Abstract
Genetic variants of PNPLA3 have been implicated in hepatocellular carcinoma (HCC) susceptibility; however, published findings have been both conflicting and inconclusive. To obtain a more precise estimate of the association of the PNPLA3 rs738409 (C > G) polymorphism with the overall risk of HCC and in patients with cirrhosis, we performed a meta-analysis of nine eligible studies identified through an online search of Ovid, PubMed, EBSCO, the Cochrane Library, the Web of Science, the China National Knowledge Infrastructure, Wanfang, and Chinese Biomedicine databases. The studies comprised 1175 patients with HCC, 876 with cirrhosis, and 3026 healthy controls. Pooled odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were calculated to assess associations, using fixed-effects models. Etiology subgroup and sensitivity analyses were also performed. Our results showed that rs738409 was associated with overall HCC risk and in patients with cirrhosis in the genetic contrast modes: G vs. C, GG + CG vs. CC, GG vs. CG + GG, and GG vs. CC. Stratification by etiology did not reveal any significant association between this polymorphism and hepatitis C virus (HCV)-related HCC in patients with HCV-related cirrhosis (P > 0.05). However, healthy individuals harboring two copies of the rs738409 G variant had a higher risk of HCC (GG vs. CC: OR = 4.40, 95% CI: 3.28-5.91) than those carrying a single G allele (CG vs. CC: OR = 1.62, 95% CI: 1.34-1.59); these significant association were also present in each subgroup. Furthermore, the association was more pronounced in alcohol vs. HCV-related HCC. The present meta-analysis suggests that the PNPLA3 rs738409 G allele is a risk factor for HCC except in patients with HCV-related cirrhosis, and that two copies of the rs738409 G variant conferred a higher risk for HCC in controls, especially for alcohol-related HCC. Further studies with a larger sample size are needed to ascertain the association in different ethnicities.
Keywords: PNPLA3, adiponutrin, genetic polymorphism, hepatocellular carcinoma, meta-analysis
Introduction
Hepatocellular carcinoma (HCC) is the most common form of primary malignancy of the liver, and the second leading cause of cancer-related mortality worldwide [1]. There are four major epidemiological and clinical characteristics of HCC: (i) in the majority of cases, HCC development is secondary to previously diagnosed liver disease, such as cirrhosis, hepatitis, non-alcoholic fatty liver disease (NAFLD) [2-8]; (ii) HCC risk is a result of multiple factors, including gender, obesity, smoking, alcohol abuse, and rare genetic disorders [1,5,9-11]; (iii) the distribution and etiology of HCC varies according to geographical location. The incidence rates of HCC are highest in sub-African and Eastern Asia (20/100,000), including China where the prevalence of hepatitis B virus (HBV) infection. Conversely, the incidence rates are lower in North America and Europe where the most common etiology is hepatitis C virus (HCV) infection [1]. However, HBV and alcohol abuse also involved in liver cancer occurrence in Western countries [12,13]. (iv) Early detection of HCC remains difficult because of lacking of effective monitoring for HCC progression [10]. Accordingly, efforts to identify specific methods for the diagnosis and prevention of HCC are required. A growing body of evidence has indicated the importance of individual susceptibility to the development of cancer and has demonstrated that genetic polymorphisms can predict cancer risk and might enable early diagnosis of cancers [14,15], such as HCC.
Patatin-like phospholipase domain-containing 3 (PNPLA3), also called adiponutrin, belongs to a novel class of patatin-like phospholipase family proteins, which have emerged as a new biomarker of human hepatic steatosis and nonalcoholic fatty liver disease [16]. A large volume of research data has demonstrated that PNPLA3 is highly expressed in hepatic stellate and hepatoma cells [17,18], suggesting a potential role in cirrhosis and hepatocarcinogenesis. Recently, several studies have established a strong link between a single nucleotide polymorphism (SNP) in the PNPLA3 gene, rs738409, and the development of NAFLD, alcoholic liver disease (ALD), and chronic hepatitis [16,19-23]; however, its association with HCC risk is less well-defined, and findings of significant association with HCC related to cirrhosis have been inconsistent [24-26]. Therefore, the current meta-analysis was performed to clarify the association of the PNPLA3 rs738409 polymorphism with HCC risk and the development of HCC in patients with cirrhosis, through the combined analysis of relevant published studies.
Materials and methods
Literature and search strategy
A comprehensive electronic online search of Ovid, PubMed, EBSCO, the Cochrane Library, the Web of Science, the China National Knowledge Infrastructure, Wanfang, and the Chinese Biomedicine Database was conducted to identify studies linking PNPLA3 polymorphisms and HCC risk and/or the development of liver cancer in cirrhosis. The search was performed independently by two investigators (Wu & Li). Studies available online involving human HCC, and written in English or Chinese, were retrieved up to November 2014. The following query terms were used to carry out the electronic database search: “hepatocellular carcinoma” or “HCC” or “liver cancer” or “liver carcinoma” and “PNPLA3” or “adiponutrin” or “rs738409” and “polymorphism” or “polymorphisms” or “SNP” or “variant” or “genotype” or “mutation”. Other potentially eligible studies were identified through individual and manual searches of reference lists of major textbooks, reviews, and included articles. In the case of overlapping studies, only the study with the largest sample size was included.
Inclusion criteria
The following eligibility criteria were set for the inclusion of studies in the current meta-analysis: (1) case-control or cohort studies evaluating the association of the PNPLA3 rs738409 polymorphism with HCC risk or the development of HCC in cirrhosis; (2) studies where sufficient evidence was presented to support the diagnosis of HCC or cirrhosis, such as confirmation by histological pathology; (3) studies where the healthy control subjects were genetically unrelated family members and without any liver disease; and (4) studies where sufficient data were provided for the calculation of odds ratios (ORs) with 95% confidence intervals (CIs).
Exclusion criteria
Studies were excluded from the meta-analysis based on the following criteria: (1) where control populations were not included; (2) where outcomes of comparative analyses were not reported or were difficult to determine; (3) where investigations were based on incomplete raw data or overlapping studies; and (4) posters, abstracts, seminars, snapshots, case reports, comments, letters, reviews, or editorials.
Data extraction
The following data were extracted from each included publication by two independent investigators (Wu & Li) using a standardized form: first author, year of publication, ethnicity, etiology of cases, source of healthy controls, genotyping method, allele or genotype frequencies, and evidence of Hardy-Weinberg equilibrium (HWE). Any discrepancies were resolved by discussion or consultation with a third investigator until a consensus was reached.
Quality assessment
because all included studies used case-control or cohort designs, we used the Newcastle-Ottawa Scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) to assess quality, which was evaluated independently by two investigators (Liu & Wang). Any disagreements were resolved by discussion. The NOS assessment comprised three items and was graded as follows: selection of study groups (4 stars), comparability between groups (2 stars), and ascertainment of exposure or outcome (3 stars). We assessed the quality of all included studies accordingly, regarding the assessment of HCC risk and risk of HCC in patients with cirrhosis.
Statistical analysis
Because the lack of current knowledge precludes determination of which genetic model might best explain the effect of rs738409 on HCC occurrence, we analyzed all possible genetic models. First, we analyzed the association between the PNPLA3 rs738409 polymorphism and HCC risk, followed by an assessment of the effect of rs738409 on HCC development subsequent to cirrhosis in a similar manner. Individual or pooled ORs with 95% CIs were calculated to assess the strength of the associations between PNPLA3 rs738409 polymorphic variants and HCC risk, as well as with the risk of development of HCC in patients with cirrhosis using Review Manager version 5.3 software (kindly provided by The Cochrane Collaboration, Oxford, England) (http://www.cochrane.org/software/revman.htm). Inter-study heterogeneity was estimated graphically by the examination of forest plots. The chi-square-based Q-statistic test of heterogeneity was used to statistically assess the presence of significant heterogeneity, and the inconsistency index (I2) was used to assess the magnitude of heterogeneity within included studies. A x2-test P-value (Ph) < 0.1 or I2 > 50% were considered indicative of substantial heterogeneity [27,28]. The significance of the pooled statistical data was determined by the Z-test, with P < 0.05 considered significant, using the fixed-effects or random-effects model in the absence (Ph > 0.1 or I2 < 50%) or presence (Ph < 0.1 or I2 > 50%) of heterogeneity [29,30]. Subgroup analysis according to etiology was also performed. The potential for publication bias was measured using Begg’s funnel plot, in which the standard error of logOR of each study was plotted against its logOR; an asymmetric plot suggested possible publication bias [31]. Funnel plot asymmetry was further assessed using Egger’s linear regression test (a P-value of < 0.05 was considered significant) [32]. Begg’s and Egger’s tests were both performed using Stata 12.0 software (Stata Corporation, College Station, Texas, USA). To enhance the credibility of the results, sensitivity analysis was also performed by sequential omission of individual studies in the analysis of various contrasts.
Results
Study characteristics
Following the initial database searches, 112 potentially relevant publications met the requirements of the search strategy. After initial review, 59 publications consisting of 41 reviews, two meta-analyses, and 16 other forms of publications such as editorials were excluded. After a careful review of the titles and abstracts, 30 additional studies not conforming to the research topic were excluded. After the full texts were reviewed, seven studies met the defined inclusion criteria and were included in the current meta-analysis [24,25,33-37] (Figure 1). Because more than one case-control study was included in the reports published by Nischalke et al. [24] and Falleti et al. [34], each paper was considered as a separate study in the meta-analysis. To obtain the publication date of the paper written by Falleti et al. [34], we consulted another article written by Trepo et al. [38]. Furthermore, as the two papers written by Valenti et al. [36,39] contained overlapping data, we included the research subjects only from the study which had the larger sample size. Therefore, a total of nine case-control or cohort studies from seven publications (six in English and one in Chinese) were used in this study, comprising 1175 patients with HCC, 876 patients with cirrhosis, and 3026 healthy controls. Detailed characteristics of the included studies are summarized in Table 1.
Figure 1.
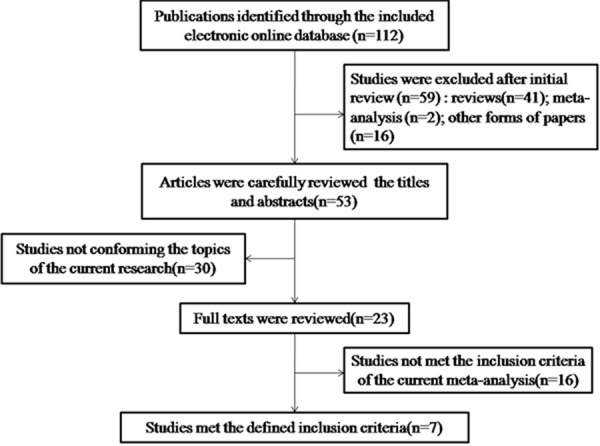
Flowchart of the current meta-analysis showing studies included.
Table 1.
Summary of characteristics of all included studies in the meta-analysis
| First author | Year | Ethnicity | Aetiology of cases | Genotype-HCC case | Genotype-cirrhosis case | Genotype-healthy control | Source of control | Genotype method | HWE test | NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| CC/CG/GG | CC/CG/GG | CC/CG/GG | NOS-Ca | NOS-Ci | |||||||
| Nischalke | 2011 | Caucasian | 57/73/31 | 77/69/15 | 112/69/9 | Volunteers | LCrt-PCR | Y | 8 | 7 | |
| HCV | 40/33/8 | 45/31/5 | - | ||||||||
| Alcoholic | 17/40/23 | 32/38/10 | - | ||||||||
| Falleti | 2011 | Caucasian | 43/60/38 | 125/160/57 | 218/175/35 | blood donors | PCR | Y | 7 | 6 | |
| HCV | 17/25/10 | 63/74/20 | |||||||||
| HBV | 11/9/3 | 24/22/7 | |||||||||
| Alcoholic | 15/26/25 | 38/64/30 | |||||||||
| Valenti | 2013 | Caucasian | 194/187/79 | 146/95/16 | blood donors | TaqMan | Y | 7 | - | ||
| HCV | 132/107/32 | ||||||||||
| HBV | 31/19/7 | ||||||||||
| B + C | 2/6/0 | ||||||||||
| Alcoholic | 18/32/30 | ||||||||||
| Others | 11/23/10 | ||||||||||
| Friedrich | 2014 | Caucasian | 33/41/12 | 1135/718/97 | blood donors | RTqPCR | Y | 6 | - | ||
| HBV,HCV | 26/19/2 | ||||||||||
| Alcoholic | 3/17/9 | ||||||||||
| Other | 4/5/1 | ||||||||||
| Ezzikouri | 2014 | North-African | HCV | 43/35/23 | 66/54/12 | Hospital | TaqMan | Y | 6 | - | |
| Gao | 2014 | Asian | Unknown | 29/26/12 | 41/24/4 | Volunteers | PCR | Y | 7 | - | |
| Guyot | 2013 | Caucasian | 73/56/30 | 179/150/44 | - | TaqMan | Y | - | 8 | ||
| HCV | 54/26/13 | 86/49/25 | |||||||||
| Alcoholic | 19/30/17 | 93/101/19 | |||||||||
HBV: Hepatitis B virus; HCV: Hepatitis C virus; B + C: HBV and HCV co-infection; LCrt-PCR: LightCycler real time PCR; NOS-Ca: NOS of HCC risk; NOS-Ci: NOS of development of HCC in patients with cirrhosis.
Quality assessment
The quality of all included studies is shown in Table 1. The average NOS score of the HCC risk was 6.8, and it was 7 for other groups. The main bias arose from comparability where the additional confounding factors were not well controlled. Besides, some bias arose from Non-Response rate in the case-control study’s exposure.
HCC risk
In total, six studies that examined the association between the PNPLA3 rs738409 polymorphism and HCC risk, including 1016 patients with HCC and 3026 healthy controls, were included in our meta-analysis. We found a significant association between rs738409 and HCC risk using multiple different genetic models: allele contrast, G vs. C: OR = 2.02, 95% CI: 1.77-2.30; dominant, GG + CG vs. CC: OR = 2.03, 95% CI: 1.70-2.41; recessive, GG vs. CG + CC: OR = 3.56, 95% CI: 2.70-4.69; and co-dominant, GG vs. CC: OR = 4.40, 95% CI: 3.28-5.91, CG vs. CC: OR = 1.62, 95% CI: 1.34-1.59 (Table 2; Figure 2).
Table 2.
Association between PNPLA3 rs748309 polymorphisms and HCC risk
| Study group | Simple size (Case/Control) | Genetic models | I2 (%) | Ph | Effect-model | OR [95% CI] | P | |
|---|---|---|---|---|---|---|---|---|
| Overall | 1016/3026 | Allele contrast | G vs. C | 0 | 0.50 | Fixed | 2.02 [1.77, 2.30] | <0.00001 |
| Dominant model | GG + CG vs. CC | 0 | 0.41 | Fixed | 2.03 [1.70, 2.41] | <0.00001 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.91 | Fixed | 3.56 [2.70, 4.69] | <0.00001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.70 | Fixed | 4.40 [3.28, 5.91] | <0.00001 | ||
| CG vs. CC | 0 | 0.42 | Fixed | 1.62 [1.34, 1.59] | <0.00001 | |||
| Over-dominant model | GG + CC vs. CG | 5 | 0.39 | Fixed | 0.84 [0.71, 1.00] | 0.05 | ||
| Hepatitis virus | 640/2957 | Allele contrast | G vs. C | 0 | 0.63 | Fixed | 1.44 [1.23, 1.68] | <0.00001 |
| Dominant model | GG + CG vs. CC | 0 | 0.85 | Fixed | 1.40 [1.14, 1.72] | 0.001 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.66 | Fixed | 2.15 [1.52, 3.04] | <0.001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.67 | Fixed | 2.35 [1.63, 3.38] | <0.00001 | ||
| CG vs. CC | 0 | 0.87 | Fixed | 1.24 [1.00, 1.54] | 0.05 | |||
| Over-dominant model | GG + CC vs. CG | 0 | 0.74 | Fixed | 0.93 [0.75, 1.14] | 0.47 | ||
| Alcohol | 255/2825 | Allele contrast | G vs. C | 0 | 0.7 | Fixed | 3.94 [3.21, 4.82] | <0.00001 |
| Dominant model | GG + CG vs. CC | 13 | 0.33 | Fixed | 4.90 [3.52, 6.81] | <0.00001 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.94 | Fixed | 7.99 [5.56, 11.48] | <0.00001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.45 | Fixed | 14.65 [9.47, 22.64] | <0.00001 | ||
| CG vs. CC | 34 | 0.21 | Fixed | 3.23 [2.27, 4.60] | <0.00001 | |||
| Over-dominant model | GG + CC vs. CG | 46 | 0.14 | Fixed | 0.74 [0.56, 0.98] | 0.03 | ||
| HCV | 505/1007 | Allele contrast | G vs. C | 0 | 0.67 | Fixed | 1.53 [1.28, 1.83] | <0.00001 |
| Dominant model | GG + CG vs. CC | 0 | 0.65 | Fixed | 1.49 [1.18, 1.88] | 0.0007 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.88 | Fixed | 2.39 [1.63, 3.50] | <0.00001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.82 | Fixed | 2.65 [1.77, 3.97] | <0.00001 | ||
| CG vs. CC | 0 | 0.58 | Fixed | 1.28 [1.00, 1.64] | 0.05 | |||
| Over-dominant model | GG + CC vs. CG | 0 | 0.51 | Fixed | 0.92 [0.73, 1.17] | 0.50 | ||
Figure 2.
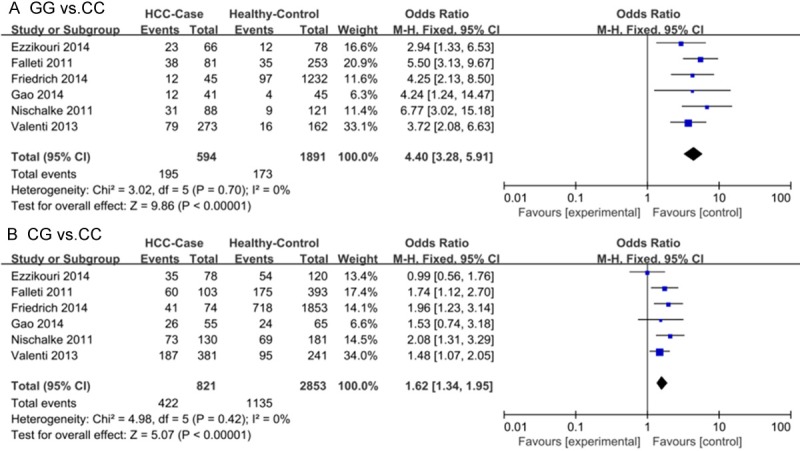
Forest plots of meta-analysis of association between the PNPLA3 rs738409 polymorphism and HCC risk without considering etiology (A GG vs. CC; B CG vs. CC).
Because the etiology of HCC in the patients reported by Gao et al. [37] was not obvious, and it was difficult to calculate the genetic frequencies of the HBV- and HCV-related HCC patients in the publication written by Friedrich et al. [35], these studies were not included in the meta-analysis subgroup stratification by etiology. In addition, due to limited sample sizes, we only were able to perform stratification analysis using hepatitis virus, alcohol, and HCV subgroups. As for the total study population, the results from the stratification analysis were significant in certain genetic models: hepatitis virus: allele contrast, G vs. C: OR = 1.44, 95% CI: 1.23-1.68; dominant, GG + CG vs. CC: OR = 1.40, 95% CI = 1.14-1.72; recessive, GG vs. CG + CC: OR = 2.15, 95% CI: 1.52-3.04; and co-dominant, GG vs. CC: OR = 2.35, 95% CI: 1.63-3.38; alcohol: allele contrast, G vs. C: OR = 3.94, 95% CI: 3.21-4.28; dominant, GG + CG vs. CC: OR = 4.90, 95% CI: 3.52-6.81; recessive, GG vs. CG + CC: OR = 7.99, 95% CI: 5.56-11.48; co-dominant, GG vs. CC: OR = 14.65, 95% CI: 9.47-22.64, CG vs. CC: OR = 3.23, 95% CI: 2.27-4.60; and over-dominant, GG + CC vs. CG: OR = 0.74, 95% CI: 0.56-0.98; HCV: allele contrast, G vs. C: OR = 1.53, 95% CI: 1.28-1.83, dominant, GG + CG vs. CC: OR = 1.49, 95% CI: 1.18-1.88; recessive, GG vs. CG + CC: OR = 2.39, 95% CI: 1.63-3.50; and co-dominant, GG vs. CC: OR = 2.65, 95% CI: 1.77-3.97) (Table 2).
Risk for development of HCC in cirrhosis
Following application of the inclusion and exclusion criteria, three studies including 461 patients with HCC related to cirrhosis and 876 patients with cirrhosis were included in our meta-analysis. We found significant overall association using several different genetic models: allele contrast, G vs. C: OR = 1.39, 95% CI: 1.17-1.64; dominant, GG + CG vs. CC: OR = 1.30, 95% CI: 1.03-1.65; recessive, GG vs. CG + CC: OR = 1.91, 95% CI: 1.41-2.58; and co-dominant, GG vs. CC: OR = 2.00, 95% CI: 1.43-2.79 (Table 3; Figure 3). However, associations were only observed in patients with HCC related to alcoholic cirrhosis when sub-grouped by etiology: allele contrast, G vs. C: OR = 1.82, 95% CI: 1.43-2.32; dominant model, GG + CG vs. CC: OR = 1.87, 95% CI: 1.28-2.73; recessive model, GG vs. CG + CC: OR = 2.65, 95% CI: 1.75-4.02; and co-dominant model, GG vs. CC: OR = 3.28, 95% CI: 2.01-5.35 (Table 3).
Table 3.
Association between PNPLA3 rs748309 polymorphisms and development of HCC in patients with cirrhosis
| Study group | Simple size (Case/Control) | Genetic models | I2 (%) | Ph | Effect-model | OR [95% CI] | P | |
|---|---|---|---|---|---|---|---|---|
| Overall | 461/876 | Allele contrast | G vs. C | 0 | 0.43 | Fixed | 1.39 [1.17, 1.64] | 0.0001 |
| Dominant model | GG + CG vs. CC | 5 | 0.35 | Fixed | 1.30 [1.03, 1.65] | 0.03 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.78 | Fixed | 1.91 [1.41, 2.58] | <0.0001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.52 | Fixed | 2.00 [1.43, 2.79] | <0.0001 | ||
| CG vs. CC | 4 | 0.35 | Fixed | 1.14 [0.89, 1.48] | 0.30 | |||
| Over-dominant model | GG + CC vs. CG | 0 | 0.54 | Fixed | 1.12 [0.88,1.41] | 0.36 | ||
| Alcohol | 212/425 | Allele contrast | G vs. C | 0 | 0.61 | Fixed | 1.82 [1.43, 2.32] | <0.0001 |
| Dominant model | GG + CG vs. CC | 0 | 0.5 | Fixed | 1.87 [1.28, 2.73] | 0.001 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.55 | Fixed | 2.65 [1.75, 4.02] | <0.00001 | ||
| Co-dominant model | GG vs. CC | 0 | 0.37 | Fixed | 3.28 [2.01, 5.35] | <0.00001 | ||
| CG vs. CC | 0 | 0.48 | Fixed | 1.45 [0.96, 2.17] | 0.08 | |||
| Over-dominant model | GG + CC vs. CG | 0 | 0.56 | Fixed | 1.13 [0.80, 1.58] | 0.49 | ||
| HCV | 226/398 | Allele contrast | G vs. C | 19 | 0.29 | Fixed | 1.10 [0.86, 1.42] | 0.44 |
| Dominant model | GG + CG vs. CC | 0 | 0.42 | Fixed | 1.09 [0.78, 1.53] | 0.62 | ||
| Recessive model | GG vs. CG + CC | 0 | 0.46 | Fixed | 1.22 [0.74, 1.99] | 0.43 | ||
| Co-dominant model | GG vs. CC | 9 | 0.33 | Fixed | 1.23 [0.73, 2.07] | 0.43 | ||
| CG vs. CC | 0 | 0.63 | Fixed | 1.05 [0.73, 1.52] | 0.78 | |||
| Over-dominant model | GG + CC vs. CG | 0 | 0.85 | Fixed | 1.01 [0.71, 1.43] | 0.97 | ||
Figure 3.
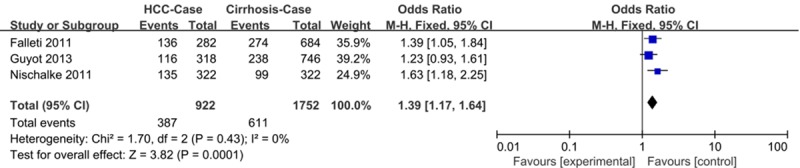
Forest plots of meta-analysis of association between the PNPLA3 rs738409 polymorphism and the risk of HCC in patients with cirrhosis using allele contrast model (G vs. C).
Sensitivity analysis and publication bias
Sensitivity analysis was performed by sequential omission of individual studies to investigate the influence of each study on the overall OR. The significance of the pooled OR in the analysis of HCC risk and the risk of development of HCC in cirrhosis were not excessively affected in any of the contrasts analyzed, indicating the robustness of the meta-analysis results. No evidence of publication bias was found in the symmetrical graphics obtained using Begg’s funnel plot for the rs738409 polymorphism and either HCC risk (Figure 4) or the development of HCC in cirrhosis (Figure 5); this finding was supported by the results of Egger’s tests (each genetic model P > 0.05).
Figure 4.
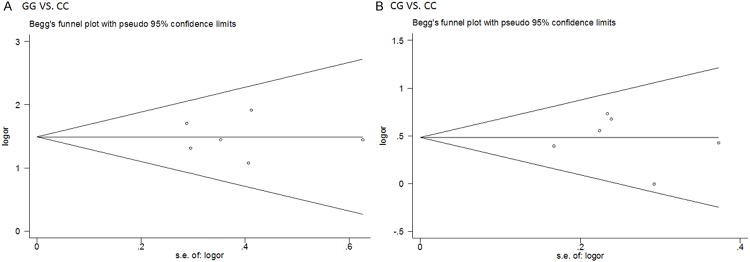
Begg’s funnel plots for publication bias test on the associations of PNPLA3 rs738409 polymorphism and HCC risk without considering etiology (A: GG vs. CC; B CG vs. CC).
Figure 5.
Begg’s funnel plots for publication bias test on risk for development of HCC in cirrhosis without considering etiology (G vs. C).
Discussion
Following the genome-wide association studies (GWAS) and gradually apply within success in the medical fields in recent years, an increasing number of studies have reported that genetic alterations might be used to predict the risk of various cancers [40-42]. Without doubt, these findings will likely contribute to the primary prevention of cancers. Some SNPs might even be regarded as prognostic factors for the success (or failure) of certain chemotherapy strategies [43]. However, only a few genetic variants have been reproducibly demonstrated to be linked to hepatocarcinogenesis [44]. To date, the effect of PNPLA3 rs738409 polymorphic variants on HCC risk and development has been investigated in numerous association studies. However, the results have been inconsistent, possibly due to limited sample sizes and ethnic variation. In order to address the inconsistencies and limitations of previously published studies, and to draw a more robust conclusion, the current meta-analysis was performed.
The key findings of this meta-analysis are: (1) the PNPLA3 rs738409 polymorphism is a risk factor for HCC occurrence in both healthy individuals and patients with cirrhosis; (2) this association is present for HCV-related HCC, alcohol-related HCC, and for the development of HCC in alcoholic cirrhosis; (3) the association is more pronounced in alcohol-than in HCV-related HCC; and (4) healthy individuals harboring two copies of the rs738409variant G allele had a high risk of HCC occurrence.
The first section of our meta-analysis included six studies that met the inclusion criteria, comprising1175 patients with HCC and 3026 healthy controls that were used to investigate the association between the PNPLA3 rs738409 polymorphism and risk of HCC without considering the etiology of the disease. Statistically significant results were found in our analysis, suggesting that the PNPLA3 rs738409 polymorphism might be a risk factor for HCC occurrence. When subgroup analysis was performed by etiology, the association was also identified, and was ascertained to be more pronounced in alcohol-related HCC than in HCV-related HCC. Upon comparison of results from the same genetic model (except for the over-dominant model), we found that the OR of the alcohol-related HCC risk was always higher than that of the HCV-related HCC risk, and that the confidence intervals did not overlap, supporting the premise that the differences were significant. Based on similar evidence, we also have sufficient reason to believe that harboring two copies of the rs738409 G variant was associated with a high risk of HCC occurrence. Similarly, the association with the GG genotype is even more pronounced in alcohol-related HCC. Three factors might account for this phenomenon: (1) both alcoholic and HCV-related liver diseases including HCC occur during the process of liver fat accumulation. However, compared to HCV-related liver disease, alcohol increases the level of patient serum triglycerides; in addition, the PNPLA3 rs738409 mutation also increase the intracellular accumulation of triglycerides [45,46], which might further amplify the HCC risk. (2) Several HCV proteins have been shown to have direct effects on hepato-carcinogenesis and to upregulate the processes of carcinogenesis [47], however, there are no such proteins that can upregulate alcohol-related hepato-carcinogenesis; to some extent, this also indirectly amplifies the effect of alcohol. (3) To some extent, we may also use gene-environment interacts to explain our findings, and in fact, our finding that may also indirectly support the role of genetic and environmental interactions in hepatocarcinogenesis. Generally, two of the studies included in our meta-analysis [24,35], in contrast to certain earlier individual studies, reported that the frequency of the Gallele did not differ between healthy controls and patients with HCC related to HCV, and that the G allele did not increase hepatitis-related HCC risk. However, other included studies [33,34,36] concluded that the G allele might increase HCC risk independent of liver disease, in line with the results of the current meta-analysis. Small sample sizes likely account for discrepancies between studies. An assessment of the association between the PNPLA3 rs738409 polymorphism and HCC risk found that more precise estimates were obtained using large sample numbers than when using individual analyses using smaller sample sizes. Moreover, smaller sample sizes serve to amplify the effects of genetic backgrounds, which may contribute to variations in the association of SNPs with HCC.
In the second part of our meta-analysis study, we examined one case-control and two cohort studies, including 461 patients that developed HCC consequent to cirrhosis, and 876 patients with cirrhosis that served as controls. In this analysis, we found that the PNPLA3 rs738409 G allele also served as a risk factor for HCC occurrence in patients with cirrhosis, suggesting that a variation in PNPLA3 rs738409 plays an incontrovertible role in the development of HCC in patients with cirrhosis. However, when the subgroups were stratified by etiology, this significant association was only identified in patients with HCC that had evolved from alcohol-related cirrhosis. We note that the results of our current study are different from those of a previous meta-analysis performed by Trepo et al. [38] due to differences in the included studies. First, in our study, the included patients with HCC were enrolled based on cirrhosis, which differed from the previous meta-analysis; second, our current study did not include three studies that were included in the previous meta-analysis. Two of these were published in the form of letters, which did not meet our criteria as we were unable to accurately assess the quality of the study. In addition, upon careful review of the paper written by Valenti et al. [39], fifty patients with HCC were identifiedin total, with genotype frequencies of 17:21:12 for CC: CG: GG; however, when Trepo et al. [38] performed theprevious meta-analysis, the frequencies of the CC:CG:GG genotypeswere reported as 12:21:17. An additional discrepancy with the results from the previous meta-analysis was observed for the 44 of 50 instances of HCC that occurred in patients with a previous diagnosis of cirrhosis, in whom the frequency of GG genotype was reported to be 10 [38]. These limitations highlight the value of the current meta-analysis based on its overall quality and stability.
Both previous [38,48] and our present meta-analyses demonstrated that the G allele of rs738409 is a risk factor for HCC, but our starting point differs and we have introduced strategic innovations. In the first section of our meta-analysis, all individuals in the control groups were healthy, and in the second meta-analysis section, we considered patients with cirrhosis as the matched control group, where in the etiology of cirrhosis was clear. Additionally, when we performed a subgroup analysis by etiology, where each individual had a clear cause of liver disease with a corresponding etiology. Finally and foremost, we found that healthy individuals carrying two copies of the rs738409 G variant allele had a higher risk of HCC than did those carrying one variant allele. As expected, among the healthy individuals carrying the rs738409 G variant allele, the HCC risk was higher in patients who had abused alcohol compared with those infected with HCV, owing to the difference in underlying pathogenesis.
Despite the considerable efforts that have been made to explore the associations between the PNPLA3 rs738409 polymorphism and overall HCC risk as well as the risk of its development in patients with cirrhosis, several limitations in this meta-analysis must be acknowledged. First, the sample size of our current meta-analysis was limited, especially regarding the development of HCC in patients with cirrhosis. Most of these patients were Caucasian, which is the reason why subgroups were not analyzed by ethnicity. Second, variations in individual susceptibility to HCC can also clearly be attributed to complex gene-gene and gene-environmental exposure interactions [9]. In addition, hepatitis virus infection, alcohol abuse, smoking, and obesity can also increase the risk of HCC [1,5,9-11]. However, we were unable to investigate the interactions of the PNPLA3 rs738409 polymorphism with other genes and environmental factors potentially involved in the development of HCC due to the lack of necessary data. We must further recognize that heterogeneity exists in any study, including our meta-analysis. Another limitation is that the results obtained in the present study were based on unadjusted estimations. Finally, additional major potential confounders, including age, gender, obesity, diabetes, and environmental factors should be taken into consideration for a more accurate analysis.
In summary, our meta-analysis combining all currently available data suggests that there are significant associations of the PNPLA3 rs738409 polymorphism with the risk of HCC and its development of HCC in patients with cirrhosis. However, further studies in different ethnic groups using larger sample sizes and well-matched controls are greatly needed to clarify any associations identified herein. Future studies considering gene-gene and gene-environment interactions are also encouraged to provide a more comprehensive understanding of the potential role of the PNPLA3 polymorphism in the pathogenesis of HCC.
Disclosure of conflict of interest
None.
References
- 1.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47(Suppl):S2–6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Beaugrand M. Hepatocellular carcinoma: present status and future prospects. J Hepatol. 2003;38(Suppl 1):S136–49. doi: 10.1016/s0168-8278(02)00432-4. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Dyson J, Jaques B, Chattopadyhay D, Lochan R, Graham J, Das D, Aslam T, Patanwala I, Gaggar S, Cole M, Sumpter K, Stewart S, Rose J, Hudson M, Manas D, Reeves HL. Hepatocellular cancer: the impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2014;60:110–117. doi: 10.1016/j.jhep.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Sanyal AJ. NASH: A global health problem. Hepatol Res. 2011;41:670–674. doi: 10.1111/j.1872-034X.2011.00824.x. [DOI] [PubMed] [Google Scholar]
- 8.Harada K, Hirohara J, Ueno Y, Nakano T, Kakuda Y, Tsubouchi H, Ichida T, Nakanuma Y. Incidence of and risk factors for hepatocellular carcinoma in primary biliary cirrhosis: national data from Japan. Hepatology. 2013;57:1942–1949. doi: 10.1002/hep.26176. [DOI] [PubMed] [Google Scholar]
- 9.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 10.Forner A, Bruix J. Biomarkers for early diagnosis of hepatocellular carcinoma. Lancet Oncol. 2012;13:750–751. doi: 10.1016/S1470-2045(12)70271-1. [DOI] [PubMed] [Google Scholar]
- 11.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 12.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Putman RK, Rosas IO, Hunninghake GM. Genetics and early detection in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;189:770–778. doi: 10.1164/rccm.201312-2219PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares S, Craveiro R, Catarino R, Breda E, Medeiros R, Bravo I. The role of nt590 P21 gene polymorphism in the susceptibility to nasopharyngeal cancer. Exp Oncol. 2014;36:44–47. [PubMed] [Google Scholar]
- 16.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirazzi C, Valenti L, Motta BM, Pingitore P, Hedfalk K, Mancina RM, Burza MA, Indiveri C, Ferro Y, Montalcini T, Maglio C, Dongiovanni P, Fargion S, Rametta R, Pujia A, Andersson L, Ghosal S, Levin M, Wiklund O, Iacovino M, Boren J, Romeo S. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014;23:4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min HK, Sookoian S, Pirola CJ, Cheng J, Mirshahi F, Sanyal AJ. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G66–76. doi: 10.1152/ajpgi.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stickel F, Buch S, Lau K, Meyer zu Schwabedissen H, Berg T, Ridinger M, Rietschel M, Schafmayer C, Braun F, Hinrichsen H, Gunther R, Arlt A, Seeger M, Muller S, Seitz HK, Soyka M, Lerch M, Lammert F, Sarrazin C, Kubitz R, Haussinger D, Hellerbrand C, Broring D, Schreiber S, Kiefer F, Spanagel R, Mann K, Datz C, Krawczak M, Wodarz N, Volzke H, Hampe J. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53:86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 20.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 21.Trepo E, Pradat P, Potthoff A, Momozawa Y, Quertinmont E, Gustot T, Lemmers A, Berthillon P, Amininejad L, Chevallier M, Schlue J, Kreipe H, Deviere J, Manns M, Trepo C, Sninsky J, Wedemeyer H, Franchimont D, Moreno C. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54:60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 22.Tong J, Guo J, Hu J, Hou S, Zhang Y, Li Q. Correlation between patatin-like phospholipase domain-containing protein 3 gene polymorphisms and liver cirrhosis in a chinese han population with chronic hepatitis B. Hepat Mon. 2014;14:e18943. doi: 10.5812/hepatmon.18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouwer WP, van der Meer AJ, Boonstra A, Pas SD, de Knegt RJ, de Man RA, Hansen BE, Ten Kate FJ, Janssen HL. The impact of PNPLA3 (rs738409 C>G) polymorphisms on liver histology and long-term clinical outcome in chronic hepatitis B patients. Liver Int. 2015;35:438–447. doi: 10.1111/liv.12695. [DOI] [PubMed] [Google Scholar]
- 24.Nischalke HD, Berger C, Luda C, Berg T, Muller T, Grunhage F, Lammert F, Coenen M, Kramer B, Korner C, Vidovic N, Oldenburg J, Nattermann J, Sauerbruch T, Spengler U. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS One. 2011;6:e27087. doi: 10.1371/journal.pone.0027087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyot E, Sutton A, Rufat P, Laguillier C, Mansouri A, Moreau R, Ganne-Carrie N, Beaugrand M, Charnaux N, Trinchet JC, Nahon P. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol. 2013;58:312–318. doi: 10.1016/j.jhep.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 26.Corradini SG, Burza MA, Molinaro A, Romeo S. Patatin-like phospholipase domain containing 3 sequence variant and hepatocellular carcinoma. Hepatology. 2011;53:1776. doi: 10.1002/hep.24244. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 30.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 31.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 32.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezzikouri S, Alaoui R, Tazi S, Nadir S, Elmdaghri N, Pineau P, Benjelloun S. The adiponutrin I148M variant is a risk factor for HCV-associated liver cancer in North-African patients. Infect Genet Evol. 2014;21:179–183. doi: 10.1016/j.meegid.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Falleti E, Fabris C, Cmet S, Cussigh A, Bitetto D, Fontanini E, Fornasiere E, Bignulin S, Fumolo E, Bignulin E, Pirisi M, Toniutto P. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31:1137–1143. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 35.Friedrich K, Wannhoff A, Kattner S, Brune M, Hov JR, Weiss KH, Antoni C, Dollinger M, Neumann-Haefelin C, Seufferlein T, Schemmer P, Schirmacher P, Stremmel W, Gotthardt DN. PNPLA3 in end-stage liver disease: alcohol consumption, hepatocellular carcinoma development, and transplantation-free survival. J Gastroenterol Hepatol. 2014;29:1477–1484. doi: 10.1111/jgh.12540. [DOI] [PubMed] [Google Scholar]
- 36.Valenti L, Motta BM, Soardo G, Iavarone M, Donati B, Sangiovanni A, Carnelutti A, Dongiovanni P, Rametta R, Bertelli C, Facchetti F, Colombo M, Fargion S, Fracanzani AL. PNPLA3 I148M polymorphism, clinical presentation, and survival in patients with hepatocellular carcinoma. PLoS One. 2013;8:e75982. doi: 10.1371/journal.pone.0075982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao C, Wang X, Li L, Zhou X. The Correlation study of PNPLA3 Gene Polymorphism and Genetic Susceptibility to Hepatocellular Carcinoma. Medical Innovation of China. 2014:12–14. [Google Scholar]
- 38.Trepo E, Nahon P, Bontempi G, Valenti L, Falleti E, Nischalke HD, Hamza S, Corradini SG, Burza MA, Guyot E, Donati B, Spengler U, Hillon P, Toniutto P, Henrion J, Franchimont D, Deviere J, Mathurin P, Moreno C, Romeo S, Deltenre P. Association between the PNPLA3 (rs738409 C > G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology. 2014;59:2170–2177. doi: 10.1002/hep.26767. [DOI] [PubMed] [Google Scholar]
- 39.Valenti L, Rumi M, Galmozzi E, Aghemo A, Del Menico B, De Nicola S, Dongiovanni P, Maggioni M, Fracanzani AL, Rametta R, Colombo M, Fargion S. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53:791–799. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Wang H. XRCC3 T241M polymorphism is associated risk of hepatocellular carcinoma in the Chinese. Tumour Biol. 2013;34:2249–2254. doi: 10.1007/s13277-013-0765-4. [DOI] [PubMed] [Google Scholar]
- 41.Mohamed FZ, Hussein YM, El-Deen IM, Sabea MS. Cyclooxygenase-2 single-nucleotide polymorphisms and hepatocellular carcinoma in Egypt. Mol Biol Rep. 2014;41:1461–1468. doi: 10.1007/s11033-013-2991-7. [DOI] [PubMed] [Google Scholar]
- 42.Hosen MB, Islam J, Salam MA, Islam MF, Hawlader MZ, Kabir Y. N-acetyltransferase 2 gene polymorphism as a biomarker for susceptibility to bladder cancer in Bangladeshi population. Asia Pac J Clin Oncol. 2014;11:78–84. doi: 10.1111/ajco.12291. [DOI] [PubMed] [Google Scholar]
- 43.Kim HJ, Im SA, Keam B, Ham HS, Lee KH, Kim TY, Kim YJ, Oh DY, Kim JH, Han W, Jang IJ, Kim TY, Park IA, Noh DY. ABCB1 Polymorphism as Prognostic Factor in Breast Cancer Patients Treated with Docetaxel and Doxorubicin Neoadjuvant Chemotherapy. Cancer Sci. 2015;106:86–93. doi: 10.1111/cas.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. 2012;57:663–674. doi: 10.1016/j.jhep.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 45.Perttila J, Huaman-Samanez C, Caron S, Tanhuanpaa K, Staels B, Yki-Jarvinen H, Olkkonen VM. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am J Physiol Endocrinol Metab. 2012;302:E1063–1069. doi: 10.1152/ajpendo.00125.2011. [DOI] [PubMed] [Google Scholar]
- 46.Pirazzi C, Adiels M, Burza MA, Mancina RM, Levin M, Stahlman M, Taskinen MR, Orho-Melander M, Perman J, Pujia A, Andersson L, Maglio C, Montalcini T, Wiklund O, Boren J, Romeo S. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol. 2012;57:1276–1282. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 47.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singal AG, Manjunath H, Yopp AC, Beg MS, Marrero JA, Gopal P, Waljee AK. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am J Gastroenterol. 2014;109:325–334. doi: 10.1038/ajg.2013.476. [DOI] [PMC free article] [PubMed] [Google Scholar]


