodia driven by EphB2–ephrinB1 signaling.
Discussion
A prerequisite to understand the mechanisms by which myosins 1 control membrane remodeling is the identification of myosin 1 membrane binding partners. Here we report the interaction of Myo1b with the EphB2 receptors. Myo1b interacts directly or indirectly with EphB2 via its Tail. This interaction requires EphB2 kinase activity. We observed a partial codistribution of endogenous EphB receptors with endogenous Myo1b in LS174T cells (unpublished data). However, EphB2 activation did not affect this codistribution, but it induced an important increase in the phosphorylation of EGFP-Myo1b (unpublished data; Fig. 1 J). Thus, Myo1b–EphB2 interaction is independent of EphB2 stimulation but required its kinase activity, whereas Myo1b tyrosine phosphorylation depends on the stimulation of the EphB2 receptors. Given the basal phosphorylation of EphB2 receptors in the cellular pools and in LS174T cells it is likely that EphB2 forms autophosphorylated dimers with the juxtamembrane domain conformation, allowing Myo1b binding to EphB2. Stimulation of EphB2 may induce the formation of trimmers and tetramers that may increase Myo1b tyrosine phosphorylation and its EphB2-mediated function (Wybenga-Groot et al., 2001; Schaupp et al., 2014).
To our knowledge this is the first time that experimental data demonstrate that Eph receptors can activate the function of an unconventional myosin beside myosin II contractibility. A serine or threonine phosphorylation of myosin 1 motor is required for chemotactic stimulation in amoeba or function of myosin 1 in yeast (Bement and Mooseker, 1995; Gliksman et al., 2001; Oberholzer et al., 2002). However, this phosphorylation site is replaced in nearly all metazoan myosins 1 by glutamic or aspartic acid (Bement and Mooseker, 1995). Our observations suggest that metazoan myosin 1 tail phosphorylation may lead to conformational changes that could regulate myosin 1 motor activity.
We demonstrated that in addition to interacting with EphB2 receptors, Myo1b is required for the function of EphB2 forward signaling. One of the main functions of EphB2–ephrinB1 signaling involves cell repulsion to form and maintain tissue boundaries during embryonic development. We showed that Myo1b regulates cell segregation mediated by EphB2–ephrinB1 signaling and cell repulsion. We confirmed that repulsive signal generated by contact between EphB2 and ephrinB1 cells involved NMM2 and membrane protrusions (Astin et al., 2010). In addition, we show that at least a part of these protrusions are EphB2-enriched filopodia that are formed at the EphB2–ephrinB1 cell interface and required for cell repulsion. These filopodia are formed in two different cell types and in primary cell culture expressing endogenous EphB receptors. Following the suggestion of Yang and Svitkina (2011) to name all the thin protrusions filopodia in outlining their function, we propose to name these EphB2-enriched filopodia, which sense ephrinB1 ligands at the surface of the neighboring cell and lead to cell repulsion, repulsive filopodia.
Recent experimental evidence suggests that Myo1b controls directed cell migration during development of zebrafish embryo (Diz-Muñoz et al., 2010). We now report that Myo1b also controls cell repulsion. Myo1b motor activity and Myo1b EphB2-dependent phosphorylation both being required for the EphB2-mediated redistribution of NMM2 suggest that Myo1b regulates cell repulsion by controlling NMM2 distribution. However, Myo1b motor activity and Myo1b EphB2-dependent phosphorylation are also both required for the formation of EphB2-mediated repulsive filopodia, suggesting that Myo1b regulates both the formation of repulsive filopodia and cell contraction mediated by EphB2–ephrinB1 signaling.
Although Myo1b controls membrane trafficking along the endocytic and exocytic pathways (Raposo et al., 1999; Salas-Cortes et al., 2005; Almeida et al., 2011) it is rather unlikely that Myo1b controls cell contraction and repulsive filopodia by controlling the delivery of EphB2 receptors to the plasma membrane. Indeed, depletion of Myo1b did not significantly affect the amount of EphB2 receptors associated with the plasma membrane (Fig. S3). The need of Myo1b motor activity and its EphB2-dependent phosphorylation for the redistribution of NMM2 but not for MRLC phosphorylation suggests that, similarly to its role for coupling the actin cytoskeleton to organelle membrane, Myo1b may couple mechanically the contractile acto-NMM2 fibers to the plasma membrane after its phosphorylation by EphB2 (Almeida et al., 2011; Yamada et al., 2014). Myo1b may also couple actin polymerization to the plasma membrane and thereby transduces the force generated by actin polymerization to the membrane to form repulsive filopodia. Alternatively, Myo1b may control the dendritic actin network required for the formation of repulsive filopodia because we have previously shown that Myo1b controls the Arp2/3-dependent dendritic actin network in the region of the trans Golgi network (Almeida et al., 2011).
Altogether, this work reveals a new function for Myo1b, which is to act as an effector of EphB2–ephrinB1 forward signaling to control the formation of repulsive filopodia and acto-NMM2 fibers driving cell repulsion, an important mechanism for cell segregation to maintain tissue border during embryonic development and in the adult hood. Studying Myo1b function during embryonic development and tissue patterning in the adult when these processes involve EphB2 signaling is an exciting future challenge.
Materials and methods
Antibodies and reagents
The following antibodies were used: anti-Myo1b polyclonal antibody (1:1,000 for Western blot; 1:50 for immunofluorescence; Almeida et al., 2011); anti-EphB2 polyclonal antibody (0.5 µg/ml; R&D Systems); anti-GFP mouse monoclonal antibody (1:1,000 for Western blot; Roche); anti-phosphotyrosine mouse monoclonal antibody (1:1,000 for Western blot; clone 4G10); anti-HA monoclonal antibody (1:400; 3f10; Roche); anti-tubulin monoclonal antibody (1:5,000; Sigma-Aldrich); anti-pMLC (ser19; 1:1,000; Cell Signaling Technology); anti-NM myosin heavy chain II (polyclonal antibody; 1:2,000; Covance); and Alexa- and horseradish peroxidase–conjugated secondary antibodies (1:500; Invitrogen; 1:5,000; Jackson ImmunoResearch Laboratories, Inc.; 1:500; Molecular Probes). Alexa-conjugated phalloidin was used to detect F-actin (1:500; Invitrogen).
Plasmids
EGFP-Myo1b, FlagHA-Myo1b-5M, and FlagHA-Myo1b-5MR plasmids generated by site-directed mutagenesis of the plasmid encoding FlagHA-Myo1b-5M with a N160A mutation have been reported previously (Almeida et al., 2011). EGFP-Myo1b motor and EGFP-Myo1b-Tail have been generated by cloning at EcoRI and XbaI or BglII and SalI sites of pEGFP-C1 (Takara Bio Inc.). DNA fragments were generated by PCR on rat Myo1b cDNA (accession no. NM_053986) with 5′ primers ATGGCCAAGAAGGAGGTAAAAT or ATTGGCCATCAAGACCTTACCTA and 3′ primers CTGATATCGCTTTTGTTGCGCGT or CCTCACTTAAGGGACAGCGACTT, respectively. GST-Myo1b-Tail was generated by cloning at BamHI and SalI sites of pGEX4T (GE Healthcare), the same DNA fragment as to generate EGFP-Myo1b-Tail. Flag-EphB2 (pJK1), flag-EphB2-Kinase–deficient (Lys660-Arg)(pJK2), and flag-EphB2-YFP (pJK12) plasmids were a gift from R. Klein (Max Planck Institute of Neurobiology, Martinsried, Germany; Zimmer et al., 2003); Cherry-ephrinB1 was generated by PCR cloning of ephrinB1 from ECFP-HA–ephrinB1 (pJK30; Zimmer et al., 2003) in M-p-cherry-C1 plasmid; plasmid encoding LifeAct-GFP and LifeAct-Cherry was a gift from G. Montagnac (Institut Curie, Paris, France; Riedl et al., 2008); and plasmids encoding MRLC-RFP were a gift from E. Paluch (Medical Research Council Laboratory for Molecular and Cell Biology, London, England; Charras et al., 2006).
siRNA
In-house–designed Myo1b siRNA (5′-GCTTACCTGGAAATCAACAAG-3′) and a nontargeting sequence designed by Dharmacon used as control siRNA have been previously described (Almeida et al., 2011). Fascin siRNA (5′-GAGCAUGGCUUCAUCGGCU-3′) was described previously (Vignjevic et al., 2007).
Cell culture
Hek293T cells and HCT116 cells were cultured at 37°C and 10% CO2 in DMEM supplemented with 10% fetal bovine serum. HUVECs (Promocell) were cultured at 37°C and 5% CO2 in endothelial cell growth medium 2 (Promocell) on flasks coated with 0.2% gelatin from bovine skin (Sigma-Aldrich) in PBS. For immunofluorescence labeling, YFP-EphB2-Hek293T cells were grown on glass coverslips coated for 2 h with 0.02 mg/ml laminin (Sigma-Aldrich) and YFP-EphB2-HCT116 cells were grown on glass coverslips coated with collagen (0.05 mg/ml). For co-culture, 0.15 × 106 YFP-EphB2-HCT116 cells were cocultivated with 0.15 × 106 Cherry-ephrinB1-HCT116 or Cherry-HCT116 cells on 12-mm-diameter coverslips coated with 0.05 mg/ml collagen. For live-cell imaging, 18 × 103 Cherry-ephrinB1 and YFP-EphB2 Hek293T cells were cultivated independently in two silicone inserts (Ibidi) on glass-bottomed dishes (Fluorodish; World Precision Instruments) coated with 0.02 mg/ml laminin. 48 h later the separation was removed and 16–20 h later the behavior of YFP-EphB2-Hek293T cells was monitored by time-lapse microscopy. For confocal microscopy 0.2 × 106 YFP-EphB2-HCT116 and Cherry-ephrinB1-HCT116 or Cherry-HCT116 cells were cocultivated on glass-bottomed dishes coated with collagen. For stimulation with ephrinB1-Fc, ephrinB1-Fc chimera (R&D Systems) were cross-linked with goat anti–human IgG Fc and used at 5 µg/ml (Jackson ImmunoResearch Laboratories, Inc.; ratio 2:1).
Transfection and selection of stable cellular pools
For recombinant protein expression, Hek293T or HCT116 cells were transfected with complementary DNA using effectene (QIAGEN), lipofectamine, or lipofectamine LTX (Invitrogen) and analyzed 24 h later. After transfection with Flag-EphB2-YFP (pJK12), Cherry-ephrinB1, or Cherry plasmids and culture in a selective medium, cellular pools were isolated with FACSVantage. For protein KD expression, YFP-EphB2-Hek293T and YFP-EphB2-HCT116 cells were transfected with 10 or 30 nM of specific or control siRNAs for Myo1b or Fascin KD, respectively, using Lipofectamine RNAiMax (Invitrogen) and analyzed after 48, 72, or 96 h. For recombinant protein expression in HUVECs, 0.6 × 106 cells were electroporated with 3 and 2 µg of plasmid encoding GFP-LifeAct and Cherry-ephrinB1, respectively, and 30 pmol siRNA using Amaxa HUVEC Nucleofector kit (Lonza) according to the manufacturer’s protocol; plated on glass-bottomed dishes coated with 0.2% gelatin from bovine skin (Sigma-Aldrich) in PBS; and analyzed after 24 h.
Drug treatments
100 µm CK666 (ChemDiv, Inc.) in 0.1% DMSO, 50 µm blebbistatin (Sigma-Aldrich) in 0.3% DMSO, and 1 µm PCIP (gift from H.-J. Knölker, Technische Universität, Dresden, Germany) in 0.1% DMSO were incubated for 30 min at room temperature. Control experiments were monitored with 0.1% DMSO for CK666 and PCIP treatments and 3% DMSO for blebbistatin treatment.
Immunoblotting
Proteins separated by SDS-PAGE were transferred to nitrocellulose membranes and processed for immunoblotting using Super Signal West Pico Chemiluminescent substrate (Thermo Fisher Scientific). Images of immunoblots were captured with an imager (Fuji LAS-3000; Fujifilm) or with CL-XPosure film (Thermo Fisher Scientific) within the linear range and quantified by densitometry using the Analyze gels function in ImageJ.
Immunoprecipitations and GFP-Trap pull-down
6 × 106 cells were incubated for 30 min on ice in 1 ml of lysis buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM ATP, 10% glycerol, 1 mM DTT, 0.1% protease inhibitor cocktail (Sigma-Aldrich), 0.15 mM sodium orthovanadate, and 1% phosphatase inhibitor cocktail (Sigma-Aldrich). For immunoprecipitation, 1 ml of supernatant collected after 20 min of centrifugation of the cell lysates at 20,000 g was incubated for 1 h with 15 µl of protein G–Sepharose beads (Thermo Fisher Scientific). After removing the beads, 2 µg of antibodies were added and incubated overnight at 4°C. Then 15 µl of protein G–Sepharose beads were added for 2 h at 4°C. For GFP-Trap pull-down, 25 µl of GFP-Trap beads (ChromoTek) were incubated directly with 1 ml of supernatants for 2 h at 4°C. Protein G–Sepharose or GFP-Trap beads were washed five times with lysis buffer, resuspended in 25 µl of Lemmli buffer supplemented with β-mercaptoethanol, and boiled 5 min before analysis by immunoblotting.
Immunofluorescence labeling
Cells were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100 or saponin before antibody incubation using standard procedures. Nuclei were labeled by DAPI (Sigma-Aldrich) and F-actin by fluorescent phalloidin. To detect EphB receptors and ephrin at the cell surface, cells were incubated for 30 min at 4°C with 5 µg/ml of recombinant mouse EphB2-Fc or ephrinB1-Fc (R&D Systems) cross-linked with goat anti–human IgG Fc. The protein complexes were then detected with fluorescently labeled donkey anti–goat antibodies.
Image acquisition
Image acquisition and image analysis were performed on workstations of the PICT-IBiSA Lhomond Imaging facility of Institut Curie. Epifluorescence microscopy (Fig. 3) was performed with a microscope (DM6000B; Leica) equipped with 10× NA 0.3 and 20× NA 0.7 dry objectives and with 63× NA 1.32 and 100× NA 1.4 oil immersion objectives and a CoolSNAP HQ camera (Photometrics). 3D deconvolution microscopy (Figs. 8 C and 9 A) was performed using an upright microscope (Eclipse 80i; Nikon) equipped with a 100× NA 1.4 oil immersion objective, a piezo-electric driver mounted underneath the objective, and a CoolSNAP HQ2 camera. Z series of images were taken at 0.2-µm increments. Deconvolution was performed by the 3D deconvolution Metamorph module with the fast iterative constrained point spread function–based algorithm 44. Video microscopy (Fig. 7 and Videos 5, 6, and 7) was performed with an Eclipse inverted microscope (Nikon) equipped with a 40× NA 1.3 oil immersion objective, CoolSNAP HQ2 camera, under 5% CO2, and at 37°C. Spinning-disc confocal microscopy (Figs. 8 B and 9 E and Videos 8 and 9) was performed with a spinning-disc head (CSU-22; Yokogawa Electric Corporation) on a microscope (TE-2000U; Nikon) equipped with a 100× NA 1.4 oil immersion objective and a CoolSNAP HQ2 camera, a NanoScanZ piezo focusing stage (Prior Scientific), and a motorized scanning stage (Marzhauser) or a spinning-disc head (CSU-X1; Yokogawa Electric Corporation) on a microscope (TI; Nikon) equipped with a 40× NA 1.3 oil immersion objective and an intensifier electron microscopy charge coupled device camera (Figs. 4, 5, and 6 and Videos 1, 2, 3, and 4) under 5% CO2 and at 37°C. Kymographs were generated using the software Fiji. Confocal imaging (Fig. 8 A) was performed with a confocal microscope (A1r; Nikon) equipped with a 100× NA 0.75 dry immersion objective. These microscopes were steered with Metamorph 7.1 (Universal Imaging Corporation).
Online supplemental material
Fig. S1 shows the interaction of GST-Myo1b-tail with EphB2-coated beads and the inhibition of EphB2 and Myo1b tyrosine phosphorylation in the presence of genistein. Fig. S2 shows expression level of YFP-EphB2 and Cherry-ephrinB1 in Hek293T and HCT116 cellular pools, their availability at the surface, and the ability of the EphB2 receptor to be stimulated in both cellular pools. Fig. S3 shows the image treatment to quantify the number of cells per islet formed in the repulsion cell experiments. Fig. S4 shows efficiency of Myo1b KD by siRNA in YFP-EphB2-HCT116 and YFP-EphB2-HEK293T cellular pools as well as the absence of effect of Myo1b depletion on EphB2 expression at the cell surface. Fig. S5 shows the formation of filopodia depending in GFP-LifeAct–expressing HUVECs when cocultivated with Cherry-ephrinB1-HUVEC. Video 1 (related to Fig. 4) shows YFP-EphB2-HCT116 cell behavior when in contact with Cherry-ephrinB1-HCT116 (A) or Cherry-HCT116 cells (B). Video 2 (related to Fig. 4) shows YFP-EphB2-HCT116 cell behavior after treatment with 50 µM blebbistatin, in the presence of 1 µM PCIP or DMSO when they contact Cherry-ephrinB1-HCT116 cells. Video 3 (related to Fig. 5) shows a HUVEC expressing EGFP-LifeAct when they contact Cherry-ephrinB1-HUVEC treated or not with DMSO and PCIP. Video 4 (related to Fig. 5) shows a HUVEC expressing EGFP-LifeAct transfected with control or Myo1b siRNA when they contact Cherry-ephrinB1-HUVEC. Video 5 (related to Fig. 6) shows control siRNA, Myo1b, and fascin siRNA–transfected YFP-EphB2-HCT116 cell behavior when in contact with Cherry-ephrinB1-HCT116 cells. Video 6 (related to Fig. 7 A) shows the behavior of YFP-EphB2-HEK293T cells cultivated on Ibidi culture inserts in front of other YFP-EphB2-HEK293T cells (A) or reaching Cherry-ephrinB1-HEK293T cells (B). Video 7 (related to Fig. 7 D) shows the behavior of YFP-EphB2-HEK293T cells transfected with control siRNA and cultivated on Ibidi culture inserts with Cherry-ephrinB1-HEK293T cells to visualize the initiation of filopodia. Video 8 (related to Fig. 7 D) shows the behavior of YFP-EphB2-HEK293T cells transfected with Myo1b siRNA cultivated on Ibidi culture inserts with Cherry-ephrinB1-HEK293T cells to visualize the initiation of filopodia. Video 9 (related to Fig. 8 A) shows the behavior of YFP-EphB2-HEK293T cells treated with EphrinB1-Fc. Video 10 (related to Fig. 8 B) shows the behavior of YFP-EphB2-HEK293T–expressing MRLC-RFP and treated with EphrinB1-Fc. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501018/DC1.
Supplementary Material
Acknowledgments
We thank V. Fraisier, F. Waharte, and L. Sengmanivong from the PICT-IBiSA Lhomond Imaging Facility of Institut Curie for their expertise in microscopy. We thank Dr. R. Klein for giving us the plasmids encoding EphB2 and ephrinB1 and Dr. Vanderberg for critically reading the manuscript.
This work was supported by Institut Curie, Centre National de la Recherche Scientifique (CNRS), and Association pour la Recherche sur le Cancer foundation (grant SFI2012205571). The E. Coudrier group belongs to the CNRS consortium CellTiss and to the Labex CelTisPhyBio 11-LBX-0038.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- coIP
- coimmunoprecipitated
- HUVEC
- human umbilical vein endothelial cell
- KD
- knockdown
- MRLC
- myosin regulatory light chain
- NMM2
- non-muscle myosin 2
- PCIP
- pentachloropseudilin
References
- Almeida C.G., Yamada A., Tenza D., Louvard D., Raposo G., and Coudrier E.. 2011. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat. Cell Biol. 13:779–789. 10.1038/ncb2262 [DOI] [PubMed] [Google Scholar]
- Astin J.W., Batson J., Kadir S., Charlet J., Persad R.A., Gillatt D., Oxley J.D., and Nobes C.D.. 2010. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat. Cell Biol. 12:1194–1204. 10.1038/ncb2122 [DOI] [PubMed] [Google Scholar]
- Batlle E., and Wilkinson D.G.. 2012. Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harb. Perspect. Biol. 4:a008227 10.1101/cshperspect.a008227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E., Henderson J.T., Beghtel H., van den Born M.M., Sancho E., Huls G., Meeldijk J., Robertson J., van de Wetering M., Pawson T., and Clevers H.. 2002. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 111:251–263. 10.1016/S0092-8674(02)01015-2 [DOI] [PubMed] [Google Scholar]
- Bement W.M., and Mooseker M.S.. 1995. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil. Cytoskeleton. 31:87–92. 10.1002/cm.970310202 [DOI] [PubMed] [Google Scholar]
- Charras G.T., Hu C.K., Coughlin M., and Mitchison T.J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490. 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinthalapudi K., Taft M.H., Martin R., Heissler S.M., Preller M., Hartmann F.K., Brandstaetter H., Kendrick-Jones J., Tsiavaliaris G., Gutzeit H.O., et al. 2011. Mechanism and specificity of pentachloropseudilin-mediated inhibition of myosin motor activity. J. Biol. Chem. 286:29700–29708. 10.1074/jbc.M111.239210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortina C., Palomo-Ponce S., Iglesias M., Fernández-Masip J.L., Vivancos A., Whissell G., Humà M., Peiró N., Gallego L., Jonkheer S., et al. 2007. EphB–ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 39:1376–1383. 10.1038/ng.2007.11 [DOI] [PubMed] [Google Scholar]
- Diz-Muñoz A., Krieg M., Bergert M., Ibarlucea-Benitez I., Muller D.J., Paluch E., and Heisenberg C.P.. 2010. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol. 8:e1000544 10.1371/journal.pbio.1000544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I.R., Renne T., Gertler F.B., and Nobes C.D.. 2007. Ena/VASP proteins mediate repulsion from ephrin ligands. J. Cell Sci. 120:289–298. 10.1242/jcs.03333 [DOI] [PubMed] [Google Scholar]
- Fagotto F., Rohani N., Touret A.S., and Li R.. 2013. A molecular base for cell sorting at embryonic boundaries: contact inhibition of cadherin adhesion by ephrin/Eph-dependent contractility. Dev. Cell. 27:72–87. 10.1016/j.devcel.2013.09.004 [DOI] [PubMed] [Google Scholar]
- Genander M., Halford M.M., Xu N.J., Eriksson M., Yu Z., Qiu Z., Martling A., Greicius G., Thakar S., Catchpole T., et al. 2009. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell. 139:679–692. 10.1016/j.cell.2009.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliksman N.R., Santoyo G., Novak K.D., and Titus M.A.. 2001. Myosin I phosphorylation is increased by chemotactic stimulation. J. Biol. Chem. 276:5235–5239. 10.1074/jbc.M008319200 [DOI] [PubMed] [Google Scholar]
- Groeger G., and Nobes C.D.. 2007. Co-operative Cdc42 and Rho signalling mediates ephrinB-triggered endothelial cell retraction. Biochem. J. 404:23–29. 10.1042/BJ20070146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie F., and Yamaguchi Y.. 2002. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat. Neurosci. 5:1117–1118. 10.1038/nn964 [DOI] [PubMed] [Google Scholar]
- Kayser M.S., Nolt M.J., and Dalva M.B.. 2008. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron. 59:56–69. 10.1016/j.neuron.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. 2012. Eph/ephrin signalling during development. Development. 139:4105–4109. 10.1242/dev.074997 [DOI] [PubMed] [Google Scholar]
- Komaba S., and Coluccio L.M.. 2010. Localization of myosin 1b to actin protrusions requires phosphoinositide binding. J. Biol. Chem. 285:27686–27693. 10.1074/jbc.M109.087270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston D.J., Dickinson S., and Nobes C.D.. 2003. Rac-dependent trans-endocytosis of ephrinBs regulates Eph–ephrin contact repulsion. Nat. Cell Biol. 5:879–888. 10.1038/ncb1044 [DOI] [PubMed] [Google Scholar]
- Martin R., Jäger A., Böhl M., Richter S., Fedorov R., Manstein D.J., Gutzeit H.O., and Knölker H.J.. 2009. Total synthesis of pentabromo- and pentachloropseudilin, and synthetic analogues—allosteric inhibitors of myosin ATPase. Angew. Chem. Int. Ed. Engl. 48:8042–8046. 10.1002/anie.200903743 [DOI] [PubMed] [Google Scholar]
- Mazerik J.N., and Tyska M.J.. 2012. Myosin-1A targets to microvilli using multiple membrane binding motifs in the tail homology 1 (TH1) domain. J. Biol. Chem. 287:13104–13115. 10.1074/jbc.M111.336313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell R.E., and Tyska M.J.. 2010. Leveraging the membrane – cytoskeleton interface with myosin-1. Trends Cell Biol. 20:418–426. 10.1016/j.tcb.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G., Xu Q., and Wilkinson D.G.. 1999. Eph receptors and ephrins restrict cell intermingling and communication. Nature. 400:77–81. 10.1038/21907 [DOI] [PubMed] [Google Scholar]
- Moeller M.L., Shi Y., Reichardt L.F., and Ethell I.M.. 2006. EphB receptors regulate dendritic spine morphogenesis through the recruitment/phosphorylation of focal adhesion kinase and RhoA activation. J. Biol. Chem. 281:1587–1598. 10.1074/jbc.M511756200 [DOI] [PubMed] [Google Scholar]
- Mohamed A.M., Boudreau J.R., Yu F.P., Liu J., and Chin-Sang I.D.. 2012. The Caenorhabditis elegans Eph receptor activates NCK and N-WASP, and inhibits Ena/VASP to regulate growth cone dynamics during axon guidance. PLoS Genet. 8:e1002513 10.1371/journal.pgen.1002513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R., McConnell R.E., and Tyska M.J.. 2009. Control of cell membrane tension by myosin-I. Proc. Natl. Acad. Sci. USA. 106:11972–11977. 10.1073/pnas.0901641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R., McConnell R.E., and Tyska M.J.. 2010. Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell. Mol. Life Sci. 67:1239–1254. 10.1007/s00018-009-0254-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U., Marcil A., Leberer E., Thomas D.Y., and Whiteway M.. 2002. Myosin I is required for hypha formation in Candida albicans. Eukaryot. Cell. 1:213–228. 10.1128/EC.1.2.213-228.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A., Cotrina M.L., Pasini A., and Wilkinson D.G.. 2008. Regulation of EphB2 activation and cell repulsion by feedback control of the MAPK pathway. J. Cell Biol. 183:933–947. 10.1083/jcb.200807151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Cordonnier M.N., Tenza D., Menichi B., Dürrbach A., Louvard D., and Coudrier E.. 1999. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol. Biol. Cell. 10:1477–1494. 10.1091/mbc.10.5.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D., Stauffer T., Chen W., Shen K., Guo S., York J.D., Sheetz M.P., and Meyer T.. 2000. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton–plasma membrane adhesion. Cell. 100:221–228. 10.1016/S0092-8674(00)81560-3 [DOI] [PubMed] [Google Scholar]
- Riedl J.A., Brandt D.T., Batlle E., Price L.S., Clevers H., and Bos J.L.. 2005. Down-regulation of Rap1 activity is involved in ephrinB1-induced cell contraction. Biochem. J. 389:465–469. 10.1042/BJ20050048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani N., Canty L., Luu O., Fagotto F., and Winklbauer R.. 2011. EphrinB/EphB signaling controls embryonic germ layer separation by contact-induced cell detachment. PLoS Biol. 9:e1000597 10.1371/journal.pbio.1000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Cortes L., Ye F., Tenza D., Wilhelm C., Theos A., Louvard D., Raposo G., and Coudrier E.. 2005. Myosin Ib modulates the morphology and the protein transport within multi-vesicular sorting endosomes. J. Cell Sci. 118:4823–4832. 10.1242/jcs.02607 [DOI] [PubMed] [Google Scholar]
- Schaupp A., Sabet O., Dudanova I., Ponserre M., Bastiaens P., and Klein R.. 2014. The composition of EphB2 clusters determines the strength in the cellular repulsion response. J. Cell Biol. 204:409–422. 10.1083/jcb.201305037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M.P. 2001. Cell control by membrane–cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2:392–396. 10.1038/35073095 [DOI] [PubMed] [Google Scholar]
- Tang N., and Ostap E.M.. 2001. Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 11:1131–1135. 10.1016/S0960-9822(01)00320-7 [DOI] [PubMed] [Google Scholar]
- Tolias K.F., Bikoff J.B., Kane C.G., Tolias C.S., Hu L., and Greenberg M.E.. 2007. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc. Natl. Acad. Sci. USA. 104:7265–7270. 10.1073/pnas.0702044104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M.J., and Nambiar R.. 2010. Myosin-1a: A motor for microvillar membrane movement and mechanics. Commun. Integr. Biol. 3:64–66. 10.4161/cib.3.1.10141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Schoumacher M., Gavert N., Janssen K.P., Jih G., Laé M., Louvard D., Ben-Ze’ev A., and Robine S.. 2007. Fascin, a novel target of β-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 67:6844–6853. 10.1158/0008-5472.CAN-07-0929 [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot L.E., Baskin B., Ong S.H., Tong J., Pawson T., and Sicheri F.. 2001. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 106:745–757. 10.1016/S0092-8674(01)00496-2 [DOI] [PubMed] [Google Scholar]
- Xu Q., Mellitzer G., Robinson V., and Wilkinson D.G.. 1999. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature. 399:267–271. 10.1038/20452 [DOI] [PubMed] [Google Scholar]
- Yamada A., Mamane A., Lee-Tin-Wah J., Di Cicco J., Prévost C., Lévy D., Joanny J.-F., Coudrier E., and Bassereau B.. 2014. Catch-bond behaviour facilitates membrane tubulation by a non-processive myosin 1b. Nat. Commun. 5:3624. [DOI] [PubMed] [Google Scholar]
- Yang C., and Svitkina T.. 2011. Filopodia initiation: Focus on the Arp2/3 complex and formins. Cell Adhes. Migr. 5:402–408. 10.4161/cam.5.5.16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M., Palmer A., Köhler J., and Klein R.. 2003. EphB–ephrinB bi-directional endocytosis terminates adhesion allowing contact mediated repulsion. Nat. Cell Biol. 5:869–878. 10.1038/ncb1045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.