BLM helicase facilitates telomere replication by resolving G-quadruplex structures that can form in the G-rich repeats during leading strand synthesis.
Abstract
Based on its in vitro unwinding activity on G-quadruplex (G4) DNA, the Bloom syndrome–associated helicase BLM is proposed to participate in telomere replication by aiding fork progression through G-rich telomeric DNA. Single molecule analysis of replicated DNA (SMARD) was used to determine the contribution of BLM helicase to telomere replication. In BLM-deficient cells, replication forks initiating from origins within the telomere, which copy the G-rich strand by leading strand synthesis, moved slower through the telomere compared with the adjacent subtelomere. Fork progression through the telomere was further slowed in the presence of a G4 stabilizer. Using a G4-specific antibody, we found that deficiency of BLM, or another G4-unwinding helicase, the Werner syndrome-associated helicase WRN, resulted in increased G4 structures in cells. Importantly, deficiency of either helicase led to greater increases in G4 DNA detected in the telomere compared with G4 seen genome-wide. Collectively, our findings are consistent with BLM helicase facilitating telomere replication by resolving G4 structures formed during copying of the G-rich strand by leading strand synthesis.
Introduction
Mammalian chromosome ends are capped by telomeres, specialized structures composed of hundreds to thousands of short, tandem repeat sequences complexed with several proteins, including the telomere-specific shelterins (de Lange, 2005). The noncoding repetitive telomeric DNA provides a buffer against genetic information loss. Telomeres also protect against deleterious repair activities by preventing chromosome ends from being perceived as broken or damaged DNA by the DNA repair machinery (de Lange, 2005).
Efficient replication of telomeric DNA is essential for the maintenance of telomere structure and function. The bulk of telomere DNA is duplicated by conventional semiconservative DNA replication (for review see Gilson and Géli, 2007). The structural organization and repetitive nature of telomeres present potential challenges to the replication machinery. Mammalian telomere termini are organized into protective structures termed t-loops, where a lariat structure is formed by invasion of the terminal-most telomere DNA, which is single-stranded, into the double-stranded region of the telomere (Griffith et al., 1999; Doksani et al., 2013). Consequently, a requisite step in the replication of telomeres is the disassembly of t-loops. Other structural elements, including secondary structures derived from the repetitive G-rich telomere sequence, could also act as potential obstacles to replication forks. In particular, the G-rich sequence can fold into G-quadruplex (G4) DNA, a stacked structure composed of highly stable planar G-quartet rings stabilized via Hoogsteen bonds (for review see Burge et al., 2006). G4 structures pose challenges to replication, requiring specialized helicases to unwind G4 DNA to maintain genomic stability of G4 motifs (for review see Maizels and Gray, 2013).
The Bloom syndrome helicase (BLM) and the Werner syndrome helicase (WRN) are two helicases that have been proposed to aid in the resolution of potential replication-impeding structures formed during telomere replication. Both of these RecQ-family helicases possess robust in vitro G4 unwinding activity (Opresko, 2008; Chavez et al., 2009; Singh et al., 2012; Croteau et al., 2014). BLM has been shown to suppress the generation of replication-dependent abnormal telomeric structures termed fragile telomeres (Sfeir et al., 2009), whereas WRN has been shown to suppress defects in telomere lagging strand synthesis (Crabbe et al., 2004), demonstrating roles for these helicases in telomere replication. Significantly, WRN does not suppress telomere fragility (Sfeir et al., 2009), indicating a functional distinction between BLM and WRN, and suggesting that BLM may play a more extensive role in telomere replication.
Additional lines of evidence support the involvement of BLM in telomere replication. BLM binds to the telomere-specific shelterin proteins TRF1, TRF2, and POT1, and its helicase activity can be stimulated by TRF2 and POT1 in vitro (Opresko et al., 2002, 2005; Lillard-Wetherell et al., 2004). However, the observation of elevated levels of telomere-to-telomere associations in BLM-deficient cells (Lillard-Wetherell et al., 2004), along with recent evidence showing BLM localization to ultra-fine bridges that can form between telomeres in anaphase (Barefield and Karlseder, 2012), suggests a recombination-based role for BLM in telomere maintenance. Moreover, data directly demonstrating BLM participation in telomere copying in vivo is lacking.
To elucidate the contribution of BLM to telomere replication, we examined the replication of individual telomeres using a single molecule approach. In previous studies using this approach, we demonstrated in human (Drosopoulos et al., 2012) and mouse cells (Sfeir et al., 2009) that telomere replication can initiate from origins within the telomere, resulting in replication of the G-rich strand by both the leading and lagging strand replication machinery. Here, we show that BLM aids in copying the G-rich strand during leading strand replication by forks progressing from telomeric origins. In addition, we demonstrate that BLM can suppress G4 structure formation both genome-wide and specifically at telomeres. Importantly, our findings are consistent with the BLM helicase facilitating telomere replication by resolving G4 structures that can form in the G-rich repeats during leading strand synthesis.
Results
We have previously applied an individual molecule approach termed single molecule analysis of replicated DNA (SMARD) to examine the replication of specific, individual human telomeres (Drosopoulos et al., 2012). Asynchronous cultures of cells are sequentially pulse-labeled with two distinguishable nucleoside analogues. Individual molecules of DNA replicated during the pulses are isolated and immunostained, then imaged by fluorescence microscopy, revealing labeling patterns. Regions replicated during the first pulse appear red and regions replicated during the second pulse appear green (Fig. S1). Using locus-specific FISH probes that produce asymmetric signal patterns (“blue bar codes”), specific pulse-labeled molecules of interest are identified and oriented. To identify telomere sequences within the labeled molecules of interest, a telomere-specific peptide nucleic acid (PNA) probe is used. Analysis of the labeling patterns allows for the position, direction, and density of the replication forks in the population of replicated molecules to be determined (Norio and Schildkraut, 2001). This information also establishes the sites of replication initiation and termination in a given genomic region. Moreover, the average speed of the DNA replication forks can be calculated from the observed proportions of differently labeled molecules of the same size (Norio and Schildkraut, 2001).
To investigate the role of BLM helicase in telomere replication, we used SMARD to examine the effect of BLM deficiency on telomere replication. BLM-deficient mouse fibroblasts were used that contained a homozygous BLM mutation that leads to expression of a truncated polypeptide lacking all catalytic domains, consisting of only the N-terminal 296 amino acids of the 1,416-residue full-length protein (Luo et al., 2000). As heterozygous cells express significant levels of wild-type BLM protein (62% of wild type cells; Luo et al., 2000; McDaniel et al., 2003), these cells served as BLM-proficient cells in these studies. Murine cells were used because the average telomere length in these cells is significantly longer (>2.5×) than in most human cells. This affords greater opportunity to detect events in the telomere by SMARD. As a model locus, we used a 320-kb genomic segment from the end of chromosome arm 14q (Ch14q) containing the telomere and associated subtelomeric region. This segment was selected based on the availability of a restriction endonuclease site distal enough from the chromosome end to allow a telomere-containing segment of suitable size for SMARD (140–400 kb) to be excised.
BLM-deficient cells exhibit slowed replication fork movement through telomere repeats
Our SMARD analysis revealed key features of the replication program of the telomere-containing Ch14q segment common to both BLM-proficient and -deficient cells. We observed initiation events in the telomere in both cell types (Figs. 1 A and 2). These were primarily detected in molecules as a red tract that does not extend beyond the telomere or extends only a few kilobases beyond the telomere/subtelomere junction (Figs. 1 A and 2), as origins that gave rise to these forks must have been located within the telomere. Due to the length of the red pulse (4 h), examples of telomeric initiation events where a red tract is flanked by green tracts (Figs. S1 B and 2) were rarely observed. In addition, almost all of the Ch14q molecules displayed continuous red labeling extending from the telomere into the subtelomere, indicating the presence of an origin within the red-labeled region. Because only one example of a subtelomeric origin was observed in the telomere-containing right half of the segment compared with nine observed telomeric initiations (Figs. 1 and 2), this strongly suggests that telomere replication in molecules with continuous red labeling from the right (telomere) end of the segment was more often initiated within the telomere. Collectively, these findings indicated that the Ch14q telomere appears to be preferentially replicated by forks originating from within the telomere. An important consequence of initiating telomere replication within the telomere is that G-rich strand replication would be accomplished by both the leading and lagging strand replication machinery (Figs. 1 and 2). We have previously found that initiations can occur in mammalian telomeres, but these were infrequent events (Sfeir et al., 2009; Drosopoulos et al., 2012). After examining four specific individual telomeres, we determined that human telomere replication initiates predominantly in the subtelomere (Drosopoulos et al., 2012). Similarly, initiations were detected at low frequency in mouse telomeres, suggesting that telomere replication initiates primarily in the subtelomere (Sfeir et al., 2009). However, in our previous mouse studies using SMARD, smaller fragments of total genomic telomeric DNA were analyzed. Because these fragments were anonymous segments from the total population of telomeres, it was not possible to obtain information regarding the replication program of specific telomeres. Moreover, these telomere fragments contained substantially shorter associated subtelomere DNA compared with the Ch14q segment, so forks emerging from the telomere could escape detection. Thus, our results for the Ch14q telomere are the first demonstration of a telomere that appears to be replicated primarily from origins within the telomere.
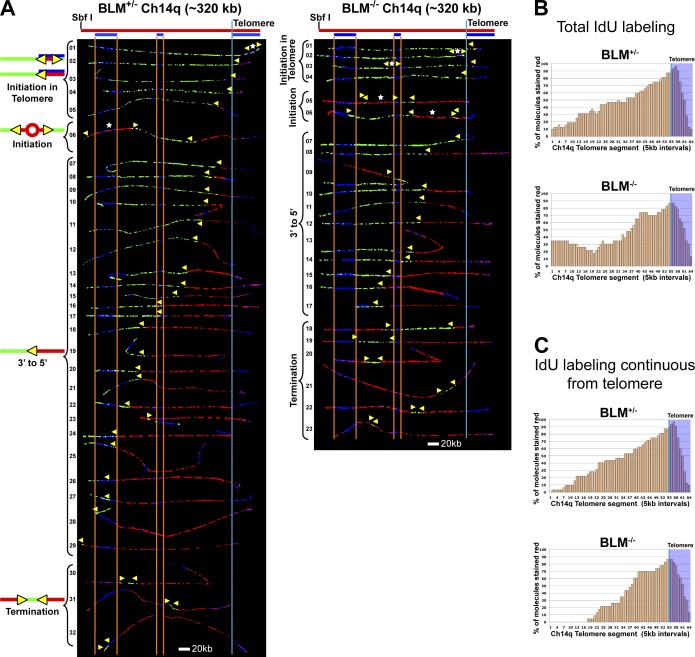
Figure 1.
Replication fork movement is slowed through the telomere in BLM-deficient cells. (A) SMARD analysis of a segment of Ch14q containing the telomere from BLM-proficient (left) and BLM-deficient (right) cells. Alignments of replicated molecules are shown, collected from 6–8 independent samples stretched on slides. Of the 136 BLM-proficient and 118 BLM-deficient fully labeled (IdU, CldU, or IdU/CldU) molecules collected, only the subset of molecules fully labeled with both IdU (red) and CldU (green; IdU/CldU) are shown. A map of the 14q locus is depicted above each alignment, with the positions of the FISH probes (blue bars below) used to identify and orient the molecules indicated. Vertical orange lines mark positions of the ends of the subtelomeric FISH signals used to align the molecules. The boundary between the subtelomere and telomere is delineated by a vertical blue line. Yellow arrowheads mark sites of transition from IdU to CldU incorporation and indicate the direction of fork progression at the moment of transition during replication (polarity of fork direction assigned relative to the polarity of the telomere G-rich strand of the segment). Initiation events (red tracts flanked by green) are seen in both telomeres and subtelomeres. Telomeric initiation events are detected in molecules where the red tract of the 3′ to 5′ fork does not extend out of the telomere or extends only a few kilobases out from the telomere (BLM-proficient, molecules 1–5; BLM-deficient, molecules 1–4). White stars indicate the centers of initiations. (B) Replication profiles, histograms of the percentage of molecules containing IdU per 5 kb interval along the segment, are shown for each alignment. The telomere portion of the segment is shaded in blue and the telomere–subtelomere boundary is indicated by a blue vertical line. (C) Replication profiles of the portion of each molecule continuously IdU labeled from telomere.
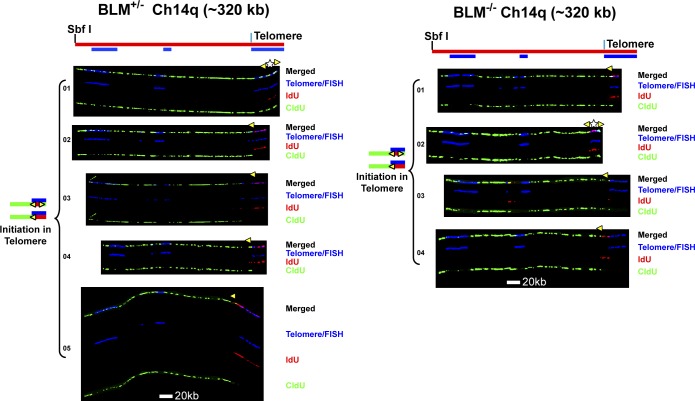
Figure 2.
Replication initiation events are detected in the Ch14q telomere in BLM-proficient and BLM-deficient cells. Replicated molecules from BLM-proficient and BLM-deficient cells that display telomeric initiation events (presented in Fig. 1) are shown. Images of individual color channels depicting telomere PNA and locus-specific FISH probes (blue), IdU (red), and CldU (green) signals are shown below the merged images to allow better visualization of telomeric initiation events partially obscured by overlapping (blue) staining in the merged images. A map of the 14q locus is depicted above the molecule images, with the positions of the telomeric PNA and FISH probes (blue bars below) used to identify and orient the molecules indicated. Symbols are as in Fig. 1. White stars indicate definitive centers of initiations, where a red signal is surrounded by green (see Fig. S1 B).
One difference seen between the BLM-proficient and -deficient cells was that continuous red (iododeoxyuridine [IdU]) labeling from the telomere end generally extended further from the telomere in BLM-proficient cells compared with BLM-deficient cells (Fig. 1 C). This indicates that forks originating in the telomere had not progressed as far in BLM-deficient cells, implying that forks emerging from the telomere travel at a reduced rate compared with BLM-proficient cells. This is further supported by the observation that when the junction from red to green signal is scored relative to the position of the short middle FISH probe, an equal number of molecules have the red/green junction at the left or on the right of the middle FISH probe. In contrast, in BLM-deficient cells the ratio is skewed to only 26% of the molecules with the red/green junction on the left as compared with 74% of the molecules on the right of the middle FISH probe. There were also more molecules containing termination events (a green tract flanked by red tracts; Fig. S1) in the BLM-deficient cells (6) compared with the BLM-proficient cells (3). Another difference between the two cell lines was that the sites of subtelomeric initiations that were seen were located closer to the telomere in the BLM-deficient cells. In addition, the ratios of the limited subtelomeric initiation events observed (3 in 23 BLM-deficient molecules vs. 1 in 32 BLM-proficient molecules) suggested that BLM-deficient cells have a higher frequency of subtelomeric initiation compared with BLM-proficient cells.
From the proportions of fully IdU-, chlorodeoxyuridine (CldU)-, and IdU/CldU-labeled molecules, we calculated the average fork rates for both cell lines. We found that forks traveled at a slightly reduced rate in the BLM-deficient cells compared with the BLM-proficient cells (Table 1). This agreed with previous observations that BLM-deficient human fibroblasts derived from Bloom syndrome patients exhibit reduced genomic replication fork speed (Hand and German, 1975; Rao et al., 2007). In addition to obtaining fork rates for an entire segment, the fork rate for a smaller region within the same segment can be derived from SMARD data on the full segment (Norio and Schildkraut, 2004). Therefore, we determined the average fork rate for the terminal-most 100 kb of the 320 kb full-length fragment, where a larger percentage of the segment was telomere DNA. When we calculated rates for the terminal-most 100 kb, we found a greater difference in fork rates between BLM-deficient and -proficient cells than in rates seen for the complete 14q segment (Table 1). These data suggest that replication forks move slower through telomere repeats in BLM-deficient cells.
Table 1. Average speed of replication forks.
| Cell type/segment | BLM+/− Ch14q (320 kb) | BLM−/− Ch14q (320 kb) | BLM+/− Ch14q PhenDC3 (320 kb) | BLM−/− Ch14q PhenDC3 (320 kb) | BLM+/− Igh PhenDC3 (235 kb) | BLM−/− Igh PhenDC3 (235 kb) |
|---|---|---|---|---|---|---|
| NR | 48 | 50 | 71 | 49 | 55 | 57 |
| NG | 56 | 45 | 68 | 57 | 82 | 62 |
| NRG | 32 | 23 | 43 | 27 | 38 | 36 |
| Total forks | 38 | 37 | 50 | 53 | 38 | 36 |
| NRG100 | 9 | 8 | 11 | 13 | – | – |
| Total forks100 | 10 | 11 | 13 | 16 | – | – |
| Average fork speed (kb/min; full segment) | 2.95 ± 0.17 | 2.54 ± 0.12 | 3.00 ± 0.06 | 2.01 ± 0.14 | 2.73 ± 0.49 | 2.59 ± 0.1 |
| Average fork speed (kb/min; terminal 100 kb) | 3.50 ± 0.24 | 2.67 ± 0.13 | 3.61 ± 0.07 | 2.06 ± 0.15 | – | – |
Fork speeds were determined from fully IdU (red) and/or CIdU (green) substituted molecules. The number of red (NR), green (NG), and red-green (NRG) substituted molecules obtained for each segment is shown. The average speed per replication fork (kb/min) was calculated using the following equation: average kb/min = [length of segment (kb)/Td (min)]/average number of replication forks in the segment, where average number of replication forks in the segment = total forks/NRG molecules and Td (min) = the time required to duplicate a particular segment. Td (min) was determined using the following equation: Td (min) = Tp1 × NRG/(NR + NRG) or Td (min) = Tp2 × NRG/(NG + NRG) (Norio and Schildkraut, 2001), where Tp1 and Tp2 = length of time (240 min) of the red and green labeling pulses, respectively; NRG = the number of molecules fully stained in both red and green; and NR or NG = the number of molecules fully stained in red or green (Figs. S3, S4, and S5). Fork speeds and standard deviations were determined from averages of values obtained from NR and NG (n = 2–4). The duplication time for the terminal 100 kb of the telomeric segment (which included the telomere) was calculated using the following equation: Td = Tp1 × NRG100/(NR + NRG) or Td = Tp2 × NRG100/(NG + NRG) (Norio and Schildkraut, 2001, 2004), where NRG100 = the number of molecules fully stained in both red and green in the terminal 100 kb of the telomeric segment containing the telomere.
Slowdown of telomeric replication forks by aphidicolin results in origin activation in the subtelomere
We observed that a greater percentage of replication forks progress further from the telomere in BLM-proficient cells compared with BLM-deficient cells. There was also an increase in the percentage of origins that were detected in the subtelomere of BLM-deficient cells. One consequence of replication fork slowing is the activation of less frequently fired (dormant) origins, to allow for timely completion of replication. This suggested that fork slowdown in BLM-deficient cells results in subtelomeric dormant origin firing. To confirm that slowing of forks emerging from the telomere results in activation of subtelomeric dormant origins, we treated BLM-proficient cells with low levels of aphidicolin to reduce replication fork speed. Previous studies have shown that reduction of fork speed by aphidicolin induces dormant origins in mammalian cells, leading to shorter inter-origin distances (Courbet et al., 2008).
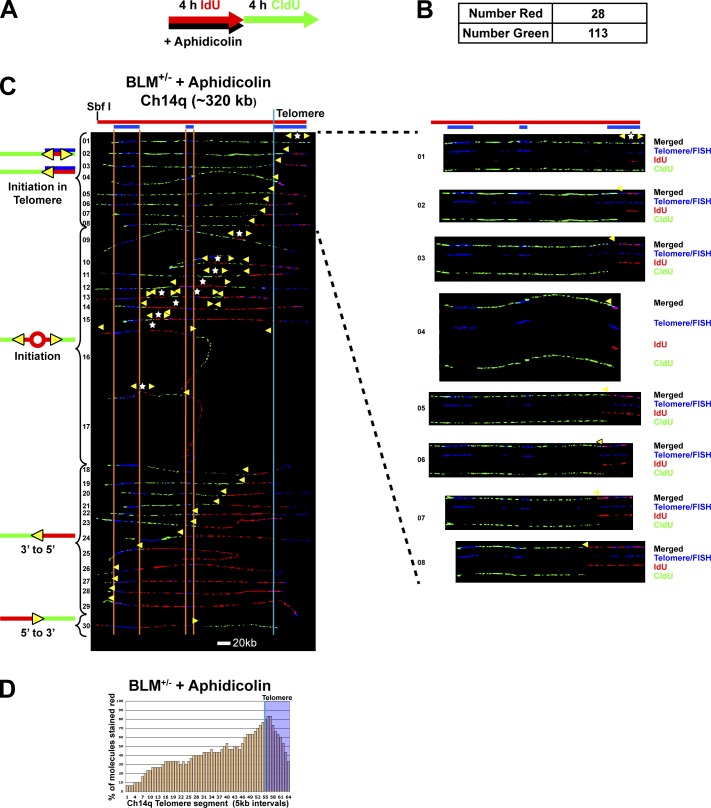
We performed SMARD on cells treated with aphidicolin (110 nM) during the IdU labeling pulse followed by CldU pulsing in the absence of aphidicolin (Fig. 3). Because recovery from aphidicolin inhibition is rapid (Levenson and Hamlin, 1993), effects on fork speed were limited primarily to the IdU pulse. Fork speed determination by SMARD assumes that average fork speeds are the same during both pulses. In practice, this is confirmed by the general observation of similar numbers of fully IdU- and CldU-labeled molecules. Because under aphidicolin treatment conditions fork speeds differed during each pulse, we could not use the standard SMARD analysis to determine the effect of aphidicolin on fork speed. Instead we determined relative fork speeds during each pulse by comparing the total numbers of fully IdU-labeled (red) and fully CldU-labeled (green) molecules that were also obtained in addition to the dual-labeled molecules (Fig. 3 B). We observed ∼1/4 as many fully red molecules as fully green molecules, indicating that replication in the presence of aphidicolin proceeded at ∼1/4 the rate as in the absence of aphidicolin.
Figure 3.
Slowdown of telomeric replication forks by aphidicolin leads to dormant origin activation in the subtelomere. SMARD analysis of the Ch14q telomere segment from BLM-proficient cells treated with aphidicolin. (A) Cells were pulse-labeled with IdU in the presence of aphidicolin (110 nM) followed by labeling with CldU in the absence of aphidicolin. (B) Shown are the number of fully stained red (NR) and green (NG) molecules collected. The ratio of red to green molecules (28/113) indicates that replication in the presence of aphidicolin proceeded at ∼1/4 the rate as in the absence of aphidicolin. (C) Alignments of replicated molecules fully labeled with both IdU (red) and CldU (green) are shown (of the 171 fully labeled [IdU, CldU, or IdU/CldU] molecules collected from three independent samples stretched on slides). Almost one third of the molecules (9/30) contain subtelomeric origins, a significantly higher percentage than in untreated cells (1/32; Fig 1). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Fig. 1. Symbols are as in Fig. 1. On the right are images of individual color channels depicting telomere PNA and locus-specific FISH probes (blue), IdU (red), and CldU (green) signals shown below the merged images of molecules that display telomeric initiation events. (D) Histogram of molecule alignment.
We found that similar to untreated cells (Fig. 1), aphidicolin-treated cells show initiation in the telomere (Fig. 3 C), indicating that the aphidicolin treatment did not prevent telomeric initiation. Importantly, we found that almost one third of the molecules (9/30) had initiations in the subtelomere (Fig. 3 C, molecules 9–17), a significantly higher percentage than seen in untreated cells (1/32; see Fig. 1). These origins activated during aphidicolin treatment were distributed throughout the subtelomere rather than at a specific site. Collectively, our results demonstrated that slowing of forks progressing from the telomere led to firing of subtelomeric origins.
Our aphidicolin experimental findings also highlight the utility of the Ch14q telomere as a model locus to study telomere replication, with the induction of dormant origins in the adjacent subtelomere providing a valuable readout of challenged replication in the telomere.
G4 structures in the leading strand impede telomere replication in BLM-deficient cells
One proposed role for BLM in telomere maintenance is the removal of G4 structures formed during replication of the G-rich telomeres. As we found that the rate of fork progression through telomere repeats was reduced in BLM-deficient cells, we sought to determine if G4 structures were involved. To address this question, we determined the effect of treating cells with the G4 stabilizer PhenDC3 on the replication of the Ch14q telomere. The bisquinolinium compound PhenDC3 is a G4 DNA ligand with strong quadruplex stabilizing ability on intramolecular quadruplexes formed from the mammalian telomeric repeat sequence, high selectivity and nanomolar affinity for G4 DNA (De Cian et al., 2007b; Monchaud et al., 2008), and the demonstrated ability to disrupt telomere stability (Piazza et al., 2010; Lopes et al., 2011). NMR spectroscopy analysis has revealed that Phen-DC3 interacts with the quadruplex through extensive stacking with the guanine bases of the top G-quartet (Chung et al., 2014).
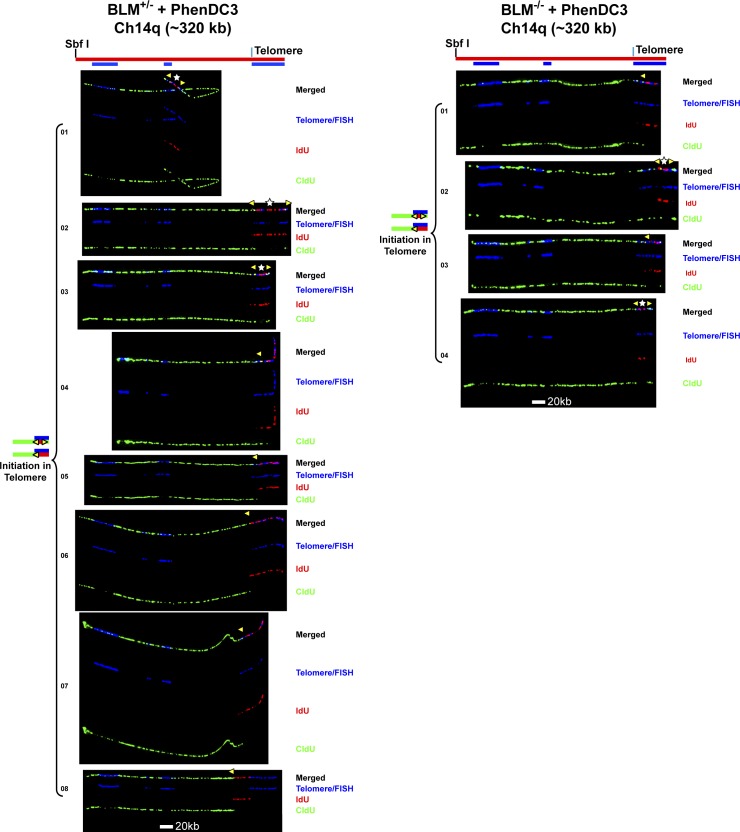
To examine the effects of G4 stabilizer treatment, cells were incubated with PhenDC3 starting 30 min before IdU labeling and continued during the IdU pulse, followed by CldU pulsing in the absence of the ligand (Figs. 4 and 5). SMARD analysis revealed that a much greater number of subtelomeric origins were fired in the BLM-deficient cells compared with BLM-proficient cells (Fig. 4). Moreover, ligand treatment increased subtelomeric origin firing only in BLM-deficient cells (compare Figs. 1 A and 4). No increase was seen in the treated BLM-proficient cells compared with untreated cells (Fig. 1 A). Because fork slowdown by aphidicolin led to subtelomeric origin induction (Fig. 3), this raised the possibility that PhenDC3 treatment may also be slowing fork progression through the telomere in BLM-deficient cells, resulting in subtelomeric origin firing. Similar to aphidicolin-induced origins, the G4 ligand-induced origins were distributed throughout the subtelomere rather than at a specific site. In fact, we found that PhenDC3 treatment caused a reduction in average fork speed in BLM-deficient cells while the BLM-proficient cells were unaffected, both for the entire analyzed segment as well as the terminal 100 kb containing the telomere (Table 1). Interestingly, we found that in BLM-deficient cells, PhenDC3 treatment reduced fork speed by ∼0.6 kb/min in both the full segment and the terminal-most 100 kb containing the telomere. That similar reductions were observed was somewhat unexpected since PhenDC3 should be targeting G4 structures and the subtelomere portion of the full segment has far less G4-forming potential than the telomere (Table S1). However the fork speed reduction seen in the subtelomere is probably an indirect effect resulting from the presence of the additional induced subtelomeric origins. It has previously been shown that forks associated with closely spaced origins move slower than those emanating from widely spaced origins (Norio et al., 2005; Conti et al., 2007; Courbet et al., 2008). Importantly, when compared with BLM-proficient cells, BLM-deficient cells showed a greater reduction in fork speed in the telomere-terminal 100 kb than in the entire 320-kb segment (Table 1). Thus, our data are consistent with the conclusion that G4 stabilization by PhenDC3 slows replication fork progression through the telomere.
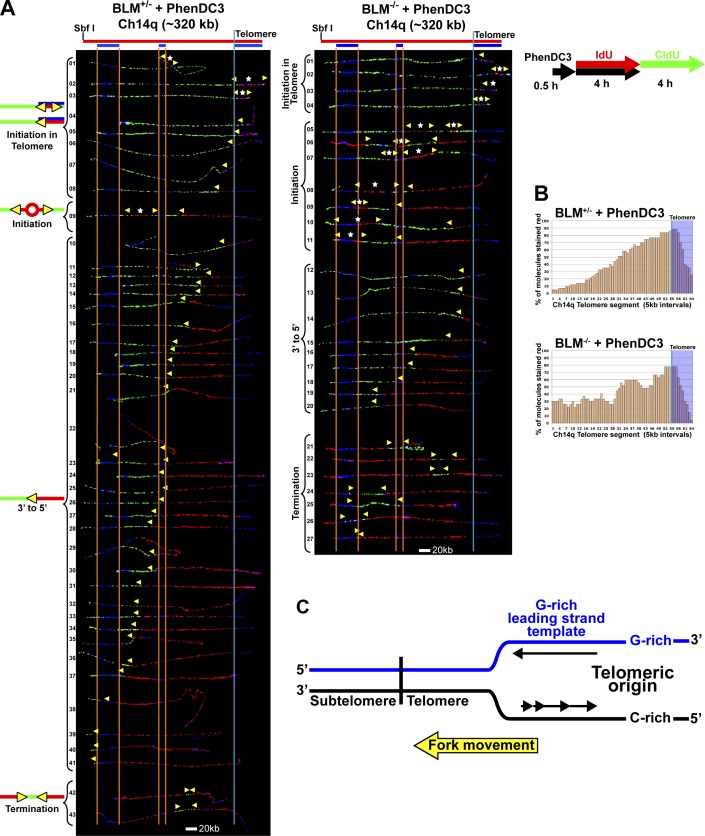
Figure 4.
G4 structures impede telomere replication in BLM-deficient cells. (A) SMARD analysis of the Ch14q telomere segment from BLM-proficient (left) and BLM-deficient (right) cells treated with G4-stabilizing ligand PhenDC3. Cells were treated with PhenDC3 (10 µM) for 30 min then pulse-labeled with IdU in the presence of the ligand followed by labeling with CldU in the absence of the ligand. Alignments of replicated molecules fully labeled with both IdU (red) and CldU (green) are shown (of the 182 BLM-proficient and 133 BLM-deficient fully labeled (IdU, CldU, or IdU/CldU) molecules collected from 5–7 independent samples stretched on slides). A greater number of subtelomeric origins are seen in the BLM-deficient cells (9) compared with BLM-proficient cells (1). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Fig. 1. Symbols are as in Fig. 1. (B) Histograms of alignments. (C) In the Ch14q telomere, the G-rich strand serves as the template for leading strand copying by the replication fork proceeding from telomere to subtelomere.
Figure 5.
Replication initiation events are detected in the Ch14q telomere in BLM-proficient and BLM-deficient cells treated with G4-stabilizing ligand PhenDC3. Replicated molecules from BLM-proficient and BLM-deficient cells treated with G4-stabilizing ligand PhenDC3 that display telomeric initiation events (presented in Fig. 4) are shown. Images of individual color channels depicting telomere PNA and locus-specific FISH probes (blue), IdU (red), and CldU (green) signals are shown below the merged images. A map of the 14q locus is depicted above the molecule images, with the positions of the telomeric PNA and FISH probes (blue bars below the map) used to identify and orient the molecules indicated. Symbols are as in Fig. 1. White stars indicate definitive centers of initiations, where a red signal is surrounded by green (see Fig. S1 B).
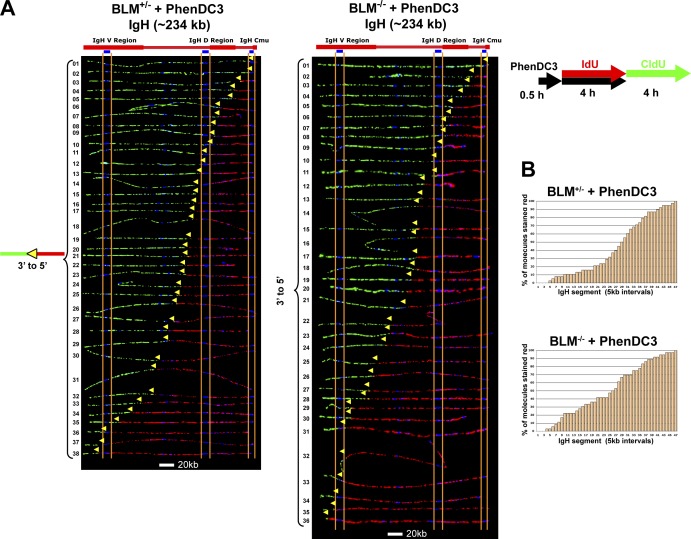
To confirm that the observed effects of the G4 stabilizer were G4 DNA–specific, we also examined the effect of ligand treatment on the replication of a 234-kb segment of the immunoglobulin heavy chain (Igh) locus. This segment represents a genomic region with much lower G4 DNA–forming potential than the telomere (Table S1), despite the presence of the ∼1.5-kb Mu switch element (which contains 9 of the 95 potential G4-forming sequences in the segment [Table S1]) within the terminal 6 kb of one end of the segment. We have previously shown that a large portion of the Igh region (>500 kb), which includes this segment, is replicated by a single fork progressing across the region (Norio et al., 2005; Guan et al., 2009). When we examined the replication of this 234-kb Igh segment by SMARD, we saw no differences in the replication programs between ligand-treated BLM-deficient and BLM-proficient cells (Fig. 6). In both cell types, we observed only molecules with a single fork and an absence of initiations, as we have previously found for this region for all non-B cells (Norio et al., 2005; Guan et al., 2009). Moreover, we found no differences in the fork rate between ligand-treated BLM-deficient and BLM-proficient cells (Table 1). Thus, in contrast to our findings with the telomere-containing Ch14q segment, PhenDC3 treatment had no apparent effects on the replication of the Igh segment.
Figure 6.
No differences are detected in the replication program of a nontelomeric locus in G4 stabilizer–treated BLM-proficient and BLM-deficient cells. SMARD analysis of a segment of the Igh locus from BLM-proficient (left) and BLM-deficient (right) cells treated with G4-stabilizing ligand PhenDC3. Cells were treated with PhenDC3 (10 µM) as in Fig. 4. Alignments of replicated molecules fully labeled with both IdU (red) and CldU (green) are shown (of the 175 BLM-proficient and 155 BLM-deficient fully labeled [IdU, CldU, or IdU/CldU] molecules collected from 4–5 independent samples stretched on slides). Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Fig. 1. Symbols are as in Fig. 1. (B) Histograms of alignments. In both cell types, only molecules with a single fork and an absence of initiation events are seen.
Importantly, because the Ch14q telomere is replicated from origins within the telomere, some of the telomere is copied by a fork progressing from the telomere to the subtelomere. Given the sequence orientation of telomeres, the G-rich strand serves as the template for leading strand copying by this fork (Fig. 4 C). Consequently, the differences we observe in fork speeds reflect, at least in part, differences in G-rich leading strand copying, i.e., copying of the G-rich strand by leading strand synthesis. While it is possible that G4 structures may form in the nascent G-rich strand produced during copying of the complementary C-rich strand by lagging strand synthesis, it is unlikely that they would impede copying because they would be behind the advancing lagging strand polymerase. Thus, our data support the conclusion that slowdown of telomeric replication fork movement by PhenDC3 in BLM-deficient cells involves impeded leading strand copying by stabilized G4 structures.
Evidence of an increased amount of G4 structures in telomeres of BLM-deficient cells compared with BLM-proficient cells
Our SMARD findings on the effect of G4 stabilizer treatment indicated a role for BLM in the resolution of G4 structures formed during telomere replication. However, a direct demonstration of BLM activity on G4 DNA in cells has yet to be shown. Therefore we sought to determine the effects on BLM deficiency on G4 occurrence in cells, both genome-wide and specifically at telomeres. To detect the presence of G4 structures in cells, we used a recently developed antibody against G4 DNA. The antibody, designated BG4, is a single-chain antibody isolated from a phage-display library (Schofield et al., 2007) via selection against a panel of intramolecular G4 structures (Biffi et al., 2013). This antibody has previously been used to visualize in G4 structures in mammalian cells (Biffi et al., 2013, 2014). To test the in vivo specificity of the antibody under our experimental conditions, we performed antibody blocking assays using prefolded G4 oligonucleotides. We found that nuclear signal was lost upon preincubation of the antibody with excess prefolded G4 oligonucleotide before cell immunostaining, whereas no signal loss occurred on preincubation with a non–G4-forming control single-stranded oligonucleotide (Fig. S2).
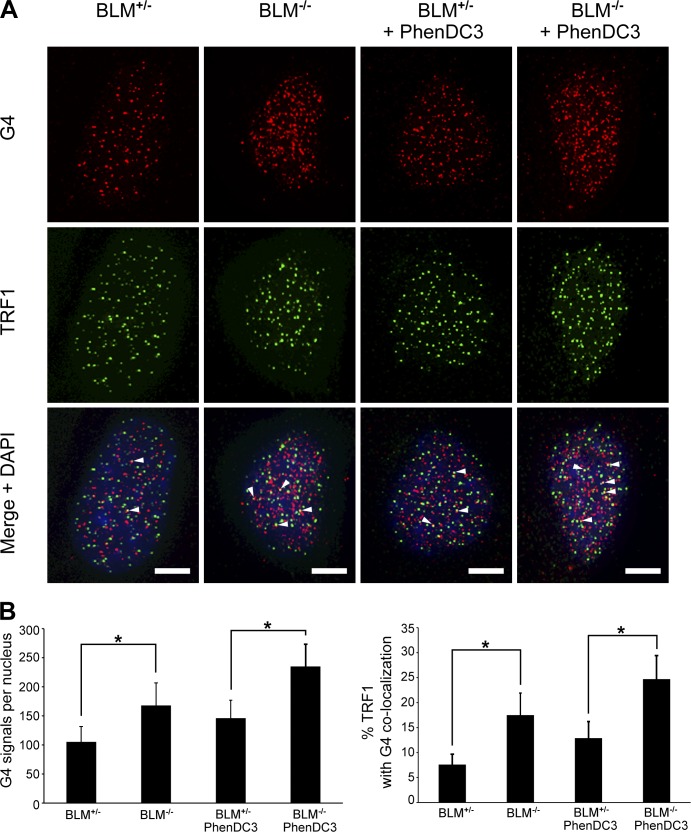
A comparison of BLM-proficient and BLM-deficient cells stained for G4 (Fig. 7) revealed that BLM-deficient cells displayed 1.6-fold more G4 antibody signals per nucleus than BLM-proficient cells. Furthermore, this difference was even greater when localizing G4 signals to the telomere. When G4 signals were colocalized with the telomere-binding protein TRF1, we found that the percentage of telomeres with overlapping G4 signals was ∼2.3-fold higher in BLM-deficient cells (Fig. 7). In addition, we observed in both cell types that the majority of the G4 signals did not localize to the telomere, corroborating previous findings indicating that genome-wide, G4 structures in mammalian cells are largely detected outside of the telomere (Biffi et al., 2013; Henderson et al., 2014). Importantly, our results show that BLM-deficient cells exhibit more G4, both genome-wide and specifically in the telomere, compared with BLM-proficient cells.
Figure 7.
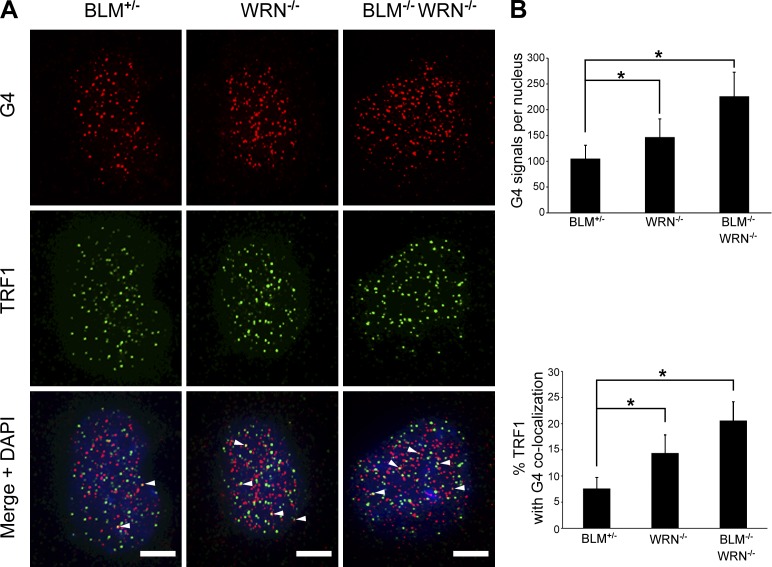
BLM-deficient cells exhibit more G4 structures in telomeres compared with BLM-proficient cells. (A) IF analysis of BLM-proficient and BLM-deficient cells stained with antibodies specific for G4 DNA (red) and the telomeric protein TRF1 (green). For cells treated with the G4 stabilizer PhenDC3 (10 µM), cells were treated for 4 h before fixation. Arrowheads indicate colocalization of G4 and TRF1 signals. Bars, 5 µm. (B) Quantification of G4 signals and the percentage of colocalization of G4 and TRF1 IF signals are shown. 50–75 cells per type/condition in each of two to three independent experiments were analyzed. Bars represent mean values with standard deviations (error bars). *, P < 0.001 via Student’s two-tailed t test.
To further confirm the participation of BLM in resolution of G4 structures in telomeres, we examined the effect of PhenDC3 treatment on the detection of G4 in BLM-deficient cells. When we compared the number of G4 signals detected in BLM-proficient and BLM-deficient cells after PhenDC3 treatment, we found that BLM-deficient cells displayed ∼1.6-fold more G4 signals per nucleus than BLM-proficient cells (Fig. 7). When G4 signals were colocalized with TRF1, we found that the percent of telomeres with overlapping G4 signals was ∼1.9-fold higher in BLM-deficient cells (Fig. 7). This reflected the findings with untreated cells, specifically that BLM-deficient cells display a larger increase (over proficient cells) in G4 signals in the telomeres compared with total nuclear G4 signals. Collectively, these findings support a role for BLM helicase in the removal of G4 structures in cells, particularly in telomeres.
WRN deficiency leads to increased firing of origins in the subtelomere and the combination of BLM and WRN deficiencies has a profound effect on telomere replication
As the above studies revealed the involvement of BLM helicase in the copying of G-rich telomere leading strand templates, we asked whether this function was shared with WRN, another RecQ-family helicase that is highly active at unwinding G4 in vitro (Fry and Loeb, 1999; Mohaghegh et al., 2001). In contrast to our findings for BLM, the WRN helicase has been suggested to be important specifically for copying of telomere G-rich lagging strands (Crabbe et al., 2004). However, studies that established the helicase’s role were performed in human cells (Crabbe et al., 2004; Barefield and Karlseder, 2012), where the G-rich strand primarily serves as a lagging strand template. Accordingly, helicase activity on telomere G-rich leading strand templates would be infrequently detected in human cells. Therefore we examined the effect of WRN deficiency on the replication of the mouse Ch14q telomere to determine if WRN can assist in the leading strand copying of telomere G-rich strands.
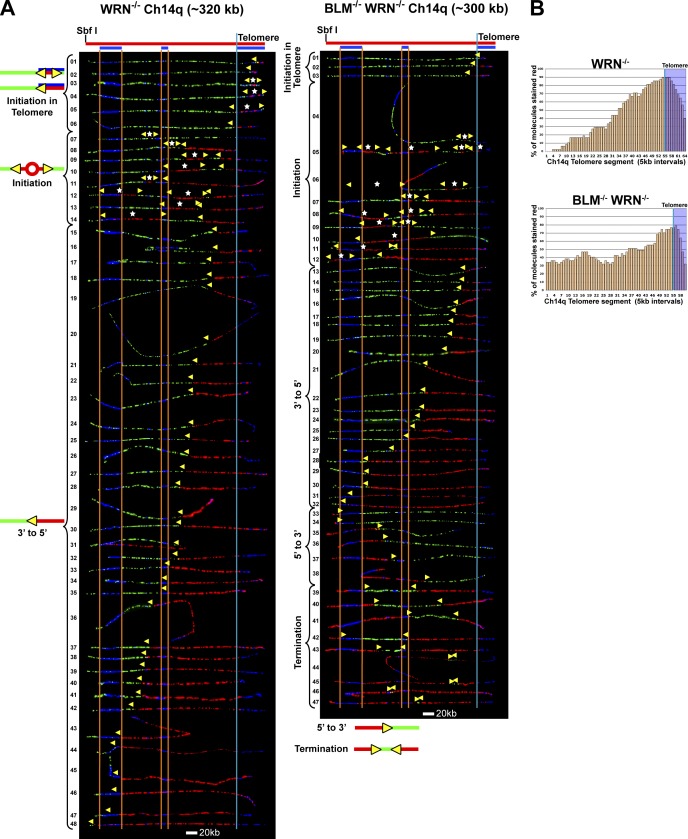
SMARD analysis revealed that Ch14q telomere replication in WRN-deficient cells, as in BLM-deficient cells, can initiate in the telomere (Fig. 8). Initiation events within the telomere were seen in six molecules (Fig. 8 A, WRN−/− molecules 1–6). Moreover, the majority of the molecules (Fig. 8 A, WRN−/− molecules 8–48) exhibited a replication fork moving away from the telomere (red signal flanked on the left by the green signal). Also similar to BLM-deficient cells, WRN-deficient cells fire origins in the subtelomere, suggesting slowed movement of the telomere-initiated fork. Unfortunately, the close proximity of subtelomeric origins to the telomere (more than half of the subtelomeric initiations [5/9] occurred within 100 kb of the telomere [Fig. 8, WRN−/− molecules 8–10, 12, and 13]) confounded the calculation of fork speed. Forks emanating from these origins quickly meet telomere-originated forks, manifesting as a single fork having replicated the region between the origins. Consequently, forks are undercounted, resulting in an inaccurate determination of fork speed, which is based on average forks per molecule (and molecule duplication time). However, a higher percentage of subtelomeric origins in the Ch14q segment was detected in the WRN−/− cells (Fig. 8) compared with BLM-deficient cells (Fig. 1), which suggests that fork movement may be slower proceeding through the telomere repeats toward the subtelomere in WRN-deficient cells compared with WRN-proficient cells.
Figure 8.
WRN deficiency leads to increased firing of origins in the subtelomere while the combination of BLM and WRN deficiencies has profound effects on telomere replication. (A) SMARD analysis of the Ch14q telomere segment from WRN-deficient (WRN−/−; left) and BLM-deficient/WRN-deficient (BLM−/−/WRN−/−; right) cells. Alignments of replicated molecules fully labeled with both IdU (red) and CldU (green) are shown (of the 300 WRN−/− and 285 BLM−/−/WRN−/− fully labeled [IdU, CldU, or IdU/CldU] molecules collected from 4–5 independent samples stretched on slides). A considerable reduction in telomere length is seen in the BLM/WRN-deficient cells. More than 20% of molecules from BLM/WRN-deficient cells show no IdU labeling in the telomere, indicating that the G-rich telomere strand is replicated solely by lagging strand copying. Vertical lines (orange and blue) demarcate the boundaries where FISH probes bind, as described in Fig. 1. Symbols are as in Fig. 1. (B) Histograms of alignments.
To further establish whether WRN may be involved in copying of G-rich telomere leading strand templates, we examined Ch14q telomere replication in cells deficient for both BLM and WRN. This analysis revealed profound effects of the combined deficiency. We found that a substantial proportion (10/47) of molecules from BLM/WRN-deficient cells (Fig. 8, BLM−/− WRN−/− molecules 4, 6, 9, 10, and 33–38) had no IdU (red) labeling in the telomere. In contrast, telomeres in all molecules from proficient cells and either single mutant, with the exception of 1 of the 48 WRN−/− molecules (Fig. 8), contained IdU labeling. This indicated that unlike in proficient or singly deficient cells, the G-rich telomere strand is replicated primarily by lagging strand synthesis that initiated in the subtelomere in a significant percentage of BLM/WRN-deficient cells. Thus, these findings point to participation of WRN in telomere G-rich leading strand copying. Furthermore, telomere length was considerably reduced in the BLM/WRN-deficient cells (Fig. 8) compared with either singly deficient line (Figs. 1 and 8), indicating a need for at least one of the helicases for proper Ch14q telomere maintenance. Collectively, our findings support some extent of functional overlap between BLM and WRN with regard to G-rich leading strand copying of telomeres.
WRN-deficient cells exhibit more G4 in the telomere compared with wild-type cells
Similar to BLM, a role for WRN helicase in the removal of G4 structures in cells has been inferred based on its in vitro G4 unwinding capacity. To date, however, direct evidence of WRN activity on G4 in cells is lacking. Thus, we probed cells with the G4-specific BG4 antibody, to determine if WRN deficiency would lead to changes in G4 detected genome-wide as well as specifically at telomeres. Immunofluorescence (IF) staining for G4 revealed that compared with WRN-proficient cells, WRN-deficient cells displayed 1.4-fold more G4 signals per nucleus. When G4 signals were colocalized with TRF1, we found that the percentage of telomeres with overlapping G4 signals was ∼1.7-fold higher in WRN-deficient cells (Fig. 9). Combined WRN and BLM deficiency led to even greater increases in detected G4. We observed that cells deficient in both WRN and BLM exhibited ∼2.2-fold more G4 signals per nucleus compared with helicase-proficient cells. Furthermore, when G4 signals were colocalized with TRF1, we found that the percentage of telomeres with overlapping G4 signals in WRN/BLM-deficient cells was ∼2.7-fold higher than in proficient cells (Fig. 9). Moreover, these findings suggest some overlapping activity in the resolution of G4 in telomeres, as the additive effect of the combining BLM and WRN deficiencies on detected G4 was partial in nature (Fig. 9). Importantly, as seen in BLM-deficient cells, WRN-deficient cells display a larger increase (over proficient cells) in G4 signals in the telomere compared with total nuclear G4 signals. Thus, these results support a role for WRN helicase in the removal of G4 structures in cells, particularly in telomeres.
Figure 9.
WRN-deficient cells exhibit more G4 in telomeres compared with WRN-proficient cells. (A) IF analysis of WRN-proficient (BLM+/−), WRN-deficient (WRN−/−), and BLM-deficient/WRN-deficient (BLM−/−/WRN−/−) cells stained with antibodies specific for G4 DNA (red) and TRF1 (green). Arrowheads indicate colocalization of G4 and TRF1 signals. Bars, 5 µm. (B) Quantification of G4 signals and the percent colocalization of G4 and TRF1 IF signals is shown. 50–75 cells per type/condition in each of two to three independent experiments were analyzed. Bars represent mean values with standard deviations (error bars). *, P < 0.001 via Student’s two-tailed t test.
Discussion
Mammalian telomeres provide an ingenious and effective cellular strategy for the protection of chromosome termini and genome integrity. One disadvantage of the G-rich composition of mammalian telomeres, however, is its underlying potential to form replication-impeding structures. Accordingly, mechanisms have evolved to alleviate these obstructions and allow for efficient telomere replication. Our studies elucidate a previously unappreciated role for BLM helicase in assisting the in vivo copying of G-rich telomere leading strand templates.
As the DNA duplex is unwound during replication fork progression, the template for leading strand synthesis has less extensive single-stranded portions than the lagging strand template. Therefore, G4 structures should have a greater opportunity for forming in G-rich lagging strand templates. Thus, G-rich sequences would predictably pose a greater replication challenge to lagging strand synthesis than to leading strand synthesis. However, our findings reveal that G-rich telomere sequences can also challenge leading strand synthesis. One possible explanation may involve increased G-rich template single-strandedness mediated by uncoupling of the replicative polymerase and helicase, due to transient polymerase slowing as the replisome progresses through the telomere. It has previously been shown that replication fork perturbation by lesions induced by UV irradiation and cis-platinum or polymerase inhibition by aphidicolin causes the functional uncoupling of the replicative helicase and polymerase (Byun et al., 2005; Pacek et al., 2006). Because of the simple repetitive nature of the telomeric sequence, it is possible that G-strand copying could lead to a transient local depletion of dCTP and dATP, creating a condition conducive to transient polymerase stalling and polymerase-helicase uncoupling (Kuzminov, 2013). As a result, the uncoupled helicase could unwind the duplex ahead of the polymerase at an increased rate, generating long stretches of single-stranded template in the telomere. These single-stranded regions promote formation of G4 structures that could obstruct and slow the polymerase further, increasing the size of the regions, leading to more G4 formation and increased replication challenge. Alternatively, it is possible that given the significant G4-forming potential across the extensive length of the telomere, fork blockage by occasional G4 structures formed during unperturbed synthesis could lead to fork slowdown to drive the uncoupling process.
Telomere fragility, the detection of aberrant telomeric structures in metaphase chromosomes, is a likely manifestation of replication challenged by structured telomeric DNA. We have previously shown a causal relationship between BLM deficiency and telomere fragility, which is evidence of the involvement of BLM in telomere replication (Sfeir et al., 2009). Our current data more clearly define the helicase’s participation in telomere replication, indicating that BLM contributes to telomere leading strand copying. Interestingly, BLM was recently found to localize to ultra-fine bridges (UFBs) that can form between telomeres in anaphase, leading to the proposal that BLM participates in telomere maintenance by processing late-replicating intermediate structures (Barefield and Karlseder, 2012). Alternatively, it is possible that telomeric UFBs may represent fully replicated, intertwined telomeres and that BLM assists in simple decatenation (Chan et al., 2007; Barefield and Karlseder, 2012). Regardless, it appears that BLM plays multiple roles in resolving telomeric structures to support telomere replication.
Despite their similar in vitro activities on telomeric substrates (Opresko et al., 2002, 2005; Lillard-Wetherell et al., 2004), distinct roles in telomere replication have been proposed for BLM and WRN based on disparate in vivo effects of helicase deficiency. In human fibroblasts, WRN deficiency leads to preferential loss of G-rich lagging strands after telomere replication, whereas no strand loss bias is seen in BLM-deficient cells (Barefield and Karlseder, 2012). In addition, BLM deficiency leads to enhanced telomere fragility in cells while WRN deficiency does not (Sfeir et al., 2009). However, the combined deficiency of both BLM and WRN in telomerase-deficient mice led to synergistic increases in telomere dysfunction, suggesting some functional redundancy between the helicases (Du et al., 2004). Consistent with this idea is our observation of an additive effect of BLM and WRN deficiencies on the efficiency of telomere G-rich leading strand copying. Thus, BLM and WRN apparently perform both unique and overlapping functions in telomere replication.
Heretofore, evidence for the participation of BLM and WRN in resolution of G4 structures in cells was indirect (Johnson et al., 2010; Aggarwal et al., 2011; Nguyen et al., 2014). The recent development of G4-specific antibodies that detect G4 structures in mammalian cells (Biffi et al., 2013; Henderson et al., 2014) has enabled the direct visualization of cellular G4 structures. Using a G4-specific antibody, we observed that deficiency of BLM or WRN led to increased detection of G4 in cells, directly demonstrating, for the first time, a contribution of these helicases in resolution of genomic G4 structures. Furthermore, combined deficiency of both helicases led to a partially additive phenotype, supporting some extent of overlapping activity on genomic G4 structures. Importantly, deficiency of either helicase resulted in greater increases in G4 detected in the telomere compared with G4 seen genome-wide, indicating that telomeres have a greater dependence on the helicases for suppression of G4 formation. This would seem to reflect the higher density of potential G4-forming sequences in telomeres relative to the genome as a whole.
Our present studies revealed that combined deficiency of BLM and WRN led to reduced efficiency of telomere G-rich leading strand copying, as indicated by an increased frequency of G-rich strand replication in the Ch14q telomere solely by lagging strand copying. BLM/WRN-deficient cells also displayed significant Ch14q telomere shortening compared with singly deficient cells, which suggests that proper leading strand copying of the G-rich strand requires at least one of these helicases. Interestingly, the replication of the G-rich strand in Ch14q telomere could still be accomplished by leading strand copying in the doubly deficient cells, implying that an additional helicase may be able to functionally substitute for BLM (or WRN). One candidate is the PIF1 helicase, which has been shown to stabilize replication of the G4-prone human CEB1 (hCEB1) minisatellite sequence in a yeast model system (Lopes et al., 2011). Significantly, deletion of PIF1 led to hCEB1 instability only when the G-rich strand was the template for leading strand replication (Lopes et al., 2011). It is also noteworthy that murine telomere homeostasis is not dependent on PIF1, which suggests genetic redundancy with other helicases, possibly BLM and/or WRN (Snow et al., 2007).
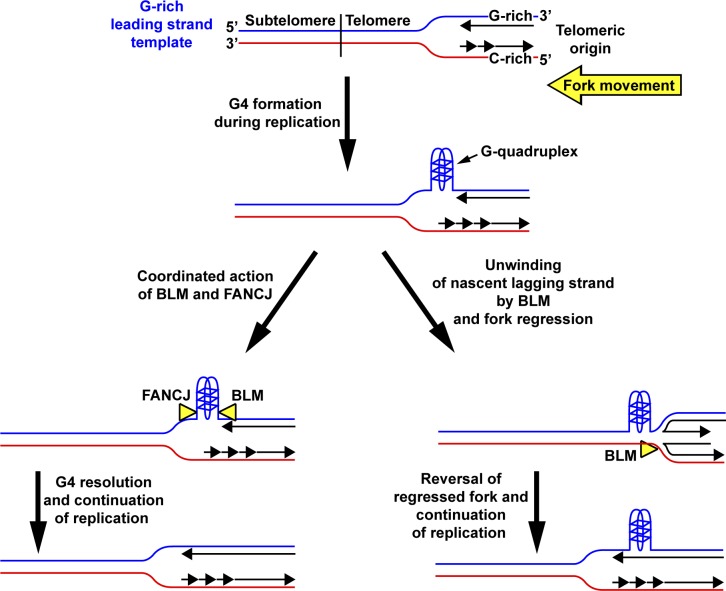
One question raised by our studies is how BLM (or WRN) assists in leading strand copying of the G-rich strands. BLM (and WRN) loads onto single-stranded (ss) DNA and translocates in a 3′ to 5′ direction to unwind DNA (Croteau et al., 2014). Thus, the helicase could simply bind adjacent to a G4 structure formed in the ss portion of the template behind the replicative helicase and unwind it. However, the polarity of BLM action would require it to bind 3′ of the structure, which may be hindered by the presence of polymerase epsilon, the leading strand polymerase. Recent data suggest an additional possible mechanism involving partnership with the FANCJ helicase, a member of the Fanconi anemia protein family with 5′ to 3′ helicase polarity (London et al., 2008; Wu et al., 2008). Studies have shown in vivo association of BLM and FANCJ, and that both helicases physically and functionally interact with each other (Suhasini et al., 2011). Significantly, FANCJ was demonstrated to act in concert with both BLM and WRN to maintain the epigenetic stability of G4-forming DNA sequences (Sarkies et al., 2012). Given that FANCJ (like BLM and WRN) has been found at telomeres (Déjardin and Kingston, 2009), it is possible that BLM (and WRN) and FANCJ act from opposite directions to collaborate on removal of G4 structures from G-rich telomere leading stand templates (Fig. 10). Alternatively, BLM and WRN may invoke a fork regression/restoration mechanism to allow the replication fork to bypass a leading strand G4 structure rather than directly unwinding the obstruction (Fig. 10). Both helicases can reverse forks in vitro (Machwe et al., 2006; Ralf et al., 2006) to generate “chicken foot”–like structures resembling Holliday junctions as well as unwind these structures to restore them to functional replication forks (Machwe et al., 2011). However, while such a mechanism would allow for replication fork continuation past G4 structures, it would also lead to loss of telomeric DNA. Importantly, it is possible for more than one of the above-proposed mechanisms to be used, as they are not mutually exclusive. Thus, future studies will be needed to determine which, if any, of the proposed mechanisms are used.
Figure 10.
Potential mechanisms of BLM participation in leading strand copying of telomeres. The G-rich telomere strand serves as the template for leading strand copying when forks from telomeric origins progress from the telomere to subtelomere. Thus, G4 structures could potentially form ahead of the fork during leading strand synthesis. One possible mechanism to remove such G4 obstructions could be for BLM to act in a coordinated manner with FANCJ to unwind the structure to allow the fork to proceed. In another potential mechanism, BLM could act to unwind the nascent strand from the lagging template strand at the obstructed fork. Template switching by the nascent leading strand to the nascent lagging strand, followed by template copying, generates a regressed fork. Subsequent unwinding of the regressed fork by BLM and reannealing of the nascent leading strand to the G-rich leading template strand results in reversal of the regressed fork, permitting the fork to bypass the G4 structure. WRN could use the same potential mechanisms as well.
In summary, we have demonstrated the participation of BLM helicase in the replication of G-rich telomere leading strands, uncovering an additional function of BLM in telomere maintenance. Our findings also indicate overlap of this function with other helicases, including WRN, suggesting that G-rich telomere leading strand templates present a significant replication challenge. The demonstrated instability of G-rich sequences when they are specifically replicated as leading strand templates in PIF1 helicase-deficient yeast cells supports this conclusion (Lopes et al., 2011). Along these lines, it will be important to determine whether BLM contributes to the faithful copying of G4-prone templates by leading strand synthesis at nontelomeric regions in mammalian cells.
Materials and methods
Cell culture
SV40-LT-immortalized BLMM3/M3 (BLM-deficient), BLM+/M3 (BLM-proficient), WRN−/−, and BLMM3/M3/WRN−/− adult mouse ear fibroblasts (Du et al., 2004; a gift from B. Johnson, University of Pennsylvania School of Medicine, Philadelphia, PA) were cultured in DMEM (HyClone) supplemented with 10% FBS, l-glutamine, and penicillin-streptomycin. The BLMM3 allele was generated by gene targeting, yielding a mutant allele that contains an extra copy of exon 3, leading to expression of a truncated polypeptide consisting of only the N-terminal 296 amino acids of the 1,416-residue full-length protein (Luo et al., 2000). BLMM3 heterozygous cells express significant levels of wild-type BLM protein (62% of wild type cells), whereas homozygous cells produce only 25% of the level of wild-type BLM protein of wild-type cells (Luo et al., 2000; McDaniel et al., 2003). When homozygous, the BLMM3 hypomorphic allele has been previously shown to induce high levels of homologous recombination, increase rates of loss of heterozygosity, and lead to a predisposition to cancer in mice (Luo et al., 2000; McDaniel et al., 2003). The reduction in BLM protein level in BLMM3/M3 cells also correlates with increased chromosome instability and tumor predisposition (McDaniel et al., 2003). The WRN– mutant allele was generated by targeted replacement of the 3′-most exon encoding the WRN catalytic helicase domain with a β geo cassette (β-galactosidase/neomycinr fusion gene; Lombard et al., 2000). No detectable WRN protein expression was seen in ear fibroblasts from WRN−/− mice using an antibody directed against the C terminus of the protein (Lombard et al., 2000). The G4 stabilizing ligand PhenDC3 (De Cian et al., 2007a,b; Monchaud et al., 2008) was generously provided by M.-P. Teulade-Fichou (Institut Curie, Paris, France).
SMARD
SMARD was performed essentially as previously described (Norio and Schildkraut, 2001, 2004; Fig. S1). Exponentially growing cells were sequentially pulse labeled with 30 µM IdU (4 h) followed by 30 µM CldU (4 h). In experiments examining the effects of aphidicolin treatment, cells were exposed to aphidicolin (Sigma-Aldrich; final [110 nM]) during the 4 h IdU labeling pulse only. In experiments examining the effects of the G4 stabilizer PhenDC3 on replication, PhenDC3 was added (final [10 µM]) to the cells 30 min before IdU labeling and was kept in the culture only during the 4-h IdU pulse. After pulsing, labeled cells were embedded in 0.5% low-melting agarose (InCert; FMC Technologies) at 106 cells per 80 µl agarose cell plug and lysed overnight at 50°C in 1% n-lauroylsarcosine and 0.5 M EDTA, pH 8, containing 20 mg/ml proteinase K. The plugs were then washed with TE (10 mM Tris, pH 8, and 1 mM EDTA), treated with 200 µM PMSF, then washed with TE, pH 8. Subsequently, the plugs were equilibrated in restriction enzyme digestion buffer (New England Biolabs, Inc.), then Sbf I (30 U/plug) was added and the DNA in the plugs digested in situ by overnight incubation at 37°C. The digested plugs were cast into 0.7% gels (SeaPlaque GTG; Lonza) and the DNA was separated by pulse field gel electrophoresis using a CHEF-DRII system (Bio-Rad Laboratories). Specific telomere/subtelomere chromosome segments within the gel were then located by performing Southern blotting on a portion of the gel using a subtelomere-specific probe (Ch14q nucleotides 124,675,539–124,676,406). After Southern blotting, pulse field gel slices containing the telomere/subtelomere segments of interest were excised and melted, and the DNA was released into the solution stretched on microscope slides coated with 3-aminopropyltriethoxysilane (Sigma-Aldrich). The stretched DNA was denatured in alkali buffer (0.1 M NaOH in 70% ethanol and 0.1% β-mercaptoethanol) and fixed in alkali buffer containing 0.5% glutaraldehyde. The denatured, fixed DNA was hybridized overnight with biotinylated probes at 37°C in humidified chamber. Biotinylated DNA FISH probes based on Ch14q subtelomeric sequence (Ch14q nucleotides 124,558,733–124,597,931 [probe 1] and nucleotides 124,668,794–124,681,022 [probe 2]) and Igh sequence (Ch 12 nucleotides 113,422,105–113,427,287 [probe 1], 113,482,544–113,490,534 [probe 2], and 113,615,750–113,624,697 [probe 3]; all map coordinates according to GRCm38.p2 reference assembly) were prepared by nick translation in the presence of biotin-16-dUTP (Roche) and used to identify the specific telomeric/subtelomeric and Igh segments. A biotin-OO-(CCCTAA)4 PNA probe (50 nM; BioSynthesis) was used to hybridize to the G-rich strand to identify the telomeric portion of Ch14q segment. After hybridization, the slides were blocked with 1% BSA for a minimum of 20 min. FISH probes were then detected by incubating with an Alexa Fluor 350–conjugated NeutrAvidin (Invitrogen) followed by two rounds of incubation first with a biotinylated anti-avidin antibody (Vector Laboratories) and then the Alexa Fluor 350–conjugated NeutrAvidin. The two incorporated, halogenated nucleosides were visualized by indirect immunostaining, during the second round of FISH detection, using a mouse anti-IdU monoclonal antibody (BD) and a rat anti-CldU monoclonal antibody (Accurate) followed by Alexa Fluor 568–conjugated goat anti–mouse (Molecular Probes) and Alexa Fluor 488–conjugated goat anti–rat antibodies. After immunostaining, coverslips were mounted on slides with antifade reagent (ProLong Gold; Invitrogen).
Photomicrograph image acquisition and analysis
Images of immunostained molecules were acquired at room temperature using a fluorescence microscope (Axioskop 2; Carl Zeiss) equipped with a Plan-Apochromatic 63× 1.4 NA oil objective lens and a charge-coupled device camera (CoolSNAP HQ; Photometrics) using IPLab software (BioVision). Images were processed with Photoshop (Adobe) and aligned according to the FISH probe pattern using Illustrator software (Adobe). Only images of molecules that were fully labeled with IdU, CldU, or both IdU and CldU, and displayed signals of both FISH probes and telomere-specific PNA probe, were collected for analysis. For analysis of the terminal 100 kb of the full-length molecules, the length of the entire segment was measured and divided by 64 to obtain the length per 5-kb unit, which was overlaid on molecules (as done for replication histograms; see Fig. S1 B) to delineate the terminal 100 kb of the segment.
G4 structure-specific antibody
The single-chain G4 structure-specific antibody BG4 was expressed from plasmid pSANG10-BG4 (Biffi et al., 2013; generous gift of S. Balasubramanian, University of Cambridge, Cambridge, England, UK). The single-chain antibody BG4 was isolated from the Sanger phage-display library (2.3 × 1010 single-chain antibody clones) by selection against a panel of intramolecular G4 structures (Biffi et al., 2013). The BG4 sequence was then cloned into the pSANG10 expression plasmid (Martin et al., 2006) to generate pSANG10-BG4, which expresses the BG4 antibody along with C-terminal hexa-histidine and 3×FLAG epitope tags. The BG4 antibody was expressed from pSANG10-BG4 in bacterial cultures grown overnight in auto-induction medium as described previously (Martin et al., 2006). After overnight culturing, the cells were pelleted and periplasmic extracts were prepared by resuspending pellets in ice-cold TES buffer (50 mM Tris, pH 8, 1 mM EDTA, and 20% sucrose), placing them on ice for 10 min followed by the addition of an equal volume of 0.2× TES buffer, and keeping them on ice for an additional 15 min. The periplasmic extracts were then centrifuged (20 min at 8,000 rpm) to remove cellular debris. The BG4 antibody was purified from the cleared extract via its hexa-histidine epitope tag by binding to Ni-NTA agarose (QIAGEN), washing with 10 mM imidazole, 100 mM NaCl in PBS, pH 8, followed by elution with 300 mM imidazole in PBS, pH 8. The eluted antibody was then dialyzed against PBS, pH 8, and stored at −80°C.
IF
Cells were grown overnight at 37°C on flame-sterilized coverslips then washed with PBS and fixed with 4% paraformaldehyde (EM Sciences). In some experiments, cells were incubated for an additional 4 h in the presence of the G4 stabilizer PhenDC3 (10 µM) before fixation. Fixed cells were permeabilized with 0.5% Triton X-100, washed with PBS, then blocked with blocking solution (5% normal goat serum and 5% normal donkey serum [Jackson ImmunoResearch Laboratories, Inc.] in PBS containing 0.05% Tween-20 [PBS-T]). After blocking, cells were incubated with anti-G4 antibody (BG4) in blocking solution for 1 h, washed with PBS-T, then incubated with anti-FLAG (M2; Sigma-Aldrich) and anti–mouse TRF1 (1449; affinity purified rabbit polyclonal; gift from T. de Lange, Rockefeller University, New York, NY) antibodies in blocking solution. Cells were then washed with PBS-T and incubated with Alexa Fluor 568–conjugated goat anti–mouse (A11031; Invitrogen) and Alexa Fluor 488–conjugated donkey anti-rabbit (A21206; Invitrogen) secondary antibodies. After antibody staining, cells were washed with PBS-T and nuclei were counterstained with DAPI (Sigma-Aldrich). Coverslips were then washed with PBS, air-dried, and mounted with antifade reagent (ProLong Gold; Invitrogen). In blocking experiments, BG4 antibody was preincubated for 1 h with an excess of prefolded G4 DNA oligonucleotide (5′-AGGGAGGGCGCTGGGAGGAGGG-3′) or single-stranded non–G4-forming DNA oligonucleotide (5′-GGCATAGTGCGTGGGCG-3′) before use in immunostaining. The G4 oligonucleotide was prefolded by heating the oligonucleotide (in 100 mM KCl, 10 mM Tris-Cl, pH 7.4) at 95°C for 5 min, then allowing the solution to slowly cool to room temperature. Z-stack images of cells were captured at room temperature using a fluorescence microscope (Axioskop 2; Carl Zeiss) equipped with a Plan Apochromatic 63× 1.4 NA oil objective lens, a motorized stage, and a charge-coupled device camera (CoolSNAP HQ; Photometrics) using IPLab software (BioVision). 50–75 cells per condition in each of two to three independent experiments were analyzed using Volocity software (PerkinElmer).
Online supplemental material
Fig. S1 depicts the SMARD methodology. Fig. S2 shows that BG4 antibody immunostaining is specifically blocked by a prefolded G4 oligonucleotide. Figs. S3, S4, and S5 depict all of the fully red and fully green molecules reported in Table 1. Table S1 details the number of G4 motifs in the DNA segments studied here. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410061/DC1.
Supplementary Material
Acknowledgements
We thank James Borowiec, Patty Opresko, Sergei Mirkin, and Barbara Birshtein for valuable suggestions in writing this manuscript.
This work was supported by National Institutes of Health/National Institute of General Medical Sciences grant 5R01-GM045751 (C.L. Schildkraut) and the Empire State Stem Cell Fund through New York State contract C024348 (C.L. Schildkraut).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CldU
- chlorodeoxyuridine
- G4
- G-quadruplex
- IdU
- iododeoxyuridine
- IF
- immunofluorescence
- SMARD
- single molecule analysis of replicated DNA
References
- Aggarwal M., Sommers J.A., Shoemaker R.H., and Brosh R.M. Jr. 2011. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. USA. 108:1525–1530. 10.1073/pnas.1006423108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield C., and Karlseder J.. 2012. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 40:7358–7367. 10.1093/nar/gks407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G., Tannahill D., McCafferty J., and Balasubramanian S.. 2013. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5:182–186. 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G., Di Antonio M., Tannahill D., and Balasubramanian S.. 2014. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 6:75–80. 10.1038/nchem.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S., Parkinson G.N., Hazel P., Todd A.K., and Neidle S.. 2006. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34:5402–5415. 10.1093/nar/gkl655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.C., Walter J.C., and Cimprich K.A.. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052. 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., North P.S., and Hickson I.D.. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397–3409. 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Tsou A.M., and Johnson F.B.. 2009. Telomeres do the (un)twist: helicase actions at chromosome termini. Biochim. Biophys. Acta. 1792:329–340. 10.1016/j.bbadis.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.J., Heddi B., Hamon F., Teulade-Fichou M.P., and Phan A.T.. 2014. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3). Angew. Chem. Int. Ed. Engl. 53:999–1002. 10.1002/anie.201308063 [DOI] [PubMed] [Google Scholar]
- Conti C., Saccà B., Herrick J., Lalou C., Pommier Y., and Bensimon A.. 2007. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 18:3059–3067. 10.1091/mbc.E06-08-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S., Gay S., Arnoult N., Wronka G., Anglana M., Brison O., and Debatisse M.. 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 455:557–560. 10.1038/nature07233 [DOI] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., and Karlseder J.. 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 306:1951–1953. 10.1126/science.1103619 [DOI] [PubMed] [Google Scholar]
- Croteau D.L., Popuri V., Opresko P.L., and Bohr V.A.. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83:519–552. 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A., Cristofari G., Reichenbach P., De Lemos E., Monchaud D., Teulade-Fichou M.P., Shin-Ya K., Lacroix L., Lingner J., and Mergny J.L.. 2007a Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. USA. 104:17347–17352. 10.1073/pnas.0707365104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A., Delemos E., Mergny J.L., Teulade-Fichou M.P., and Monchaud D.. 2007b Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 129:1856–1857. 10.1021/ja067352b [DOI] [PubMed] [Google Scholar]
- Déjardin J., and Kingston R.E.. 2009. Purification of proteins associated with specific genomic Loci. Cell. 136:175–186. 10.1016/j.cell.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Doksani Y., Wu J.Y., de Lange T., and Zhuang X.. 2013. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 155:345–356. 10.1016/j.cell.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos W.C., Kosiyatrakul S.T., Yan Z., Calderano S.G., and Schildkraut C.L.. 2012. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J. Cell Biol. 197:253–266. 10.1083/jcb.201112083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Shen J., Kugan N., Furth E.E., Lombard D.B., Cheung C., Pak S., Luo G., Pignolo R.J., DePinho R.A., et al. . 2004. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 24:8437–8446. 10.1128/MCB.24.19.8437-8446.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M., and Loeb L.A.. 1999. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 274:12797–12802. 10.1074/jbc.274.18.12797 [DOI] [PubMed] [Google Scholar]
- Gilson E., and Géli V.. 2007. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8:825–838. 10.1038/nrm2259 [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., and de Lange T.. 1999. Mammalian telomeres end in a large duplex loop. Cell. 97:503–514. 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Guan Z., Hughes C.M., Kosiyatrakul S., Norio P., Sen R., Fiering S., Allis C.D., Bouhassira E.E., and Schildkraut C.L.. 2009. Decreased replication origin activity in temporal transition regions. J. Cell Biol. 187:623–635. 10.1083/jcb.200905144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R., and German J.. 1975. A retarded rate of DNA chain growth in Bloom’s syndrome. Proc. Natl. Acad. Sci. USA. 72:758–762. 10.1073/pnas.72.2.758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr., Sen D., and Lansdorp P.M.. 2014. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 42:860–869. 10.1093/nar/gkt957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Cao K., Ryvkin P., Wang L.S., and Johnson F.B.. 2010. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 38:1114–1122. 10.1093/nar/gkp1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. 2013. Inhibition of DNA synthesis facilitates expansion of low-complexity repeats: is strand slippage stimulated by transient local depletion of specific dNTPs? BioEssays. 35:306–313. 10.1002/bies.201200128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson V., and Hamlin J.L.. 1993. A general protocol for evaluating the specific effects of DNA replication inhibitors. Nucleic Acids Res. 21:3997–4004. 10.1093/nar/21.17.3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., and Groden J.. 2004. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 13:1919–1932. 10.1093/hmg/ddh193 [DOI] [PubMed] [Google Scholar]
- Lombard D.B., Beard C., Johnson B., Marciniak R.A., Dausman J., Bronson R., Buhlmann J.E., Lipman R., Curry R., Sharpe A., et al. . 2000. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol. Cell. Biol. 20:3286–3291. 10.1128/MCB.20.9.3286-3291.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., and Hiom K.. 2008. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283:36132–36139. 10.1074/jbc.M808152200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., and Nicolas A.. 2011. G-quadruplex-induced instability during leading-strand replication. EMBO J. 30:4033–4046. 10.1038/emboj.2011.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., and Bradley A.. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424–429. 10.1038/82548 [DOI] [PubMed] [Google Scholar]
- Machwe A., Xiao L., Groden J., and Orren D.K.. 2006. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 45:13939–13946. 10.1021/bi0615487 [DOI] [PubMed] [Google Scholar]
- Machwe A., Karale R., Xu X., Liu Y., and Orren D.K.. 2011. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry. 50:6774–6788. 10.1021/bi2001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N., and Gray L.T.. 2013. The G4 genome. PLoS Genet. 9:e1003468 10.1371/journal.pgen.1003468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.D., Rojas G., Mitchell J.N., Vincent K.J., Wu J., McCafferty J., and Schofield D.J.. 2006. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 6:46 10.1186/1472-6750-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L.D., Chester N., Watson M., Borowsky A.D., Leder P., and Schultz R.A.. 2003. Chromosome instability and tumor predisposition inversely correlate with BLM protein levels. DNA Repair (Amst.). 2:1387–1404. 10.1016/j.dnarep.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Mohaghegh P., Karow J.K., Brosh R.M. Jr., Bohr V.A., and Hickson I.D.. 2001. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29:2843–2849. 10.1093/nar/29.13.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchaud D., Allain C., Bertrand H., Smargiasso N., Rosu F., Gabelica V., De Cian A., Mergny J.L., and Teulade-Fichou M.P.. 2008. Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 90:1207–1223. 10.1016/j.biochi.2008.02.019 [DOI] [PubMed] [Google Scholar]
- Nguyen G.H., Tang W., Robles A.I., Beyer R.P., Gray L.T., Welsh J.A., Schetter A.J., Kumamoto K., Wang X.W., Hickson I.D., et al. . 2014. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. USA. 111:9905–9910. 10.1073/pnas.1404807111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P., and Schildkraut C.L.. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 294:2361–2364. 10.1126/science.1064603 [DOI] [PubMed] [Google Scholar]
- Norio P., and Schildkraut C.L.. 2004. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2:e152 10.1371/journal.pbio.0020152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N.M., Thomas S., Riblet R., and Schildkraut C.L.. 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 20:575–587. 10.1016/j.molcel.2005.10.029 [DOI] [PubMed] [Google Scholar]
- Opresko P.L. 2008. Telomere ResQue and preservation—roles for the Werner syndrome protein and other RecQ helicases. Mech. Ageing Dev. 129:79–90. 10.1016/j.mad.2007.10.007 [DOI] [PubMed] [Google Scholar]
- Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., and Bohr V.A.. 2002. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277:41110–41119. 10.1074/jbc.M205396200 [DOI] [PubMed] [Google Scholar]
- Opresko P.L., Mason P.A., Podell E.R., Lei M., Hickson I.D., Cech T.R., and Bohr V.A.. 2005. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 280:32069–32080. 10.1074/jbc.M505211200 [DOI] [PubMed] [Google Scholar]
- Pacek M., Tutter A.V., Kubota Y., Takisawa H., and Walter J.C.. 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 21:581–587. 10.1016/j.molcel.2006.01.030 [DOI] [PubMed] [Google Scholar]
- Piazza A., Boulé J.B., Lopes J., Mingo K., Largy E., Teulade-Fichou M.P., and Nicolas A.. 2010. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 38:4337–4348. 10.1093/nar/gkq136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C., Hickson I.D., and Wu L.. 2006. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281:22839–22846. 10.1074/jbc.M604268200 [DOI] [PubMed] [Google Scholar]
- Rao V.A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z.H., Davies S.L., Saccá B., Hickson I.D., Bensimon A., and Pommier Y.. 2007. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 5:713–724. 10.1158/1541-7786.MCR-07-0028 [DOI] [PubMed] [Google Scholar]
- Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., and Sale J.E.. 2012. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 40:1485–1498. 10.1093/nar/gkr868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield D.J., Pope A.R., Clementel V., Buckell J., Chapple S.Dj., Clarke K.F., Conquer J.S., Crofts A.M., Crowther S.R., Dyson M.R., et al. . 2007. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 8:R254 10.1186/gb-2007-8-11-r254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., and de Lange T.. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 138:90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Ghosh A.K., Croteau D.L., and Bohr V.A.. 2012. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 736:15–24. 10.1016/j.mrfmmm.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow B.E., Mateyak M., Paderova J., Wakeham A., Iorio C., Zakian V., Squire J., and Harrington L.. 2007. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell. Biol. 27:1017–1026. 10.1128/MCB.01866-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini A.N., Rawtani N.A., Wu Y., Sommers J.A., Sharma S., Mosedale G., North P.S., Cantor S.B., Hickson I.D., and Brosh R.M. Jr. 2011. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 30:692–705. 10.1038/emboj.2010.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Shin-ya K., and Brosh R.M. Jr. 2008. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28:4116–4128. 10.1128/MCB.02210-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.