Abstract
Beclin 1 is a promoter gene for autophagy as well as a key factor for regulating tumor cell growth and death. Allelic deletion of Beclin 1 has been observed in certain triple-negative breat cancer (TNBC) cells, and it might be associated with increased proliferation and invasion in TNBC cells. In this study we investigated the relationship between Beclin 1 expression and prognosis for TNBC patients, as well as the influence on cell growth by Beclin 1 overexpression in different cultural conditions. Beclin 1 expression in TNBC tissues was measured by immunohistochemical staining and correlated with clinicopathologic parameters for TNBC patients. The plasmid of pDS-RED-C1-Beclin 1 was transfected to BT-549 and MDA-MB-231 cells and autophagy, proliferation, apoptosis, cell cycle and Epithelial-mesenchymal transition (EMT) process were measured. Results indicated that high level of Beclin 1 expression was correlated with more lymph nodes and distant metastasis but unrelated to survival rates in 5 years for TNBC patients. In vitro, overexpression of Beclin 1 improved cellular autophagy in both BT-549 and MDA-MB-231 cells, inhibited cell proliferation at normal cultural condition and increased cell survival in starvation, hypoxia or with doxorubicin stimulation. Besides, Beclin 1 overexpression decreased cell apoptosis, induced cells to be in G0/G1 phase and promoted EMT process through Wnt/β-catenin pathway in starvation. Thus, Beclin 1 overexpression plays a double role in BT-549 and MDA-MB-231 cell growth by elevating the capability of autophagy. These findings might be useful for searching a proper method for clinical therapy of TNBC from the aspect of autophagy in future.
Keywords: Triple-negative breast cancer, autophagy, Beclin 1 gene
Introduction
Triple-negative breast cancer (TNBC) is a kind of breast tumor in which estrogen receptor (ER), progestogen receptor (PR) and human epidermal growth receptor 2 (Her-2) expression are all negative [1]. Because of this special characteristic TNBC is refractory to endocrine therapy and Herceptin treatment. The major therapy for TNBC includes surgery plus chemotherapy, but prognosis is relatively poor comparing to other types of breast cancer [1]. Although studies suggest neoadjuvant chemotherapy is excellent for patients who can obtain a complete pathological response, for other patients with a residual tumor the prognosis is very poor [1]. Another TNBC therapy uses target agents such as angiogenesis inhibitor, monoclonal antibody targeting against epithelial growth factor receptor (EGFR) and poly (adenosine diphosphateribose) polymerase (PARP) inhibitor. But overall effects of those drugs need further evaluation [2,3]. Thus there is still lack of an available method to cure TNBC in clinic [3].
Autophagy is an evolutionarily conserved lysosomal pathway for degrading cytoplasmic organelles and macromolecules [4]. Autophagy can inhibit tumor cells because reduced capability of autophagy provides an oncogenic stimulus with elevated risk of malignant transformation and spontaneous tumor formation. In addition, autophagy offers a protective cell survival mechanism against environmental and cellular stress [5]. The current literatures have shown contradictory roles of autophagy not only in facilitating cell survival and delaying apoptotic death in stress, but also in promoting a specific form of cell death called autophagic cell death [5]. Therefore, either induction or inhibition of autophagy for tumor cells may provide therapeutic benefits, depending on different cellular features [5-7].
Beclin 1 is essential for inducing autophagy and is considered to be a suppressor gene [8-10]. In vivo, Beclin 1 deletion causes less survival and tumorigenesis. Mice with loss of Beclin 1 (Beclin 1-/-) die early in embryogenesis [11] while others with monoallelic deletion of Beclin 1 (Beclin 1-/+) are frequently prone to conduct mammary neoplasia [11,12]. Beclin 1 monoallelic deletion is also related to malignant proliferation and high capability of metastasis in ovarian cancerand prostate cancer cells [13].
It is worthwhile to note that Beclin 1 is frequently deleted in sporadic breast cancer patients, and Beclin 1 levels in breast cancer cells are significantly lower than those in normal breast epithelial cells [14]. It is also important to note that allelic deletion of Beclin 1 and depressed Beclin 1 expression exist in some TNBC cell lines [15], resulting in incomplete level of autophagy and different capability of proliferation and invasion. Therefore, we intended to study the relationship between Beclin 1 expression and prognosis for TNBC patients as well as the effects of altered Beclin 1 expression on TNBC cell growth in various conditions.
Materials and methods
Patients and tissue samples
Tissue samples from 53 cases of TNBC were surgically resected at Zhujiang Hospital from 2005 to 2009. For each case 3 representative cores were taken from a representative area of tumor tissue. The age of patients ranged from 45 to 76 years (median age, 53.4 years). Tumor size ranged from 0.5 to 9.5 cm (median size, 3.05 cm). Clinical parameters including patients’ ages, tumor size, existence of lymph node or distant organ metastasis were also collected. All patients agreed and provided written informed consent with full understanding of the study aims.
Immunohistochemical staining
Beclin 1 expression was examined by immunohistochemical staining. In brief, 4-μm-thick sections of formalin-fixed, paraffinem bedded tissue were deparaffinized and an antigen retrieval procedure was performed for 20 minutes at 100°C. Endogenous peroxidases were quenched by incubation of tissue with hydrogen peroxide for 5 minutes. Sections were incubated for 15 minutes at ambient temperate with primary polyclonal antibodies for Beclin-1 (1:100, Santa Cruise, USA). Beclin 1 levels were evaluated according to intensity defined in three groups: no expression, low expression and high expression. All slides were evaluated independently by 2 investigators without knowledge of patients and clinical outcomes.
Cell culture
BT-549 and MDA-MB-231 cell lines were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). BT-549 cells with Beclin 1 allelic deletion and MDA-MB-231 cells with no Beclin 1 deletion [15], both cells were routinely maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), cultured in a 37°C humidified atmosphere containing 95% air and 5% CO2. For the experiment of starvation, hypoxia and chemical drug stimulation, BT-549 and MDA-MB-231 cells were maintained in serum-deprived 1640 medium, oxygen concentration less than 10 PPM and medium containing doxorubicin (2 mg/ml), respectively.
Cell transfection
The plasmids of pDS-RED-C1-Beclin 1 and pDsRed-C1 were gifts from Professor Qing Zhong (University of California at Los Angeles, USA). BT-549 and MDA-MB-231 cells were transfected with pDS-RED-C1-Beclin 1 and pDsRed-C1 with Lipofectamine™ 2000 (Invitrogen, USA) according to the instruction of Invitrogen. Fluorescence (IX71 Olympus, Japan) was used to observe the expression of red fluorescent protein in BT-549 and MDA-MB-231 cells after the transfection of plasmids.
Quantitative PCR (qPCR) analysis
BT-549 and MDA-MB-231 cells were harvested and total RNA was isolated using Trizol reagent (Invitrogen, USA), 1 mg RNA was reverse transcribed using a cDNA synthesis kit (TaKaRa, Japan). Primer sequences were as follows: Beclin 1, forward, 5’-GGTGTCTCTCGCAGATTCATC-3’, reverse, 5’-TCAGTCTTCGGCTGAGGTTCT-3’ (120 bp); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward, 5’-GCACCGTCAAGGCTGAGAAC-3’, reverse, 5’-TGGTGAAGACGCCAGTGGA-3’ (137 bp). qPCR was performed using Light Cycler 480 Sequence Detection System (Roche, Switzerland). Results were expressed as fold differences in expression of Beclin 1 related to that of GAPDH.
Western blot assay
BT-549 and MDA-MB-231 cells were harvested and protein quantitation were conducted. Each sample of proteins (20 µg) was separated on a 10% SDS polyacrylamide gel, transferred to polyvinylidene difluoride Polyscreen membrane (millipore, Germany) and then incubated with 1:500 anti-Beclin 1, anti-E-cadherin, anti-vimentin, anti-N-cadherin, anti-Snail, anti-P-GSK-3β(Santa Cruise, USA) or 1:1000 anti-β-actin (Cell signal Technology, USA) for overnight at 4°C. Secondary anti-rabbit antibody (Santa Cruise, USA) was added according to primary antibody and incubated for 2 h. Protein signals were visualized using Odyssey Scan (LI-COR, USA) and band intensities were quantified by image j1.44. Fold changes in the intensity of the protein signals were the mean value of the results.
Acridine orange staining
Cells were washed with PBS and stained by acridine orange (Sigma, USA) for 15 min, repelled by light. Subsequently, acidic vesicular organelles (AVO) was observed by fluorescence inverted-microscope (IX71 Olympus, Japan).
Cell viability assay
Cell viability was assessed by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Amresco, USA) assay. Cells were cultured in 96-well plates, 20 µl MTT (5 mg/ml in PBS) was added to each well at special time points. After incubation for 4h in a 37°C humidified atmosphere containing 95% air and 5% CO2, 200 µl DMSO (Sigma, USA) was added to dissolve the formazan crystals. Absorption (optical density, OD value) was measured at 570 nm using microplate spectrophotometer (Molecular Devices, USA).
Cell apoptosis and cell cycle assay
Cell apoptosis and cell cycle distribution were detected by flow cytometric analysis. For cell apoptosis analysis, cells were harvested and washed by cold PBS for once, then cells were stained with an apoptosis kit (KeyGen Biotech, China) according to the manufacturer’s instructions and analyzed by flow cytometry (BD FACS Canto II, USA). For cell cycle analysis, cells were harvested and fixed with 70% ethanol at -20°C for at least 12 h, then cells were stained with 50 µg/mL propidium iodide (PI) and 100 µg/mL Rnase I in PBS at 37°C for 30 min. Cell cycle distribution was also analyzed by flow cytometry.
Statistical analysis
One-way ANOVA was used to determine the differences between groups for the results of MTT, qPCR, western blot, apoptosis and cell circle. The Kaplan-Meier method was used to determine the probability of survival. The Pearson’s x2 test was used to evaluate the association between Beclin 1 expression and clinicopathologic variables. Data were processed by SPSS 13.0 software. P<0.05 was considered as significant. The data are presented as mean ± standard error for at least 3 separate determinations.
Results
Beclin 1 expression was associated with clinical prognosis of patients in triple-negative breast cancer
Totally, 53 patients had been evaluated since 2005 to 2009. Clinical parameters including age, tumor size, lymph node metastasis and distant metastasis were collected. 17 patients showed no Beclin 1 expression. 18 patients had low expression of Beclin 1 and 18 patients were detected with high expression (Figure 1A). Further study showed that Beclin 1 expression was correlated with patients’ age, lymph node metastasis and distant organ metastasis while unrelated to tumor size. Patients with high expression of Beclin 1 had more lymph node metastasis and distant organ metastasis compared to patients with no expression of Beclin 1 (Figure 1B).
Figure 1.
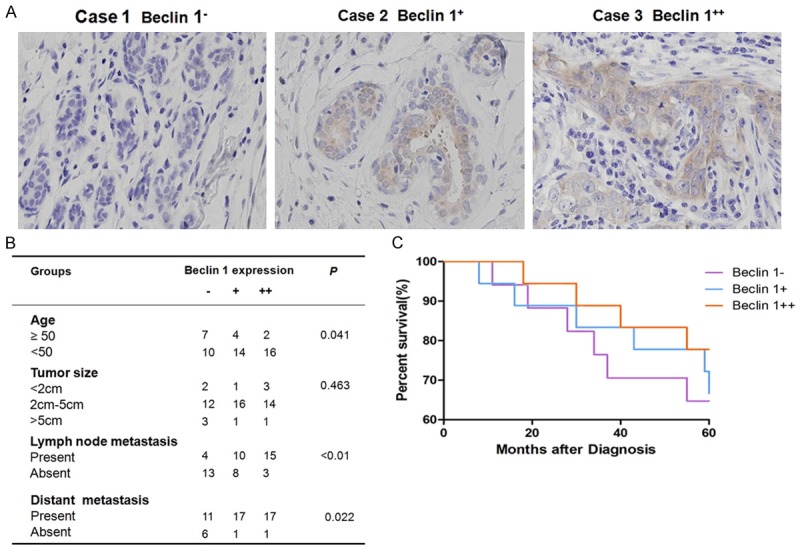
Association of Beclin 1 expression with patients’ prognosis. A. Beclin 1 expression in tumor cells of TNBC patients. B. The relationship between Beclin 1 expression and clinical parameters in TNBC patients. C. The percentage of survival in 5 years for TNBC patients with altered expression of Beclin 1.
Survival analysis was performed to evaluate the prognosis of patients with different levels of Beclin 1 (Figure 1C). The survival rate for patients with Beclin 1- was 65%, for patients with Beclin 1+ was 72% and for patients with Beclin 1++ was 78% at 5 years after the diagnosis. No significant difference in survival ratewas detected among these three groups (P=0.847).
Transfection of Beclin 1 increased capability of autophagy
In order to understand the effects of Beclin 1 overexpression on TNBC cell growth, the plasmid of pDS-RED-C1-Beclin 1 was transfected into BT-549 and MDA-MB-231 cells. Western blot and qPCR indicated that Beclin 1 protein and mRNA levels were significantly higher after transfection (P<0.05, Figure 2A).
Figure 2.
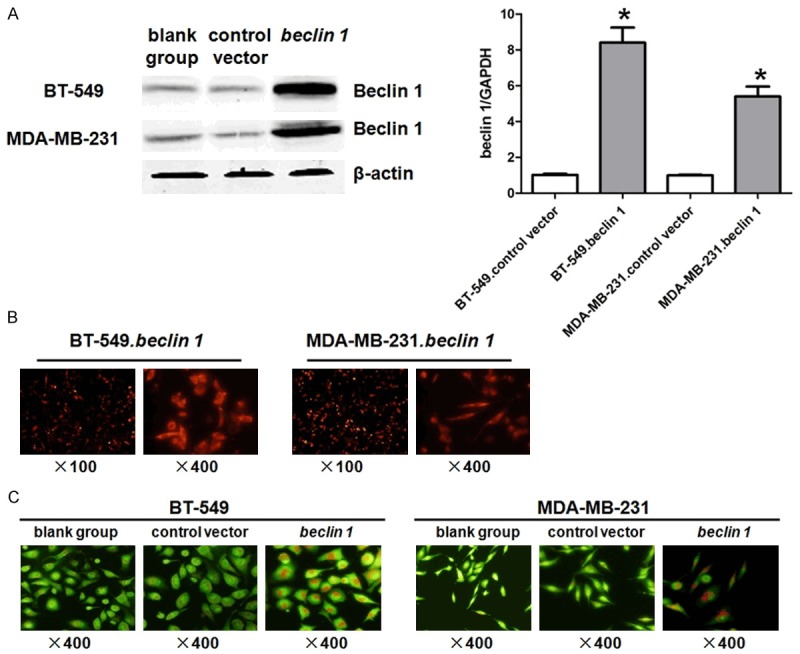
Transfection of Beclin 1 for BT-549 and MDA-MB-231 cells. A. BT-549 and MDA-MB-231 cells were tranfected with pDS-RED-C1-Beclin 1 or pDsRed-C1 plasmid, cells were harvested and analyzed by western blot and qPCR. B. Morphology of BT-549 and MDA-MD-231 cells post transfection of pDS-RED-C1-Beclin 1 plasmid were observed under fluorescence microscopy. C. Morphology of BT-549 and MDA-MD-231 cells post transfection of pDS-RED-C1-Beclin 1 or pDsRed-C1 plasmid were observed under fluorescence microscopy, staining by acridine orange. *P<0.05 vs. blank group. The results are representative of three different experiments.
Cell morphology was observed using fluorescence microscopy (Figure 2B). Autophagy was determined by the red appearance of AVO within cells stained by acridine orange (Figure 2C). There were few cells with red cytoplasm in blank group and control vector group of BT-549 and MDA-MB-231 cells. However, the number of cells with red cytoplasm was obviously elevated in both BT-549 and MDA-MB-231 cells after the transfection of pDS-RED-C1-Beclin 1, conforming the increased levels of autophagy in both cells (P<0.05).
Transfection of Beclin 1 influenced cell growth in various cultural conditions
MTT assay was performed to evaluate the effect of Beclin 1 overexpression on cell growth. BT-549 and MDA-MB-231 cells were transfected with Beclin 1 and cultured in 1640 medium plus 10% FBS, serum-deprived 1640 medium, hypoxia medium or medium contaminating doxorubicin. At the cultural condition of 1640 medium plus 10% FBS, cell growth was inhibited both in Beclin 1 overexpressing BT-549 and MDA-MB-231 cells at 96 hours compared to the controls (P<0.05, Figure 3A). In serum-deprived 1640 medium, cell survival rates have decreased since 24 hours in both cell types but were significantly higher in Beclin 1 overexpression group than other two at 48 hours (P<0.05, Figure 3B). The same trend was also observed in hypoxia environment and doxorubicin contamination (P<0.05, Figure 3C, 3D).
Figure 3.
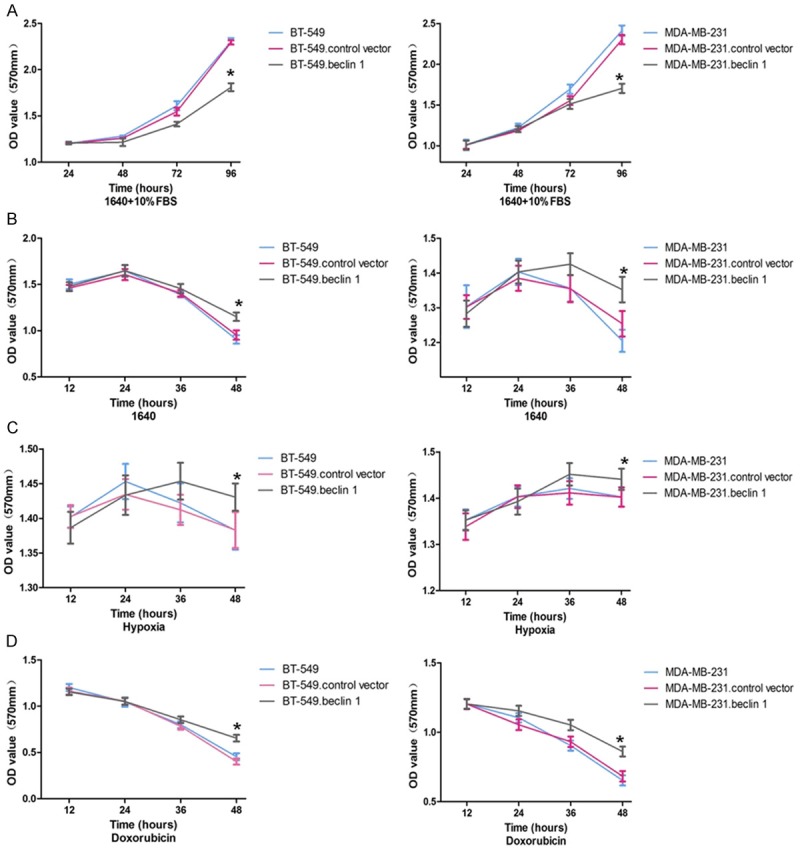
MTT results of BT-549 and MDA-MB-231 cells cultured in different condintions. A. Cells were cultured in normal condition. B. Cells were cultured in low nutritional condition. C. Cells were cultured in hypoxia. D. Cells were cultured in doxorubicin (2 mg/ml). *P<0.05 vs. blank group. The results are representative of three different experiments.
Transfection of Beclin 1 influenced cell apoptosis and cell circle at different conditions
BT-549 and MDA-MB-231 cells were cultured at different conditions and cell apoptosis was detected by flow cytometric analysis (Figure 4A). There was no significant difference when overexpressing Beclin 1 in both BT-549 and MDA-MB-231 cells cultured in 1640 plus 10% FBS. In 1640 medium with no serum supplemented, the percentages of cell apoptosis in BT-549. Beclin 1 and MDA-MB-231. Beclin 1 groups were 5.99±0.43% and 4.99±0.32%, respectively, which were significantly lower than the controls (P<0.05). The same phenomenon were also observed when the cells were cultured in hypoxia or doxorubicin.
Figure 4.
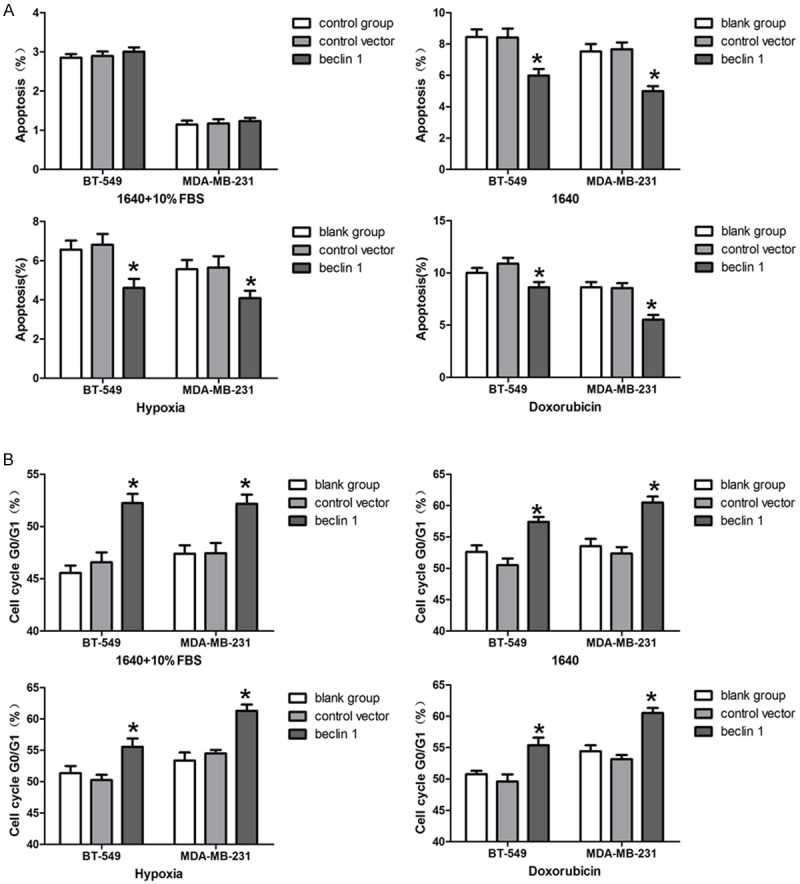
Apoptosis and cell cycle of BT-549 and MDA-MB-231 analysed by flow cytometry. A. Apoptosis was analysed as cells were cultured in different conditions. B. Cell cycle was analysed as cells were cultured in different conditions. *P<0.05 vs. blank group. The results are representative of three different experiments.
Furthermore, cell cycle was examined by flow cytometric analysis (Figure 4B). In 1640 medium plus 10% FBS, the percentages of cells in G0/G1 phase in BT-549. Beclin 1 and MDA-MB-231. Beclin 1 groups were 52.24±0.51% and 52.18±0.51%, respectively, which were significantly higher than those in the control groups (P<0.05). The same observation was obtained when cells were cultured in 1640 medium with no serum, hypoxia or doxorubicin.
Transfection of Beclin 1 induced EMT process through Wnt pathway
After culturing in serum-deprived 1640 medium for 48 h, western blot indicated that protein levels of E-cadherin were significantly lower while N-cadherin and vimentin expressions were significantly higher both in BT-549. Beclin 1 and MDA-MB-231. Beclin 1 cells compared to the controls (P<0.05, Figure 5). Further study showed that levels of β-catenin, phosphorylated-GSK-3β and Snail were also elevated when overexpressing Beclin 1, indicating the involvement of Wnt pathway in the process (P<0.05, Figure 5).
Figure 5.
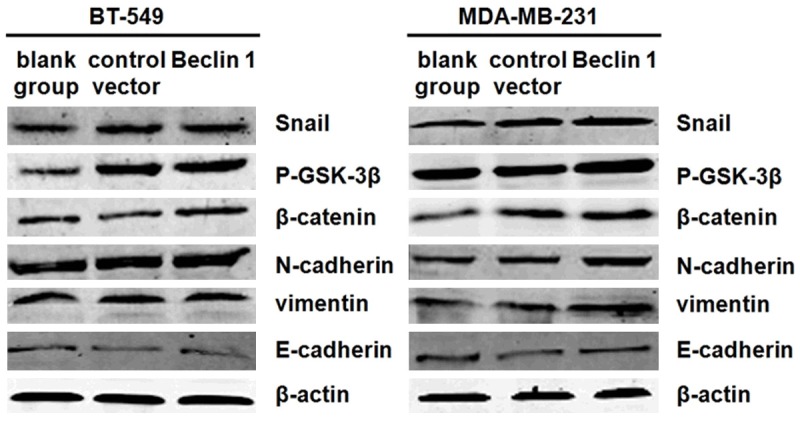
Protein levels of BT-549 and MDA-MB-231 analysed by western blot as cells cultured in starvation for 48 h.
Discussion
Beclin 1, which has a similar structure to yeast autophagy gene Atg6, encodes a Bcl-2-interacting, coiled-coil protein [14]. Beclin 1 is the first gene linking autophagic machinery and human cancer as reported by Liang et al., whose study demonstrated Beclin 1 promoted autophagy in human MCF-7 breast cancer cells with inhibition of cellular proliferation and tumorigenesis in nude mice [14]. The monoallelic deletion of Beclin 1 commonly exists in many tumor cells, such as breast cancer, ovarian cancer and prostate cancer cells. There is a trend of associating Beclin 1 loss with TP53 and PTEN mutations [15]. These findings confirm that decreased capability of autophagy regulated by Beclin 1 contributes to the development of many human malignancies [16-18].
In breast cancer patients, Beclin 1 monoallelic deletion also exists and Beclin 1 protein expression is lower in tumor cells than in normal breast epithelial cells [19], demonstrating that down regulation of Beclin 1 was correlated to malignant proliferation and metastasis in breast cancer cells. It is worthwhile to note that Beclin 1 is also deleted in certain TNBC cells which are refractory to ordinary treatment, such as chemotherapy and radiation [15]. From those studies we assume that Beclin 1 expression may be associated with the prognosis of TNBC patients, and overexpression of Beclin 1 may affect cell growth and cell survival at different cultural conditions. In this study, we investigated the relationship between Beclin 1 levels and clinical parameters for TNBC patients. Besides, TNBC cells were cultured in ordinary conditions and stress environments to detect the effect of altered Beclin 1 expression on cell growth.
From the clinical study we found that Beclin 1 expressions were variable in TNBC patients and higher levels of Beclin 1 were associated with more lymph node metastasis and distant metastasis. To further explore Beclin 1 function in TNBC, in vitro experiments were performed. BT-549 and MDA-MB-231 cells were transfected with plasmid pDS-RED-C1-Beclin 1 and improved levels of autophagy were observed in those cells. Cell growth rates of BT-549 and MDA-MB-231 were inhibited after the transfection of Beclin 1 in ordinary conditions while survival rates were preserved in stress environments such as starvation, hypoxia and doxorubicin addition. This phenomenon could be explained by fewer cells in apoptosis and more cells in G0/G1 phase, in which cells tended to be dormant in order to maintain essential nutrition for sustaining survival in stress. In this way Beclin 1 played a protective role both for BT-549 and MDA-MB-231 cells. Besides, we discovered that overexpression of Beclin 1 could improve EMT process through Wnt/β-catenin pathway, demonstrating that inducing autophagy is an important mechanism for metastasis.
Our results are consistent with other studies which have proved that autophagy serves as a protective mechanism against environmental and cellular stresses. In these studies tumor cells were cultured in conditions supplemented with radiation [20], drugs [21-23], or in starvation [24]. Thus inhibiting autophagy may be a modified method for treating tumors by disturbing the protective aspect of autophagy for tumor cells [25,26]. For example, using siRNA targeting against Beclin 1 to inhibit autophagy may promote cell death of breast cancer cells [27,28]. However, the concern that inhibiting autophagy would lead to increased proliferation of tumor cells should be considered as autophagy can prohibit cellular invasions and control the stability of genome in tumor cells [23].
In the end, our study proved a double effect of Beclin 1 on TNBC cell growth and survival both in normal condition and in stress. However, whether this phenomenon exists in vivo is still uncertain. Besides, simply increasing or decreasing the capability of autophagy in TNBC cells may not be competent enough to kill all cells.The way to control the degree of autophagy in order to make a benefit for TNBC treatment requires further exploration.
Conclusions
In conclusion, we demonstrated that higher expression of Beclin 1 in tumors was associated with more lymph node metastasis and distant metastasis in TNBC patients. Overexpression of Beclin 1 significantly improved the level of autophagy in BT-549 and MDA-MB-231 cells in which cell growth was inhibited at normal cultural condition while cell survival was increased at stress conditions. This phenomenon was associated with the fact that BT-549 and MDA-MB-231 cells were prone to maintain in G0/G1 phase post the transfection of Beclin 1. In addition, overexpression of Beclin 1 induced EMT process through Wnt/β-catenin pathway which may contribute to a more progressive characteristic for BT-549 and MDA-MB-231 cells. Collectively, results in this research provide basic knowledge for further study of Beclin 1 in the therapy of triple-negative breast cancer.
Acknowledgements
This work was supported by grant from the Natural Science Foundation of Guangdong in China (S2011010004033, S2012010009276), we thank Professor Qing Zhong for the plasmids of pDS-RED-C1-Beclin 1 and pDsRed-C1.
Disclosure of conflict of interest
None.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;36320:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Gucalp A, Traina TA. Triple-negative breast cancer: adjuvant therapeutic options. Chemother Res Pract. 2011;2011:696208. doi: 10.1155/2011/696208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhutia SK, Dash R, Das SK, Azab B, Su ZZ, Lee SG, Grant S, Yacoub A, Dent P, Curiel DT, Sarkar D, Fisher PB. Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res. 2010;70:3667–3676. doi: 10.1158/0008-5472.CAN-09-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John S, Nayvelt I, Hsu HC, Yang P, Liu W, Das GM, Thomas T, Thomas TJ. Regulation of estrogenic effects by Beclin 1 in breast cancer cells. Cancer Res. 2008;68:7855–7863. doi: 10.1158/0008-5472.CAN-07-5875. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 11.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 12.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 15.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of Beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 16.Negri T, Tarantino E, Orsenigo M, Reid JF, Gariboldi M, Zambetti M, Pierotti MA, Pilotti S. Chromosome band 17q21 in breast cancer: significant association between Beclin 1 loss and HER2/NEU amplification. Genes Chromosomes Cancer. 2010;49:901–909. doi: 10.1002/gcc.20798. [DOI] [PubMed] [Google Scholar]
- 17.Vanderlaag K, Su Y, Frankel AE, Burghardt RC, Barhoumi R, Chadalapaka G, Jutooru I, Safe S. 1,1-Bis(3’-indolyl)-1-(p-substituted phenyl) methanes induce autophagic cell death in estrogen receptor negative breast cancer. BMC Cancer. 2010;10:669. doi: 10.1186/1471-2407-10-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the Beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson EN, Bristol ML, Di X, Maltese WA, Koterba K, Beckman MJ, Gewirtz DA. A switch between cytoprotective and cytotoxic autophagy in the radiosensitization of breast tumor cells by chloroquine and vitamin D. Horm Cancer. 2011;2:272–285. doi: 10.1007/s12672-011-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang PM, Liu YL, Lin YC, Shun CT, Wu MS, Chen CC. Inhibition of autophagy enhances anticancer effects of atorvastatin in digestive malignancies. Cancer Res. 2010;70:7699–7709. doi: 10.1158/0008-5472.CAN-10-1626. [DOI] [PubMed] [Google Scholar]
- 22.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, Janji B. The acquisition of resistance to TNF alpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7:760–770. doi: 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 23.Schoenlein PV, Periyasamy-Thandavan S, Samaddar JS, Jackson WH, Barrett JT. Autophagy facilitates the progression of ERalpha-positive breast cancer cells to antiestrogen resistance. Autophagy. 2009;5:400–403. doi: 10.4161/auto.5.3.7784. [DOI] [PubMed] [Google Scholar]
- 24.Esteve JM, Armengod ME, Knecht E. BRCA1 negatively regulates formation of autophagic vacuoles in MCF-7 breast cancer cells. Exp Cell Res. 2010;316:2618–2629. doi: 10.1016/j.yexcr.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Godbole AM, Purushottamachar P, Martin MS, Daskalakis C, Njar VC. Autophagy inhibition synergistically enhances anticancer efficacy of RAMBA, VN/12-1 in SKBR-3 cells, and tumor xenografts. Mol Cancer Ther. 2012;11:898–908. doi: 10.1158/1535-7163.MCT-11-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Medina P, Silvente-Poirot S, Poirot M. Tamoxifen and AEBS ligands induced apoptosis and autophagy in breast cancer cells through the stimulation of sterol accumulation. Autophagy. 2009;5:1066–1067. doi: 10.4161/auto.5.7.9820. [DOI] [PubMed] [Google Scholar]
- 27.Thomas S, Thurn KT, Biçaku E, Marchion DC, Münster PN. Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res Treat. 2011;130:437–447. doi: 10.1007/s10549-011-1364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G, Ozpolat B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]


