Abstract
Zinc finger protein X-linked (ZFX) is a zinc finger transcription factor and plays a significant role in the self-renewal ability of embryonic stem cells and various cancers. However, its expression and function in colorectal cancer (CRC) remain unclear. In the present study, we evaluated the expression of ZFX in CRC using quantitative polymerase chain reaction (qPCR), western blot analysis and immunohistochemistry (IHC), and further explored its potential functions in CRC cell lines using cell counting kit-8 and Transwell invasion assays. qPCR and western blot analysis revealed that ZFX was significantly upregulated in CRC tissues; IHC further confirmed this finding, revealing that higher expression of ZFX was significantly associated with larger tumor size (P=0.01), higher pathological stage (P=0.02), depth of invasion (P=0.047), lymph node invasion (P=0.02) and higher American Joint Committee on Cancer (AJCC) stage (P=0.04). CRC patients with higher ZFX expression also exhibited significantly shorter survival times (P=0.019). Moreover, knockdown of ZFX significantly suppressed proliferation and invasion in CRC cell lines HCT116 and LoVo. These results suggest that ZFX plays a notable role in CRC tumorigenicity and may serve as a novel marker and therapeutic target for CRC.
Keywords: zinc finger protein X-linked, colorectal cancer, prognosis
Introduction
Colorectal cancer (CRC) is the third most common type of cancer worldwide, with over 1.4 million new cancer cases and 50,000 mortalities reported in 2013 (1). However, the biological and molecular mechanisms underlying CRC development remain largely unknown.
Zinc finger protein X-linked (ZFX), a zinc finger protein of the Zfy family, is a transcriptional factor encoded on the mammalian X chromosome which is highly conserved in vertebrates. The full-length ZFX protein contains a DNA-binding domain, an acidic transcriptional activation domain and a nuclear localization sequence (2). Previous studies have demonstrated elevated levels of ZFX in pluripotent embryonic and hematopoietic stem cells, which is required for self-renewal ability (3,4). Overexpression of ZFX has also been demonstrated to confer self-renewal capability in hepatocellular carcinoma (5). In addition, previous studies have revealed that ZFX plays a notable role in regulating the transcription of genes necessary for cell proliferation, cell cycle and cell death (6,7). Therefore, it is speculated that ZFX may also be active in cancers and may represent anti-cancer targets. Notably, ZFX is associated with a number of human cancers, including glioblastoma (8,9), breast cancer (10), osteosarcoma (11,12), lung cancer (13,14), gallbladder cancer (15), prostate cancer (16), laryngeal squamous cell carcinoma (17) and gastric cancer (18). Amini et al also identified that ZFX was significantly upregulated in colon cancer tissues and cell lines (19). However, the biological function of ZFX in CRC remains unclear.
In this study, we firstly detected the expression pattern of ZFX in CRC and revealed that the upregulation of ZFX is strongly associated with advanced tumor stage and shorter survival in CRC patients. Next, we explored the potential functions of ZFX in CRC by depletion of ZFX using small interfering RNA (siRNA) and observed that knockdown of ZFX significantly suppressed CRC cell growth and invasion. Using these approaches we aim to elucidate the role of ZFX in the development of CRC and evaluate ZFX as a potential biomarker for the prognosis of CRC.
Materials and methods
Human CRC specimens
A total of 30 CRC specimens were obtained for use in this study from the First Hospital of Jiaxing, China, between January 2013 and May 2014. The study was approved by the ethics committee of the same hospital, and written informed consent was obtained from all participants in accordance with the provisions of the Helsinki Declaration of 1975. One tissue microarray including 90 pairs of CRC and corresponding normal tissues was purchased from BioChip (Shanghai, China).
Primary antibodies
Anti-ZFX antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA) for immunohistochemical analysis and western blot analysis, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA).
Quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from 30 pairs of matched CRC tissues using TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA). All tissues were examined histologically and all matched non-tumor tissues were confirmed to be normal. First-strand cDNA was synthesized using the PrimeScript™ RT reagent kit (Takara, Shiga, Japan) according to the manufacturer's instructions. qPCR was performed using an ABI Prism 7900HT sequence detection system (Applied Biosystems Life Technologies, Foster City, CA, USA) with SYBR® Premix Ex Taq™ II (Takara). 18s RNA was used as an internal control. The primer sequences were as follows: ZFX, 5′-GGCAGTCCACAGCAAGAAC-3′ (forward) and 5′-TTGGTATCCGAGAAAGTCAGAAG-3′ (reverse); 18s, 5′-CGGACAGGATTGACAGATTGATAGC-3′ (forward) and 5′-TGCCAGAGTCTCGTTCGTTATCG-3′ (reverse).
Western blot analysis
Total proteins were harvested from tissues or cells, and standard western blot analysis was performed using anti-ZFX and anti-GAPDH antibodies. All primary antibodies were used at a 1:1000 dilution. Peroxidase-conjugated anti-rabbit IgG secondary antibody was purchased from Kangchen Biotechnology (Shanghai, China) and used at a 1:5000 dilution.
Immunohistochemistry (IHC)
IHC was conducted using a standard streptavidin-biotin-peroxidase complex method. Briefly, paraffin-embedded slices were deparaffinized and rehydrated. Endogenous peroxidase was blocked with 0.3% H2O2 for 15 min at room temperature, then the slides were microwaved for antigen retrieval. Following antigen retrieval, the sections were blocked with sheep serum for 30 min at room temperature and then treated with rabbit anti-ZFX (1:300) at 4°C overnight. Samples incubated with phosphate-buffered saline (PBS) were used as negative controls. Immunolabeling was detected using a diaminobenzidine detection kit (Maixin biotechnology, Fuzhou, China).
Cell culture
Human CRC cell lines HCT116 and LoVo were maintained in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum and cultured in a humidified incubator at 37°C under 5% CO2.
RNA interference
All siRNAs specifically targeting ZFX were synthesized by GenePharma (Shanghai, China) using the following sequence: 5′-GACGGGUGAUUCUAUACAUTT-3′, 5′-GUCGGAAAUUGAUCCUUGUTT-3′ and 5′-GCAACAGAGUGAGCUUAAATT-3′. The three siRNAs were used as a pool for siRNA transfection. Non-specific siRNAs (GenePharma) were used as a negative control.
Cell proliferation assay
HCT116 or LoVo Cells were seeded in 96-well plates at 5,000 cells/well following transfection. Cell proliferation was evaluated after 24, 48 and 72 h using cell counting kit-8 (Dojindo, Tokyo, Japan) according to the manufacturer's instructions.
Transwell invasion assay
Transwell cell invasion assays were performed using Boyden chambers with filter inserts (pore size, 8 µm) coated with 40 µg Matrigel in 24-well plates. Twenty-four hours after transfection with or without siRNA, cells were harvested, and 100 µl serum-free media containing 2×105 cells was added into the upper chamber, while 600 µl medium with 20% fetal bovine serum was placed in the lower chamber. Following incubation for 24 h, the cells were fixed in 4% formaldehyde and stained with 0.1% crystal violet in PBS for 30 min at room temperature. Cells on the lower side of the filters were counted under light microscopy in ten randomly selected fields of each filter.
Statistical analysis
Data from at least three independent experiments are presented as the means ± standard deviation. Differences between two groups were compared using Student's paired t-test. The expression of ZFX in CRC tissues was analyzed using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
ZFX is highly expressed in CRC tissues
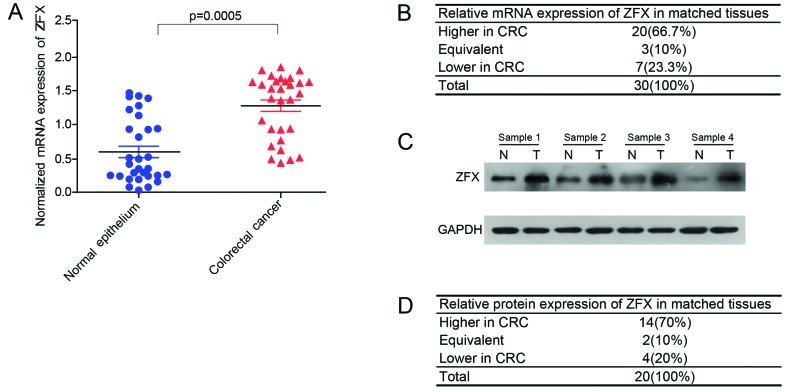
First, the ZFX expression level was investigated in normal colorectal tissues and CRC tissues by qPCR and western blot analysis. The results suggested that ZFX mRNA was highly expressed in 66.7% of CRC tissues (P=0.0005, Fig. 1A and B). Western blot analysis of 20 paired CRC tissues further confirmed this finding (Fig. 1C and D).
Figure 1.
Expression of zinc finger protein X-linked (ZFX) in colorectal cancer (CRC) tissues. (A and B) mRNA expression of ZFX in 30 pairs of CRC tissues and corresponding normal tissues was analyzed by quantitative polymerase chain reaction and revealed that the mRNA expression of ZFX was significantly upregulated in the majority of CRC tissues compared with matched normal tissues. (C and D) Protein expression of ZFX in 20 pairs of CRC tissues and corresponding normal tissues was analyzed by western blot analysis and revealed that the protein expression of ZFX was significantly upregulated in the majority of CRC tissues compared with matched normal tissues.
Next, ZFX expression in 90 paired CRC tissues was analyzed by IHC, which revealed that ZFX was significantly upregulated in CRC tissues compared with the corresponding non-tumor tissues (P<0.001, Fig. 2A and B). As shown in Fig. 2, ZFX was readily detected in the plasma and part of the nuclei of CRC tissues, whereas little or no staining was observed in the normal colorectal tissues.
Figure 2.
Immunohistochemistry (IHC) analysis of zinc finger protein X-linked (ZFX) protein expression in 90 pairs of colorectal cancer (CRC) tissues. (A) Representative images of IHC of ZFX in CRC and normal tissues (magnification, x200). (B) Statistics of ZFX protein expression levels in CRC and normal tissues. The level of ZFX protein was estimated according to immunostaining intensity and compared between cancer and normal tissues.
ZFX expression is correlated with severity of CRC
To test whether ZFX expression was correlated with various clinicopathological features of CRC, 90 CRC cases were stratified by ZFX expression as determined by IHC. ZFX upregulation was observed to be significantly associated with larger tumor size (P=0.01), higher pathological stage (P=0.02), depth of invasion (P=0.047), lymph node invasion (P=0.02) and higher American Joint Committee on Cancer (AJCC) stage (P=0.04), as detailed in Table I.
Table I.
Correlation of zinc finger protein X-linked expression and clinicopathological characteristics of colorectal cancer.
| Patients with low ZFX expression | Patients with high ZFX expression | P-value | |
|---|---|---|---|
| Gender | |||
| Male | 17 | 31 | |
| Female | 15 | 27 | 0.980 |
| Age, years | |||
| <60 | 8 | 15 | |
| ≥60 | 24 | 43 | 0.930 |
| Tumor size, cm | |||
| <5 | 22 | 24 | |
| ≥5 | 10 | 34 | 0.010 |
| Pathological stage | |||
| I–II | 20 | 21 | |
| III–IV | 12 | 37 | 0.020 |
| Depth of invasion | |||
| T1-T2 | 10 | 8 | |
| T3-T4 | 22 | 50 | 0.047 |
| Lymph node invasion | |||
| No | 20 | 21 | |
| Yes | 12 | 37 | 0.020 |
| AJCC stage | |||
| 1–2 | 21 | 25 | |
| 3–4 | 11 | 33 | 0.040 |
| Distant metastasis | |||
| No | 31 | 54 | |
| Yes | 1 | 4 | 0.650 |
ZFX, zinc finger protein X-linked; AJCC, American Joint Committee on Cancer.
Given that ZFX is highly expressed in CRC tissues and is associated with various clinicopathological features of CRC, we further tested whether ZFX expression was correlated with poor prognosis in CRC patients. As shown in Fig. 3, CRC patients with higher ZFX expression exhibited a significantly shorter survival time (P=0.019). These results strongly suggest that ZFX expression is correlated with poor prognosis in CRC.
Figure 3.
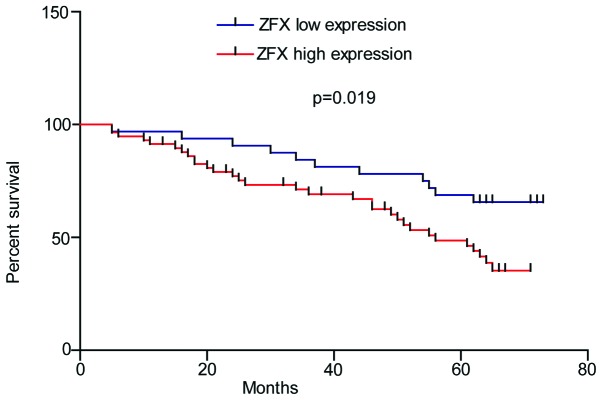
Association between zinc finger protein X-linked (ZFX) expression and overall survival in colorectal cancer (CRC) patients. The 90 CRC patients were stratified into ZFX high and ZFX low expression groups according to the immunostaining intensity of ZFX. CRC patients with higher ZFX expression exhibited a significantly shorter survival time (P=0.019).
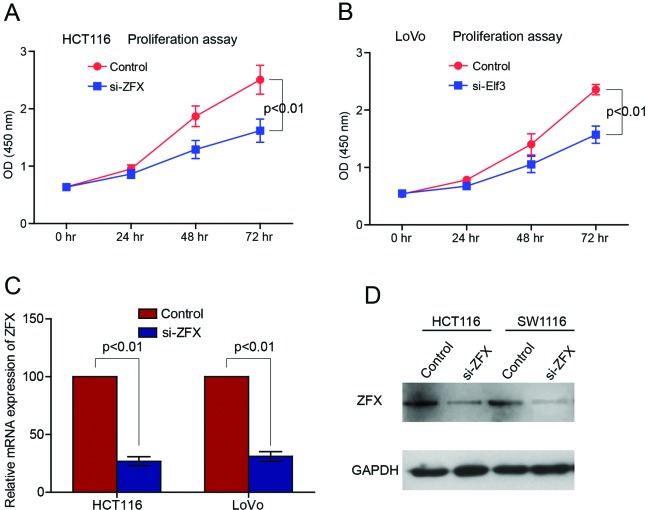
Knockdown of ZFX suppresses CRC cell proliferation and invasion
As ZFX was significantly upregulated in CRC tissues and associated with the severity of CRC, we next explored its potential function in CRC cells. To test this, specific siRNAs were employed to knock down ZFX expression in the human CRC cell lines HCT116 and LoVo. qPCR and western blot analysis revealed efficient knockdown of ZFX in HCT116 and LoVo cell lines (Fig. 4C and D). The cell counting kit-8 assay was adopted to explore the influence of proliferation by depletion of ZFX and revealed that suppression of ZFX significantly decreased the proliferation rate of HCT116 and LoVo cell lines (Fig. 4A and B). Transwell assay was used to further study the association of ZFX with cell invasion. Knockdown of ZFX significantly decreased the number of penetrated HCT116 and LoVo cells, which suggested a substantial loss of cell invasion ability by depletion of ZFX (Fig. 5A and B).
Figure 4.
Knockdown of zinc finger protein X-linked (ZFX) significantly suppressed colorectal cancer (CRC) cell proliferation. Proliferation curves of HCT116 (A) and LoVo (B) cells were determined by cell counting kit-8 assay. Knockdown of ZFX by ZFX siRNA significantly suppressed CRC cell proliferation. Quantitative polymerase chain reaction (C) and western blot analysis (D) revealed efficient knockdown of ZFX in HCT116 and LoVo cell lines.
Figure 5.
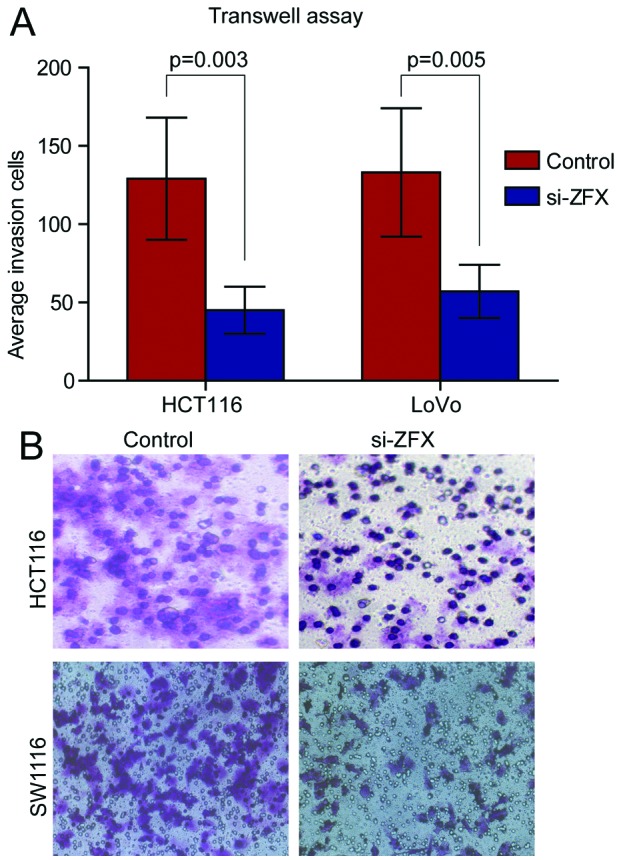
Knockdown of zinc finger protein X-linked (ZFX) significantly suppressed invasion of colorectal cancer (CRC) cells. HCT116 and LoVo cells were transfected with ZFX siRNAs, and Transwell assay was applied to evaluate cell invasion ability. (A) Statistics of average invasion cells in ZFX short interfering RNA (siRNA) group and control siRNA group in HCT116 and LoVo cells based on three independent experiments; (B) Representative images of invasion cells in ZFX siRNA group and control siRNA group in HCT116 and LoVo cells (magnification, x200).
Discussion
It was reported that zinc finger-containing transcription factors are involved in various physiological functions including differentiation, migration and proliferation, thus contributing to cancer development (20). As ZNF proteins contain an acidic transcriptional activation domain, ZFX may function as a transcription factor which promotes the transcription of growth factors or oncogenes (21). In fact, a number of previous studies have suggested ZFX as a transcriptional regulator and oncogene in various cancer types. As examples, a study by Wu et al demonstrated that knockdown of ZFX inhibited gastric cancer growth by downregulating the ERK-MAPK pathway (18), Yang et al observed that knockdown of ZFX suppressed breast cancer cell proliferation by targeting Akt and ERK2 (10), and Li et al revealed that ZFX knockdown inhibited the growth and migration of lung cancer cells by suppressing MMP-2 and Ki-67 (13). However, the function of ZFX in CRC remains unclear.
By investigating the expression pattern and function of ZFX in the context of CRC, the present study provides evidence that ZFX may function as an oncogene in CRC progression. ZFX was demonstrated to be upregulated in CRC and was associated with tumor size, pathological stage, lymph node invasion, depth of invasion and AJCC stage. More significantly, it was noted that CRC patients with higher ZFX expression levels exhibited a significantly shorter survival time. These clinicopathological findings suggest that ZFX may contribute to the malignant phenotypes of CRC. In cultured CRC cell lines, knockdown of ZFX inhibited CRC cell proliferation and invasion. These results were consistent with previous studies in breast, gastric and lung cancers (10,13,18). The strong correlation between ZFX and CRC tumor size, invasion and AJCC stage highlights the potential value of ZFX as a novel biomarker for CRC.
Previous studies have identified that ZFX regulates the expression and signaling pathway function of various genes, including ERK-MAPK, ERK2, MMP-2, CyclinD1, Stat3, Akt and c-myc. In fact, in these genes, signaling pathways along with certain other pathways, including wnt/β-catenin, play essential roles in CRC progression (22–26). The correlation between ZFX and these genes and pathways in CRC progression requires further study.
In summary, our study confirmed that ZFX was overexpressed in CRC, and was associated with tumor size, lymph node invasion, depth of invasion and overall survival time. Knockdown of ZFX in CRC cell lines inhibited cell proliferation and invasion. These results highlight the potential role of ZFX as a novel biomarker as well as a sensitive therapeutic target for CRC.
Acknowledgements
This study was funded by the Natural Science Foundation of Zhejiang (LY14H60009).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Schneider-Gädicke A, Beer-Romero P, Brown LG, Mardon G, Luoh SW, Page DC. Putative transcription activator with alternative isoforms encoded by human ZFX gene. Nature. 1989;342:708–711. doi: 10.1038/342708a0. [DOI] [PubMed] [Google Scholar]
- 3.Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cellot S, Sauvageau G. Zfx: at the crossroads of survival and self-renewal. Cell. 2007;129:239–241. doi: 10.1016/j.cell.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Lai KP, Chen J, He M, et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135:1790–1799. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang X, Huang Z, Zhou W, et al. The zinc finger transcription factor ZFX is required for maintaining the tumorigenic potential of glioblastoma stem cells. Stem Cells. 2014;32:2033–2047. doi: 10.1002/stem.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z, Li K, Xu D, Liu Y, Tang H, Xie Q, Xie L, Liu J, Wang H, Gong Y, et al. ZFX regulates glioma cell proliferation and survival in vitro and in vivo. J Neurooncol. 2013;112:17–25. doi: 10.1007/s11060-012-1032-z. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Lu Y, Zheng Y, Yu X, Xia X, He X, Feng W, Xing L, Ling Z. shRNA-mediated silencing of ZFX attenuated the proliferation of breast cancer cells. Cancer Chemother Pharmacol. 2014;73:569–576. doi: 10.1007/s00280-014-2379-y. [DOI] [PubMed] [Google Scholar]
- 11.Jiang R, Gao ZL, Sun M, Zhang XY, Wang JC, Wu H. Zinc finger X-chromosomal protein promotes growth and tumorigenesis in human osteosarcoma cells. Pak J Med Sci. 2013;29:997–1002. doi: 10.12669/pjms.294.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang R, Wang JC, Sun M, Zhang XY, Wu H. Zinc finger X-chromosomal protein (ZFX) promotes solid agar colony growth of osteosarcoma cells. Oncol Res. 2012;20:565–570. doi: 10.3727/096504013X13775486749290. [DOI] [PubMed] [Google Scholar]
- 13.Li K, Zhu ZC, Liu YJ, Liu JW, Wang HT, Xiong ZQ, Shen X, Hu ZL, Zheng J. ZFX knockdown inhibits growth and migration of non-small cell lung carcinoma cell line H1299. Int J Clin Exp Pathol. 2013;6:2460–2467. [PMC free article] [PubMed] [Google Scholar]
- 14.Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8:e74175. doi: 10.1371/journal.pone.0074175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Z, Zhang S, Li M, Wu X, Weng H, Ding Q, Cao Y, Bao R, Shu Y, Mu J, et al. Regulation of cell proliferation and migration in gallbladder cancer by zinc finger X-chromosomal protein. Gene. 2013;528:261–266. doi: 10.1016/j.gene.2013.06.064. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Zhang L, Liu J, Chen Z, Na R, Ding G, Zhang H, Ding Q. Knockdown of zinc finger protein X-linked inhibits prostate cancer cell proliferation and induces apoptosis by activating caspase-3 and caspase-9. Cancer Gene Ther. 2012;19:684–689. doi: 10.1038/cgt.2012.53. [DOI] [PubMed] [Google Scholar]
- 17.Fang J, Yu Z, Lian M, Ma H, Tai J, Zhang L, Han D. Knockdown of zinc finger protein, X-linked (ZFX) inhibits cell proliferation and induces apoptosis in human laryngeal squamous cell carcinoma. Mol Cell Biochem. 2012;360:301–307. doi: 10.1007/s11010-011-1069-x. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Lao XY, Sun TT, Ren LL, Kong X, Wang JL, Wang YC, Du W, Yu YN, Weng YR, et al. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett. 2013;337:293–300. doi: 10.1016/j.canlet.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Amini S, Fathi F, Mobalegi J, Sofimajidpour H, Ghadimi T. The expressions of stem cell markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3, Dppa4, and Esrrb in bladder, colon, and prostate cancer, and certain cancer cell lines. Anat Cell Biol. 2014;47:1–11. doi: 10.5115/acb.2014.47.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnabl B, Valletta D, Kirovski G, Hellerbrand C. Zinc finger protein 267 is up-regulated in hepatocellular carcinoma and promotes tumor cell proliferation and migration. Exp Mol Pathol. 2011;91:695–701. doi: 10.1016/j.yexmp.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider-Gädicke A, Beer-Romero P, Brown LG, Nussbaum R, Page DC. ZFX has a gene structure similar to ZFY, the putative human sex determinant, and escapes X inactivation. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Ma H, Wang Y, Cao Z, Graves-Deal R, Powell AE, Starchenko A, Ayers GD, Washington MK, Kamath V, et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J Clin Invest. 2014;124:2172–2187. doi: 10.1172/JCI71103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang MJ, Jung SA, Jung JM, Kim SE, Jung HK, Kim TH, Shim KN, Yi SY, Yoo K, Moon IH. Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res. 2011;31:575–584. [PubMed] [Google Scholar]
- 24.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y, Xu C, Liu S, Wang S, Wang L, et al. The IκB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 2013;5:280–282. doi: 10.1093/jmcb/mjt020. [DOI] [PubMed] [Google Scholar]
- 26.Pandurangan AK. Potential targets for prevention of colorectal cancer: A focus on PI3K/Akt/mTOR and Wnt pathways. Asian Pac J Cancer Prev. 2013;14:2201–2205. doi: 10.7314/APJCP.2013.14.4.2201. [DOI] [PubMed] [Google Scholar]