Abstract
Tim-3 is considered as one of the T-cell immunoglobulin mucin (TIM) gene family members, which contributes to the activating or silencing genes, but the mechanism of Tim-3 function in mediating SLE or tumor metastasis has not been well explored. Here, we reported Tim-3 was high expressed in the peripheral blood mononuclear cells (PBMCs) of patients with SLE, detected by RT-PCR, significantly, GATA-3 mRNA expression also increased in patients with SLE, compared with the healthy control groups. The bioinformatics used to detect the TCGA database indicated the abnormal expression of Tim-3 was involved in several different cancer types. Further, the higher expression of Tim-3 in kidney renal clear cell carcinoma TCGA database indicated it was a marker for worse 5-year survival. The high expression of Tim-3 in different ccRCC cell lines was detected in both RNA level and protein level. Further, two kinds of relative Tim-3 siRNAs in ccRCC cell lines inhibit cell migration and invasion in vitro, However, the inhibition could be partially rescued by the additional GATA3 knockdown. Further, the down regulation in the RNA and protein levels of GATA3, and the negative correlation between Tim-3 and GATA3 implied that suppression of downstream GATA3 was an important mechanism by which Tim-3 triggered metastasis in ccRCC cell lines. Together, our experiments reveal the role for Tim-3 in facilitating SLE or invasive potential of ccRCC cells by either activating GATA3 or inhibiting GATA3, suggesting that Tim-3 might be a potential therapeutic target for treating SLE or clear cell renal cell carcinoma.
Keywords: Tim3, GATA3, SLE, ccRCC
Introduction
T cell immunoglobulin mucin (TIM) family was considered as critical checkpoint proteins in the regulation of multiple immune response phases and in the maintaining immune homeostasis [1]. The TIM gene family was proved by different groups to have large impact on the clinical consequences of sterile inflammation, tumor immune surveillance, immune evasion, antimicrobial defense and chronic autoimmune diseases, such as multiple sclerosis and rheumatoid arthritis (RA) [2-5].
Among the members, TIM-3 was the first transmembrane protein that specifically identifies Th1 cells in both mice and human, and expressed on many cells including CD8 T-cells, monocytes, and dendritic cells [6-9]. However, the expression and function of Tim-3 in multiple human autoimmune diseases and tumor biology was limited only by a small number of studies, and the molecular mechanisms modulating Tim-3 function were not well understood.
Systemic lupus erythematosus (SLE) was a prototypic systemic autoimmune disease, the cause of which was unknown, often with the influence on multiple organ systems such as kidney. Defects in the balance regulation of T-helper 1 (Th1)/Th2 cell function has been implicated in the pathophysiology of SLE. Most SLE patients would develop kidney disease, and lupus nephritis (LN) is one of its most severe manifestations [10].
Clear cell renal cell carcinoma (ccRCC) was a major pathologic form of kidney cancer, which accounted for about 90% of kidney malignancies [11]. About 5-10% of ccRCCs would extend into the renal vein or the IVC, when this happened, patients with metastasis have a poor prognosis with a 5-year survival rate of less than 20% [12].
The GATA transcription factor families which typically bind to the element A/TGATA A/G are considered as essential regulators in numerous tissues [13,14]. They all share 2 highly conserved zinc fingers of the C2H2 type, with the function of DNA binding and majority of protein interactions. GATA3 was first identified in a screen for GATA factors in the T cell lineage [15,16], and played an essential role in early T cell development and the specification of the Th2 cells.
More and more evidence suggests that Tim-3 is involved in to maintain the malignant phenotype of autoimmune diseases such as SLE and might also has a role contribute to the tumor progression, but the mechanisms between them still need further discussion.
Materials and methods
Cell culture
Normal human kidney epithelial cell line HK-2, human clear cell renal cell carcinoma cell line 786-O and Caki-2 cells were purchased from American Tissue Type Collection (Manassas, VA). All cell lines are suitable transfection hosts. All cells were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (HyClone), 100 IU/ml penicillin and 100 IU/ml streptomycin. These cell lines were grown at 37°C in a humidified atmosphere with 5% CO2. For the transfections, cells were grown to 70% and transfected with plasmids using Lipofectamine2000 (Invitrogen) or transfected with the indicated siRNA using Lipofectamine RNAimax according to the manufacturer’s recommendation.
Patients and specimens
60 female patients diagnosed as SLE with biopsy evidence of lupus nephritis were collected in the hospital from the year of 2008 to 2010, their age were from 24 to 55, the average was (32±8). The disease activity of the SLE patients was evaluated using the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI), SLEDAI ≥10 was considered as activity. The SLEDAI score (mean ± SE) was 14.5±9.3.
Their blood samples were collected in sterile containers, 30 healthy blood samples were as controls. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density-gradient centrifugation; cells from the interface were collected and washed three times, prepared for RNA extraction and ELISA.
The 32 samples of grade III ccRCC tissues were obtained from patients in the Affiliated Hospital of Qingdao University. Samples were selected from patients with complete clinicopathologic information. The study was approved by the ethics committee of the hospital; all patients were informed with written consent to understand their tissue would be used for research purposes.
Reagents
Rabbit anti-human Tim-3 antibody, β-actin antibody, GATA3 antibody and secondary antibody were all purchased from Santa Cruz (CA, USA). Specific siRNA targeting Tim-3, GATA3 were from Sigma-Aldrich (CA, USA). A negative control siRNA (NC) was also used as control (Sigma-Aldrich). Matrigel was purchased from BD Biosciences (CA, USA). ELISA kits for Tim-3 or GATA3 detection were from NeoBioscience (Beijing, China).
Small interfering RNA-mediated RNA interference
The target sequences used were Tim-3 siRNA#1 (SASI_Hs01_00114252), Tim-3 siRNA#2 (SASI_Hs01_00114255), GATA3 siRNA (SASI_Hs01_00153934) and negative control siRNA. 100 pmol siRNA was transfected into A704 and ACHN cells using Lipofectamine RNAimax following the manufacturer’s instructions, after 72 h post-transfection, total RNA was prepared and RT-PCR was performed to analyze mRNA levels of target genes.
Quantitative real-time PCR (qRT-PCR)
Total RNA of cell lysates were extracted with Trizol solution (Invitrogen, Carlsbad, CA, USA) after 48 h-72 h post infection, the RNA were reverse transcribed to cDNA using 1 μg of total RNA with M-MLV Reverse Transcriptase, according to the manufacturer’s instructions. Real-time RT-PCR primers were as follows: Tim-3, forward 5-GCTACTACTTACAAGGTCCTCAG-3, reverse 5-ATTCACATCCCTTTCATCAGTC-3; β-actin, forward 5-TGGCACCCAGCACAATGAA-3, reverse 5-CTAAGT CATAGTCCGCCTAGAAGCA-3; GATA3, forward 5-GCTTCACAATATTAACAGACCC-3, reverse 5-GTTAAACGAGCTGTTCTTGG-3, the Real-time PCR was performed using SYBR (Roche) on an ABI 7900 sequence detection system (Applied Biosystems). All experiments were performed in three times; β-actin was used as a normalization control.
Western blot and ELISA
Cells were harvested 48 h-72 h after infection in each group and protein concentration was determined using the bicinchoninic acid (BCA). Subsequently, 30 μg protein was run on 10% SDS-PAGE gel and then transferred to NC membranes. NC membranes were blocked by 5% skim milk for 1 h at 4°C, and then incubated overnight at 4°C with primary antibodies, including Tim-3 (1:1000), GATA3 (1:1000) and β-Actin (1:500). Goat anti-Rabbit secondary antibodies (1:5000) and Western blotting Luminal reagent (Santa Cruz Biotechnology) was used to visualize the protein bands.
The Tim-3 and GATA3 levels were quantified by ELISA in accordance with the manufacturer’s instructions (Neobioscience Beijing, China).
Wound-healing assay
786-O cells were seeded in six-well plates at a density of 3×105 cells per well and cultured to form a confluent monolayer. Wounds of 2-mm width were made in the central area of the confluent culture with a sterile plastic scriber and the floating cells were washed away thrice with phosphate buffered saline (PBS). Photographs were taken at indicated time points after wounds were made. After incubation in a serum free medium for 24 hours, cultures were observed and photos were taken under a microscope. At least five randomly chosen areas were measured; phase-contrast pictures were taken of the wounded area using an inverted Leica microscope (Leica, Wetzlar, Germany).
Transwell invasion assay
The invasive ability of the cells was investigated using Transwells (8-μm pore size; for 24-well plate, Millipore) put into the 24-well plates. First, 100 μl Matrigel (50 μg/mL; BD Biosciences, CA) was added onto the surface of the chamber, incubated for 2 hours for solidification. Caki-2 cells were digested with trypsin, resuspended in media without FBS and counted; a total of 100 μl of the cell suspension (Fifty thousand cells) were then seeded into the upper chamber. The FBS in the lower chamber served as the chemoattractant. After incubation for 24 hours in the Matrigel, the cells on the upper surface were removed with cotton swabs, while the cells that invaded into the lower surface were fixed with 2% paraformaldehyde, stained with crystal violet. Images were taken under an inverted microscope (Leica, Wetzlar, Germany) at 40 magnification, each well with three random fields. Each experiment was performed in triplicate, representative photos well shown.
Statistical analysis
All observations were confirmed by at least three independent experiments. The data were presented as mean ± SD and were analyzed using the statistical package SPSS17.0. The significance of differences between two groups was determined using a two-sided Student’s t-test. ANOVA was used to evaluate the statistical significance of the mean values in case of multiple tests. Cox proportional hazards regression was used to test the prognostic significance of factors in univariate and multivariate models. P<0.05 was considered significant.
Results
The high expression of GATA3 is correlated with Tim-3 in PBMCs of SLE
First, the expression levels of Tim-3 were examined in 60 SLE samples and 30 healthy blood samples collected in our hospital and using qRT-PCR. The results showed that the mean expression levels of Tim-3 in the peripheral blood mononuclear cells of SLE (median: 0.48916, max: 2.1724, min: -1.0748) were significantly higher than those in the control groups (median: -0.51322, max: 0.42135, min: -1.16) (P=0.0003414<0.01, Figure 1A, left panel) considering the activity group and remission group as whole. The expression of GATA3 were also higher in the SLE groups (Figure 1A, right panel) (median: 0.51322, max: 3.42135, min: -1.1549) compared with the control (median: -0.8516, max: 0.87172, min: -2.0748). Further, a relative correlation was observed between Tim-3 and GATA3 in the 32 SLE activity groups using ELISA (R=0.4771; P<0.001; Figure 1B), while in the 28 remissive SLE, they were not significant correlation, datas were not shown.
Figure 1.
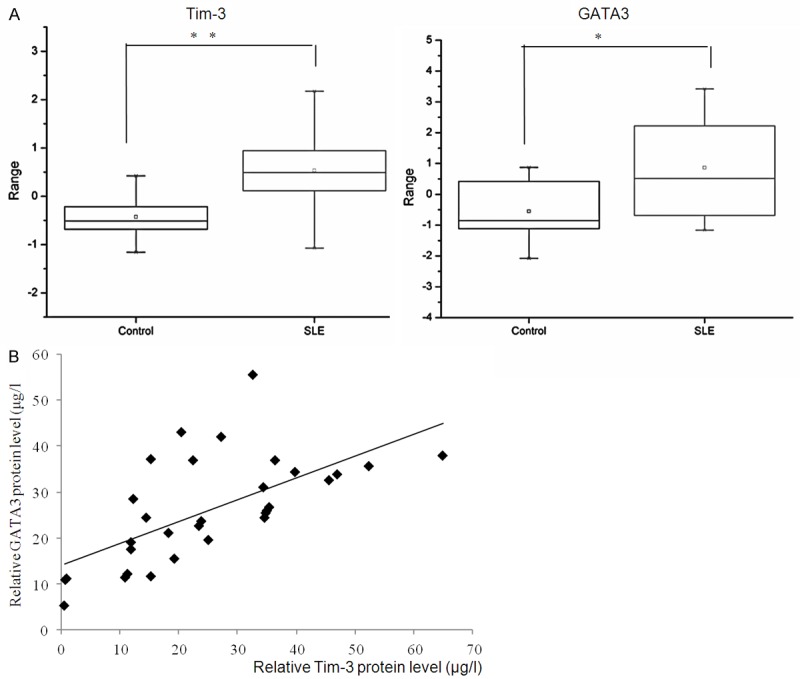
The high expression of GATA3 is correlated with Tim-3 in PBMCs of SLE. A. The relative expression level of Tim-3 was determined in 60 SLE samples and 30 control groups using Quantitative real-time PCR. Experiments were repeated three times. For all experiments n=3, Error bars represent average ± SD, Student’s t-test, *P<0.05, **P<0.01. The expression of GATA3 was determined by real-time PCR in 60 SLE samples, and 30 health control groups. Error bars represent standard error of the mean Student’s t-test, **P<0.01 (right panel). B. Correlation was found between expression of Tim-3 and GATA3 in the 32 active SLE samples (R=0.4771; P<0.001).
Bioinformatics indicated Tim-3 expression was associated with tumor grades in the ccRCC patients
In order to understand whether Tim-3 take apart in the cancer progress, we first use the cBioPortal (http://www.cbioportal.org/public-portal/) to get cross-cancer alteration summary for Tim-3 (HAVCR2), we observed that the expression of Tim-3 was abnormal in several TCGA database, high expression of Tim-3 was observed in Kidney Renal Clear Cell Carcinoma (Nature 2013, or provisional data), in Metastatic Prostate Adenocarcinoma by Michigan (Nature 2012), while in NCI-60 Cell Lines (NCI, Cancer Res. 2012), Skin Cutaneous Melanoma (Yale, Nature Genetics 2012), Uterine Corpus Endometrioid Carcinoma (TCGA, Nature 2013), Tim-3 mutation was found.
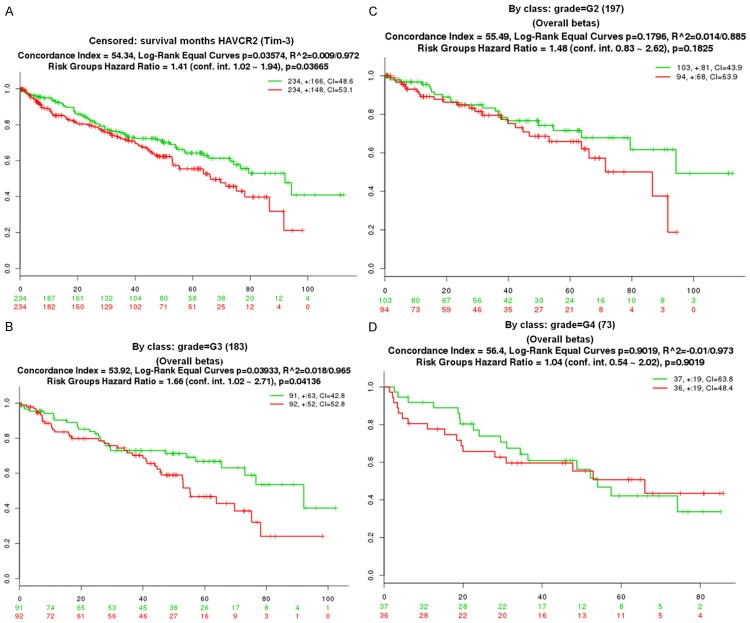
Further, we choose the kidney renal clear cell carcinoma TCGA database, including 468 samples, to investigate the role of Tim-3 contribute to the survival, grade, stage.
As shown in Figure 2A, Patients with ccRCC were divided into two groups, high and low, based on the median Tim-3 expression level. Kaplan-Meier survival analyses of the different group demonstrated that higher Tim-3 expression (red colour) was associated with a worse overall survival (Hazard Ratio =1.41, P=0.03665). Further, the patients were grouped by the tumor grade, in G3, Hazard Ratio was 1.66, P=0.04136<0.05 (Figure 2B), in G2 (n=197), Hazard Ratio =1.48, P=0.1825 (Figure 2C); in G4 (n=73), Hazard Ratio was 1.04, P=0.9019 (Figure 2D); although it was considered no obviously significant in G2 and G4, while, in G3, P was significant.
Figure 2.
Bioinformatics indicated Tim-3 expression was associated with tumor grades in the ccRCC patients. A. The kidney renal clear cell carcinoma TCGA database were used to plot Kaplan–Meier curves, and the overall 5-year survival rate was compared using the Cox log-rank test (**P<0.001). The y-axis represents the survival probability, and the x-axis represents the survival in months. B. The patients from TCGA database were further grouped by the tumor grade, in G3 (n=183), Kaplan-Meier curves were drawn; C. In G2 (n=197) Kaplan-Meier curves were drawn; D. In G4 (n=73), Kaplan-Meier curves were drawn; Cox proportional hazards regression was used to test the prognostic significance of factors in univariate and multivariate models. P<0.05 was considered significant.
The abnormal expression of Tim-3 and GATA3 in ccRCC cell lines and tissue
First, we analyzed the expression level of Tim-3 and GATA3 in two different renal cell carcinoma cell lines 786-O and Caki-2 cells and normal human kidney epithelial cell line HK-2. As shown on Figure 3A left panel, upregulation of Tim-3 was observed in the two examined ccRCC cell lines, including 786-O and Caki-2 cells, when compared to HK-2 cells, mRNA level were detected by qRT-PCR and protein level were detected by western blotting (Figure 3B, left panel). However, GATA3 was down regulated in the 786-O and Caki-2 cells compared with HK-2 (Figure 3A and 3B, right panel). Moreover, an inverse correlation was observed between Tim-3 and GATA3 (R=0.7669; P<0.001; Figure 3C) in the 32 ccRCC cancer samples collected using ELISA, the samples classified as grade III were selected for analysis, as TCGA database of ccRCC showed in the grade III, the expression of Tim-3 could be indicated as survival marker. The expression level of Tim-3 increased in the ccRCC cell lines, indicating the up regulation of Tim-3 may play critical role in it. In order to further understand the contribution of Tim-3 in the ccRCC progression, two relative siRNA was transfected into the 786-O cells. As the data shown, the efficiency of the siRNA was measured by Quantitative real-time PCR and Western Blotting. Whereas regarding the mRNA level, Tim-3 siRNA#1 was about fifteen percent that of the control group, while Tim3-siRNA#2 was about 20% compare with the control (P<0.001, Figure 3D, left panel). For protein level (Figure 3D, right panel), this inhibitory ratio was siRNA#1 12.5%, siRNA#2 20% according to the result of WB analysis.
Figure 3.
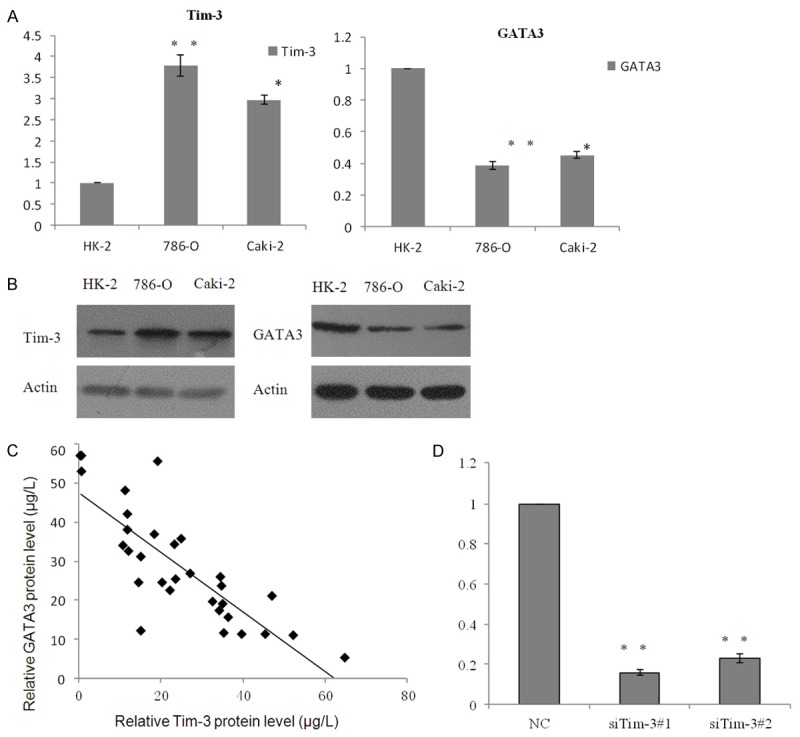
The abnormal expression of Tim-3 and GATA3 in ccRCC cell lines and tissue. A. Expression of Tim-3 was analyzed in two ccRCC cell lines 786-O and Caki-2, and normal HK-2 cells. Data are represented as mean ± S.D. Inversely, expression of GATA3 was also measured in these cell lines using RT-PCR. B. The relative protein level of Tim-3 and GATA3 were detected by western blotting. C. Negative correlation was found between RNA expression of Tim-3 and GATA3 in 32 ccRCC tumor samples (R=0.7669; P<0.001). D. The knockdown efficiencies of Tim-3 were confirmed by Quantitative real-time PCR (left panel) and western blotting (right panel).
Tim-3 effected ccRCC cell migration by down-regulation of GATA3
While Tim-3 was knockdown, GATA3 increased in the protein level (Figure 4A), respectively, in Tim-3 overexpressed 786-O cells, the protein (Figure 4B) expression of GATA3 decreased, which further supporting the notion that Tim-3 is required for the repression of GATA3.
Figure 4.
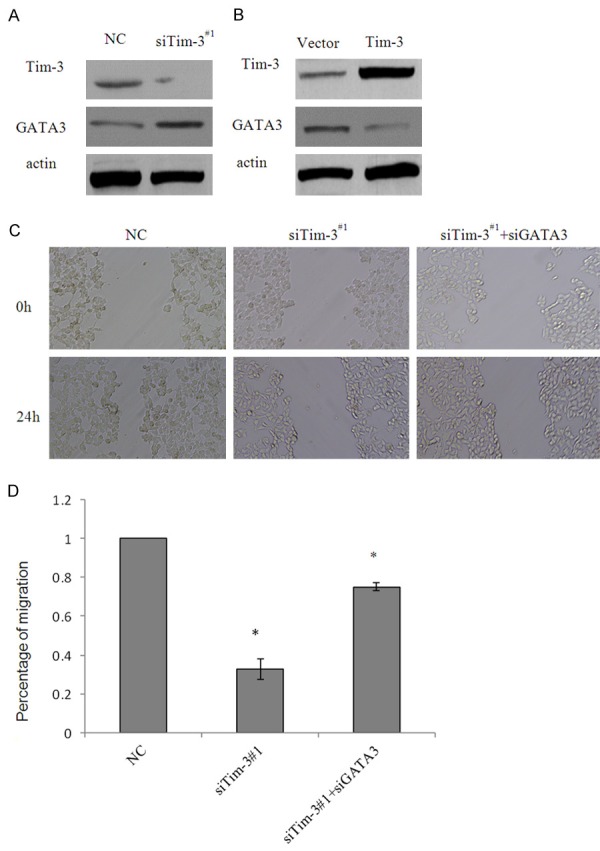
Tim-3 effected ccRCC cell migration by down-regulation of GATA3. A.786-O cells transfected with control siRNA or siTim-3#1. Tim-3 depletion led to an elevation in GATA3 protein level as measured by western blotting, β-actin was used as negative control. B. 786-O cells transfected with vector or Tim-3 recombinant plasmid. Tim-3 overexpression led to decrease in GATA3, protein level was detected by western blotting, β-actin was used as negative control. C. Cells transfected with control siRNA, siTim-3#1 or siTim-3#1plus siGATA3 were subjected to wound-healing analysis. Wound recovery was photographed 24h post infection. Representative photos were showed. D. The data shown were from a representative of three independent experiments. Error bars represent the SD from three independent experiments. *indicated P<0.05, **P<0.01.
In order to explore the role that knockdown of Tim-3 impact in the migration and invasion, wound-healing assay was then conducted, as an approach to study migration. As shown in Figure 4C, 24 h post infection in the group of control siRNA or siTim-3#1 groups, wound were manufactured and measured. 24 h later, 786-O cells depleted of Tim-3 by the relative siRNA were significantly impaired in wound recovery, compared with the control cells. Considering GATA3 as a key regulator in tumor metastasis [17,18], and as a potential target of Tim-3, in our study, we further proved the effect of loss-of-function of GATA3 as downstream target genes on the invasive potential in 786-O cells. Accord with our prospection, cells treated with Tim-3 siRNA plus GATA3 loss of function were resistant to wound healing in certain degrees, compared with siTim-3 only, the representative photos and the overall tendency was shown (Figure 4C and 4D).
Tim-3 promoted ccRCC cell invasion due to the inhibition of GATA3
To further assess the effects of Tim-3 knockdown on cell invasion, transwell assay was performed in Caki-2 cells. Cells were pretreated with either control siRNA or siTim-3#1, cells transmigrated to lower chamber were significantly decreased after infection with siTim-3#1. The migrated cells were photographed in five fields and counted, the data shown that migrated cells infected with Tim-3 loss of function counts for 25.6% of those infected with non-specific siRNA. Altogether, knockdown of Tim-3 in Caki-2 cells caused a notable decrease in migrated cells, and the additional knock down of GATA3 partially rescued the effect of knockdown Tim-3 on tumor migration and invasion (Figure 5A and 5B). As the data shown above, we might make a conclusion that Tim-3 mediated ccRCC metastasis might be linked with the inhibition of GATA3. The aim of the present study was to further investigate the clinical significance of the correlation between Tim-3 and GATA3 in more ccRCC samples and to discuss the possible mechanism responsible for Tim-3 induced EMT and metastasis.
Figure 5.
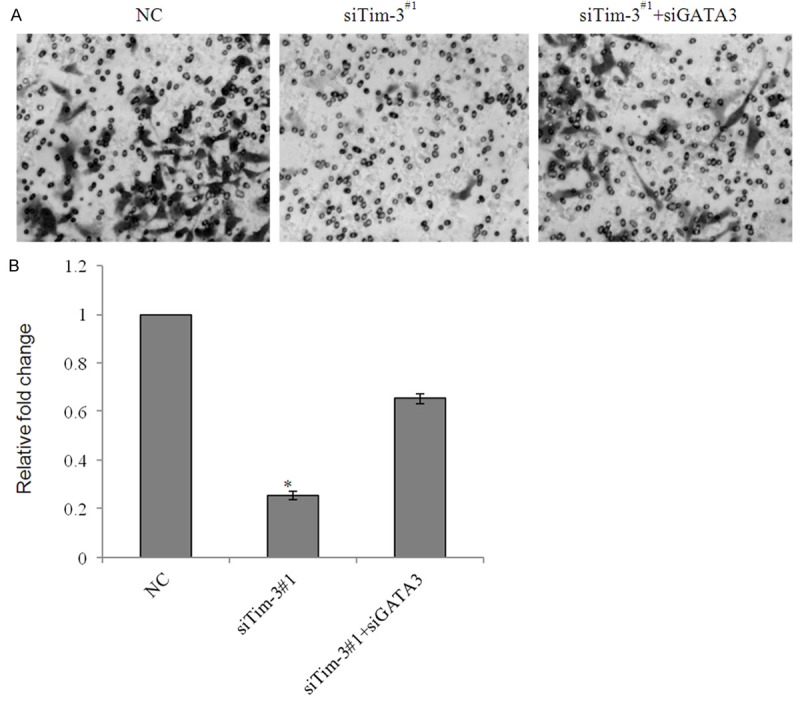
Tim-3 promoted ccRCC cell invasion due to the inhibition of GATA3. A. Caki-2 cells were transfected with control siRNA, siTim-3#1 or siTim-3#1 plus siGATA3, After 48 h of transfection, cells were starved for 18 h before cell invasion assays were performed, The invaded cells were stained and counted. Representative photos were showed under microscopy in each group. B. Statistically analyzed were represented as fold change over vector. Error bars represent the mean ± SD for triplicate measurements. *P<0.05; **P<0.01.
Discussion
Tim3 played important roles in modulating immune reaction and was regarded as a potential target molecule for immunotherapy. To our knowledge, in antitumor immunity activation, Tim3 inhibited the activity of Th1 cells and production of IFN-γ [19,20]. With the interaction of its ligands phosphatidylserine and galectin-9 [21,22], Tim3 could up-regulated apoptotic cell-associated antigens [9,22]. Further, galectin-9 could induce the death of Th1 cells, which resulted in the inhibition of Th1 autoimmunity and the generation of Th17 cells [5,21].
Although Li et al [23] reported polymorphisms of the TIM-1 and TIM-3 genes are not associated with systemic lupus erythematosus in a Chinese population. The increased expression TIM-3 ligand (galactin-9) mRNA in SLE patients (Pan et al., 2010) and the similar data from our study implied that TIM proteins might be involved in the pathogenesis of SLE. Cai et al [24] reported TIM-3 polymorphisms were susceptibility to renal cell carcinoma (RCC) in the Chinese population. To our observation of the TCGA database, TIM-3 was high expressed in the ccRCC, and the higher expression often indicated for a worse 5-year survival, especially in the grade3.
Considering GATA3 performed critical functions outside of the hematopoietic system, such as in the development of the epithelial structures of the mammary gland, skin, inner ear, central nervous system, and kidney, reviewed in [13]. Here, we reported in two ccRCC cell lines, GATA3 was down-regulated compared with the normal HK-2 cell, however, Tim-3 was overexpressed. The data from TCGA database in Kidney Renal Clear Cell Carcinoma (Nature 2013, or provisional data) also indicated Tim-3 was partially amplified. While in SLE, GATA3 was up-regulated, and the tendency of GATA3 overexpression goes against with that of Tim-3. So we here hypothesized that the correlation between Tim-3 and GATA3 was diverse in different disease models, such as SLE and ccRCC. Further, we proved that Tim-3 effected ccRCC cell migration and invasion by down-regulation of GATA3, but we are unsure whether Tim-3 has other targets related to invasion. It has been well-understood that the changes in the development of ccRCC should not be attributed as the alternations of a small number of genes. Thus, it is necessary for further exploration of the potential role of Tim-3 contributes to the ccRCC. In future, Tim-3 and GATA3 pathway that we studied might be exploited in a therapeutic approach for the treatment of ccRCC cancers. Although the expression of TIM-3 in renal tissue had been detected in patients with SLE, and was associated with the diseases’ activity [25], the mechanism of Tim-3 and GATA3 on the influence of SLE still needs further discussion.
Disclosure of conflict of interest
None.
References
- 1.Baghdadi M, Jinushi M. The impact of the TIM gene family on tumor immunity and immunosuppression. Cell Mol Immunol. 2014;11:41–48. doi: 10.1038/cmi.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuchroo VK, Meyers JH, Umetsu DT, DeKruyff RH. TIM family of genes in immunity and tolerance. Adv Immunol. 2006;91:227–249. doi: 10.1016/S0065-2776(06)91006-2. [DOI] [PubMed] [Google Scholar]
- 3.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev. 2010;235:172–189. doi: 10.1111/j.0105-2896.2010.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koguchi K, Anderson DE, Yang L, O’Connor KC, Kuchroo VK, Hafler DA. Dysregulated T cell expression of TIM3 in multiple sclerosis. J Exp Med. 2006;203:1413–1418. doi: 10.1084/jem.20060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seki M, Oomizu S, Sakata KM, Sakata A, Arikawa T, Watanabe K, Ito K, Takeshita K, Niki T, Saita N, Nishi N, Yamauchi A, Katoh S, Matsukawa A, Kuchroo V, Hirashima M. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin Immunol. 2008;127:78–88. doi: 10.1016/j.clim.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T, Kuchroo VK, Gutierrez-Ramos JC, Coyle AJ, Strom TB. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol. 2003;4:1093–1101. doi: 10.1038/ni987. [DOI] [PubMed] [Google Scholar]
- 7.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA, Freeman GJ, Kuchroo VK. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536–541. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 8.Oikawa T, Kamimura Y, Akiba H, Yagita H, Okumura K, Takahashi H, Zeniya M, Tajiri H, Azuma M. Preferential involvement of Tim-3 in the regulation of hepatic CD8+ T cells in murine acute graft-versus-host disease. J Immunol. 2006;177:4281–4287. doi: 10.4049/jimmunol.177.7.4281. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama M, Akiba H, Takeda K, Kojima Y, Hashiguchi M, Azuma M, Yagita H, Okumura K. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood. 2009;113:3821–3830. doi: 10.1182/blood-2008-10-185884. [DOI] [PubMed] [Google Scholar]
- 10.Sterner RM, Hartono SP, Grande JP. The Pathogenesis of Lupus Nephritis. J Clin Cell Immunol. 2014;5 doi: 10.4172/2155-9899.1000205. pii. 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, Kiemeney LA. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–621. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 12.Klatte T, Pantuck AJ, Riggs SB, Kleid MD, Shuch B, Zomorodian N, Kabbinavar FF, Belldegrun AS. Prognostic factors for renal cell carcinoma with tumor thrombus extension. J Urol. 2007;178:1189–1195. doi: 10.1016/j.juro.2007.05.134. [DOI] [PubMed] [Google Scholar]
- 13.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 15.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng R, Blobel GA. GATA Transcription Factors and Cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper SJ, Zou H, LeGrand SN, Marlow LA, von Roemeling CA, Radisky DC, Wu KJ, Hampel N, Margulis V, Tun HW, Blobe GC, Wood CG, Copland JA. Loss of type III transforming growth factor-beta receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene. 2010;29:2905–2915. doi: 10.1038/onc.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Reviews Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 19.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–1146. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabatos CA, Chakravarti S, Cha E, Schubart A, Sanchez-Fueyo A, Zheng XX, Coyle AJ, Strom TB, Freeman GJ, Kuchroo VK. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat Immunol. 2003;4:1102–1110. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 21.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 22.DeKruyff RH, Bu X, Ballesteros A, Santiago C, Chim YL, Lee HH, Karisola P, Pichavant M, Kaplan GG, Umetsu DT, Freeman GJ, Casasnovas JM. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J Immunol. 2010;184:1918–1930. doi: 10.4049/jimmunol.0903059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li WX, Chen GM, Yuan H, Yao YS, Li RJ, Pan HF, Li XP, Xu JH, Tao JH, Ye DQ. Polymorphisms of the TIM-1 and TIM-3 genes are not associated with systemic lupus erythematosus in a Chinese population. Mutagenesis. 2011;26:507–511. doi: 10.1093/mutage/ger009. [DOI] [PubMed] [Google Scholar]
- 24.Cai C, Wang L, Wu Z, Li M, Chen W, Sun Y. T-cell immunoglobulin- and mucin-domain-containing molecule 3 gene polymorphisms and renal cell carcinoma. DNA Cell Biol. 2012;31:1285–1289. doi: 10.1089/dna.2012.1625. [DOI] [PubMed] [Google Scholar]
- 25.Guo L, Yang X, Xia Q, Zhen J, Zhuang X, Peng T. Expression of human T cell immunoglobulin domain and mucin-3 (TIM-3) on kidney tissue from systemic lupus erythematosus (SLE) patients. Clin Exp Med. 2014;14:383–8. doi: 10.1007/s10238-013-0264-3. [DOI] [PubMed] [Google Scholar]