Abstract
Objective: Coronary heart disease (CHD), the most severe form of coronary artery disease (CAD), is a complex disease that involves a variety of genetic and environmental factors. Recently, multiple single nucleotide polymorphisms (SNPs) have been associated with CAD in Caucasians by genome-wide association (GWA) studies.However, the association of these SNPs with CHD in Asian populations has not yet been established. Here, we aim to investigate the genetic etiology of CHD in a Chinese population by genotyping SNPs previously been associated with CHD in other ethic origin in GWAS or candidate gene studies. Methods: Five SNPs, rs17114036, rs9369640, rs515135, rs579459 and rs8055236, from 5 different loci were genotyped using a sequenom Mass array system in 545CHD patients and 1008 unrelated controls from a Chinese population. Results: Our study showed that SNP rs515135 is strongly associated with CHD in a Chinese Han population (P-value=0.00333, OR=1.48). We also detected significant difference of SNP rs579459 in APOB gene in patients withsevere CAD compared to patients with mild CAD. Conclusion: SNP rs515135 is associated with the susceptibility of CHD in Chinese Han population. The location of rs515135 in the APOB gene supports its potential involvement in the pathogenesis of CAD. Our study data also support that SNP rs579459 may be associated with the severity of CHD.
Keywords: Coronary heart disease, genetic variants, case-controlled study, single nucleotide polymorphism
Introduction
Coronary artery disease (CAD) is one of the leading causes of death in the populations both in developed and developing countries. According to the World Health Organization, it was estimated that more than 700,000 people die from CAD each year in China. Coronary heart disease (CHD) is the most severe clinical manifestation of CAD and is the leading cause of death worldwide [1,2]. CHD is a complex disease characterized by the inheritance of multiple genetic variants in addition to environmental factors which worsen the disease state. Epidemiological studies have identified many risk factors for coronary heart disease, including plasma lipid concentrations, blood pressure, smoking, diabetes and markers of inflammation. Among them, a causal role has been proven only for some factors (for example, low-density lipoprotein (LDL) cholesterol and blood pressure) primarily through randomized clinical trials of drug therapy. Twin and family studies have demonstrated that a significant proportion (40-50%) of susceptibility to CHD is inherited [3]. A previous linkage study and candidate gene study have identified genetic factors predisposing to CHD. However, the results of CHD genetic studies have not been very satisfactory. Recently, GWAS have proven to be a powerful tool to identify susceptibility genes for common diseases. To date, GWAS studies in Caucasians identified multiple single nucleotide polymorphisms (SNPs) that associated with coronary artery disease (CAD) [4-12]. However, no independent studies of these SNPs have been reported in a Chinese population with CHD. Here, we intend to identify the associations between genetic variants of CHD, specifically, the associations of the SNPs rs17114036, rs9369640, rs515135, rs579459 and rs8055236 with CHD in a Chinese Han population.
Materials and methods
Patients and controls
A total of 545 blood-unrelated Chinese patients with coronary heart disease and 1008 healthy controls were recruited consecutively from outpatients at the Department of Cardiovascular Internal Medicine at Anhui Provincial Hospital. All subjects were of self-reported Chinese Han ancestry. Coronary heart disease was defined as the following: ① clinical symptoms and an electrocardiogram consistent with the diagnosis of coronary heart disease criteria formulated by the WHO in 1979. ② Coronary angiography revealed that at least 1 coronary artery stenosis was more than or equal to 50%. ③ There was no relationship between cases. Exclusion criteria: ① Associated with other cardiac diseases, such as dilated cardiomyopathy, rheumatic heart disease ② severe disease in the liver and kidney. ③ Previously received intravenous thrombolysis, coronary artery stenting and coronary artery bypass grafting patients. Control subjects were recruited from healthy adult visitors to the hospitals without a history of cardiovascular disease and family history of coronary artery disease (including first-, second- and third-degree relatives), matched for age (up to 6 years older or younger) and sex with the cases. Participants who consented to the study completed a structured questionnaire, attended a health examination, and had a venous blood sample taken. Information about demographic factors, socioeconomic status, lifestyle (smoking, leisure time, physical activity, and dietary patterns), personal and family history of cardiovascular disease, hypertension and diabetes mellitus was obtained. After written informed consent was obtained, peripheral blood samples were collected from all patients and matched healthy controls. All participants provided their written informed consent. At the same time, we promised reporting of the research results to the participants. The study was approved by the Ethical Committee of the Anhui Provincial Hospital and was performed according to the Declaration of Helsinki Principles.
Definitions
The patients were categorized as the following: the severity of coronary disease was determined to be mild (≤20 points) or severe (>20 points) according to the Gensini scoring system [13]. The family history was considered to be positive if the patient’s first-, second-, and/or third-degree relatives had coronary heart disease; the family history was considered to be negative otherwise.
Genomic DNA extracting and SNP genotyping
After informed consent, genomic DNA was isolated from peripheral blood of the patients using a Qiagen kit (Hilden, Germany). In addition, genomic DNA of 1008 unrelated healthy individuals was extracted as a control. We selected only SNPs in CAD-related genes with minor allele frequencies of 0.05 in Chinese patients in the HapMap database. We used the SNPs effect size in the references, minor allele frequencies of Chinese (based on HapMap database) and the sample size of this study to calculate the power for detecting a positive association. Excluding SNPs with power less than 0.20, we selected five SNPs (rs17114036, rs9369640, rs515135, rs579459 and rs8055236), which have been shown to significantly associate with CAD, but have not yet been tested in a Chinese population.
All SNPs were genotyped using the Sequenom iPlex platform (Sequenom, Inc., San Diego, CA, USA) at the State Key Laboratory Incubation of Dermatology, Ministry of Science and Technology, Hefei, Anhui, China. Each genotyping cluster of five selected SNPs is good, especially the genotyping cluster of rs515135 as shown in Figure 1. Besides, the primers designed for sequencing of this study are shown below (Table 1).
Figure 1.
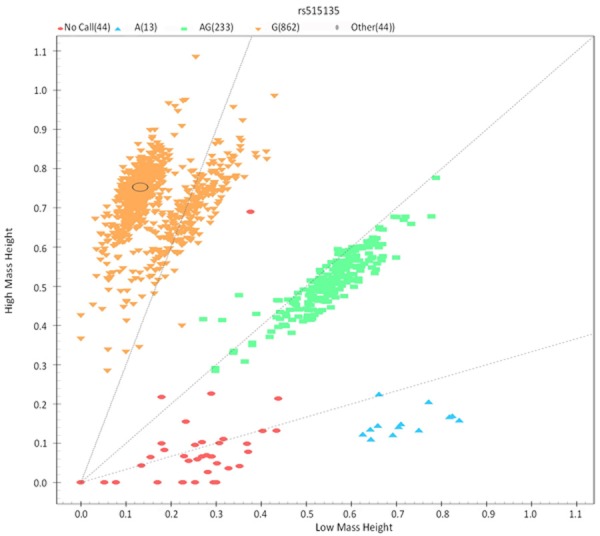
The genotyping cluster of rs515135.
Table 1.
Primer sequences of amplification and extension
| SNP | Amplification primersequences | Primer sequences of extension |
|---|---|---|
| rs17114036 | 1) ACGTTGGATGCCTATCAGCAAGAGCTGAAG | aTAAGTTTTCAGGAAAAAGTAGTC |
| 2) ACGTTGGATGCGAGAACCTGGTTTCTTGAC | ||
| rs9369640 | 1) ACGTTGGATGTGGGCTACTGGAGAATAAGG | gACTGTTGCAAAATCACTGTTAG |
| 2) ACGTTGGATGCGGTGTGTAGATACTGTTGC | ||
| rs515135 | 1) ACGTTGGATGGAACCATCTTGTTACTGCAC | tAAAAACAGCCAAAATGGAACCAAAGA |
| 2) ACGTTGGATGAGGGCTTACAGCCAAGTAAC | ||
| rs579459 | 1) ACGTTGGATGTTCTGGTTTGCATGTGTTGG | cGGTTTCTTTTCGCTACACCA |
| 2) ACGTTGGATGGAAGTAAAAGTGACTTGCTG | ||
| rs8055236 | 1) ACGTTGGATGGCCAGGCTATTTGTGCATCT | gtCTTTGTTTTTTTTCATCTTCCACTCG |
| 2) ACGTTGGATGTCCATACAGTCATTGAGGC |
Statistical analysis
Hardy Weinberg equilibrium tests of the genotyping data were performed using PLINK (version 1.07). The power of the study population was estimated using the power calculatorfor genome wide association studies program (http://www.sph.umich.edu/csg/abecasis/CaTS/). For allelic association between a SNP and CHD, the p-value and corresponding odds ratio (OR) with a 95% confidence interval were computed by Chi-square tests using Pearson’s 2X2 contingency tables as implemented in PLINK version 1.07.
For the stratified analysis, differences between the cases and controls in terms of the genotype frequency were tested using the chi squared test and by calculating the odds ratios (ORs) and 95% confidence intervals (CIs).
Results
Characters of the study cohorts
The demographic details for the patients and control subjects are shown in Table 2. Except the means of sex and the frequency of presence of alcohol intake, other characteristics in Table 2, CHD patient were significantly different from those in controls.
Table 2.
Characters of the study cohorts
| CHD patient | Control | |
|---|---|---|
|
|
||
| Characteristic | (n=545) | (n=1008) |
| Age (SD)* | 61.5 | 57.5 |
| Sex (Male/Female) | 354/191 | 605/403 |
| BMI | 24.5 | 23.8 |
| History of HT (%) | 40.18% | 21.53% |
| History of T2D (%) | 12.29% | 3.08% |
| Current Smoking (%) | 57.25% | 40.18% |
| Alcohol intake (%) | 38.35% | 37.10% |
| HDL-C mmol/L* | 1.02 | 1.13 |
| LDL-C mmol/L* | 2.53 | 2.27 |
| TG mmol/L* | 1.63 | 1.65 |
| TC mmol/L* | 4.55 | 4.59 |
CHD, Coronary heart disease; BMI, body mass index (kg/m2); HT, hypertension; T2D, type 2 diabetes; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; TC, total cholesterol;.
Data are means (SD).
Results of the case-controlled study
To demonstrate the genetic susceptibility of SNPs with CHD in a Chinese population, 5 SNPs were genotyped in 545 CHD patients and 1008 controls. Allele distribution and minor allele frequency in these patients and controls are shown in Figure 2. All of the SNPs were in Hardy-Weinberg equilibrium. Association between the SNPs and CHD is shown in Table 3. In five SNPs, rs515135 was significantly associated with CHD in a Chinese population. The scatter plots of the suggestive association evidence within 2p24-p23 (APOB) for CHDare shown in Figure 3. There is still significant association after adjustment of age, sex and BMI. There was no statistically significant association of rs17114036, rs9369640 Z, rs579459 and rs8055236 with CHD in the Chinese population.
Figure 2.
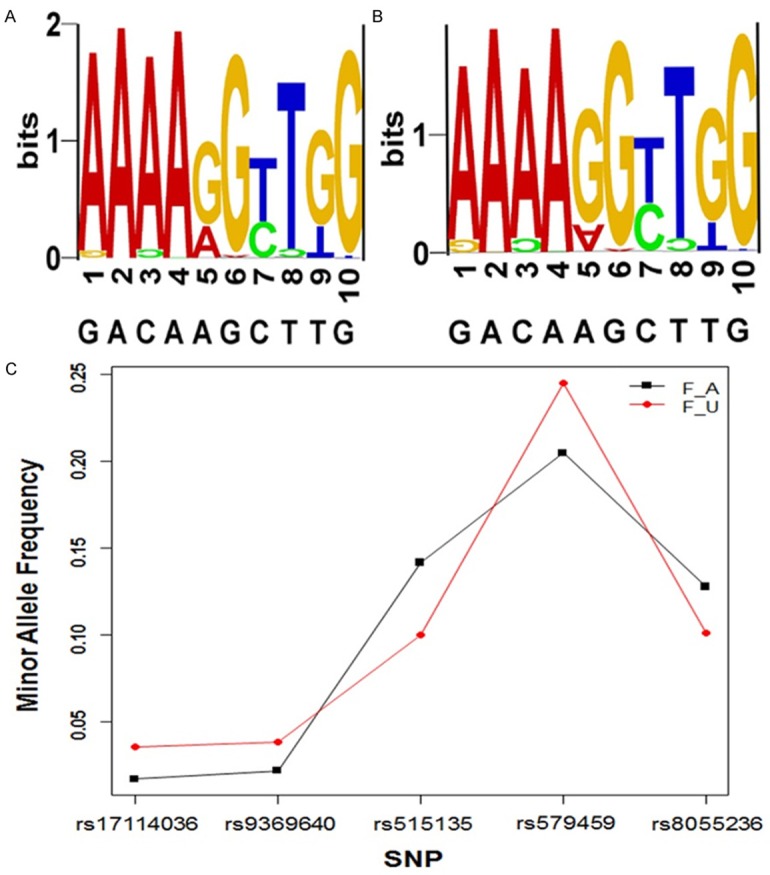
Allele distribution and minor allele frequency of 5 SNPs in the study patients and controls. A and B are SNP LOGO of 545 blood-unrelated Chinese patients and 1008 healthy controls respectively. (1, 3, 5, 7, 9 shows the minor allele proportion of five SNPs, and 2, 4, 6, 8, 10 shows the major one). C. Minor allele frequency of five SNPs in patients (F_A) and controls (F_U).
Table 3.
Association between single-nucleotide polymorphisms and coronary artery diseasesusceptibility in Chinese population coronary artery disease patients (all) versus controls
| SNP | Allele(risk) | Mode | Odds ratio (95% CI) | P-value | power |
|---|---|---|---|---|---|
| rs17114036 | G/A (A) | Dominant | 1.003 (0.999-1.007) | 0.523 | 5 |
| Recessive | 2.040 (1.124-3.701) | 0.017 | 67 | ||
| Allele | 2.093 (1.165-3.761) | 0.01167 | 41.7 | ||
| rs9369640 | C/A (A) | Dominant | 1.284 (0.116-14.208) | 1 | 4.7 |
| Recessive | 1.846 (1.062-3.209) | 0.028 | 49.6 | ||
| Allele | 1.787 (1.049-3.044) | 0.03043 | 32 | ||
| rs515135 | G/A (A) | Dominant | 1.638 (1.227-2.187) | 0.001 | 91.4 |
| Recessive | 0.698 (0.214-2.281) | 0.776 | 8.2 | ||
| Allele | 1.48 (1.138-1.925) | 0.00333 | 54.3 | ||
| rs579459 | C/T (T) | Dominant | 1.477 (0.861-2.533) | 0.155 | 28.9 |
| Recessive | 1.285 (0.999-1.652) | 0.05 | 49.9 | ||
| Allele | 1.261 (1.024-1.554) | 0.02917 | 33.6 | ||
| rs8055236 | G/T (T) | Dominant | 1.575 (0.453-5.472) | 0.471 | 87.1 |
| Recessive | 1.337 (0.998-1.791) | 0.051 | 8.1 | ||
| Allele | 1.305 (0.9978-1.707) | 0.05139 | 28.4 |
Figure 3.
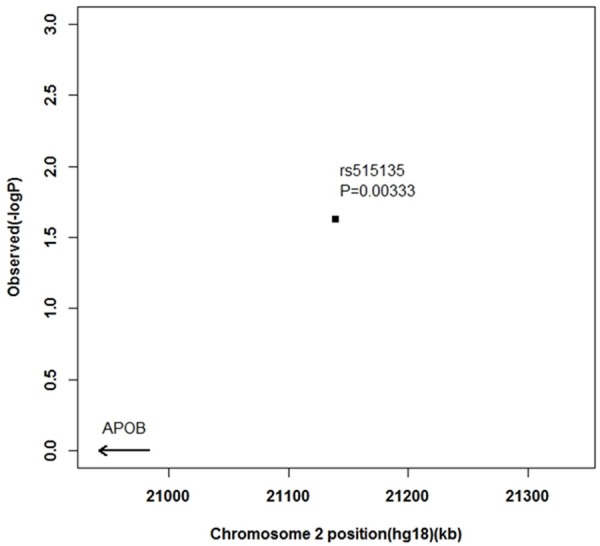
The scatter plots of the suggestive association evidences within 2p24-p23 (APOB) for CHD.
Stratified analysis
Stratified analysis revealed that rs579459 is strongly associated with severe coronary heart disease. Associations between coronary heart disease severity relative to the controls were not observed for the other four SNPs (Table 4).
Table 4.
Associations between single-nucleotide polymorphisms and coronary artery disease susceptibility in the Chinese population relative to the controls
| SNP | Allele (risk) | Mode | Odds ratio (95% CI) | P-value | Power |
|---|---|---|---|---|---|
| 1) Mild- moderatecoronary artery disease vs control | |||||
| rs17114036 | G/A (A) | Dominant | 1.00 (0.999-1.007) | 1 | NA |
| Recessive | 3.49 (1.070-11.379) | 0.032 | 70 | ||
| Allele | 3.55 (1.099-11.490) | 0.02386 | 49 | ||
| rs9369640 | C/A (A) | Dominant | 1.00 (0.999-1.007) | 1 | NA |
| Recessive | 1.59 (0.706-3.582) | 0.259 | 39 | ||
| Allele | 1.63 (0.734-3.631) | 0.2253 | 27 | ||
| rs515135 | G/A (A) | Dominant | 1.64 (1.082-2.49) | 1.90E-02 | 98 |
| Recessive | 0.52 (0.065-4.126) | 1 | 12 | ||
| Allele | 1.47 (1.005-2.145) | 0.04601 | 67 | ||
| rs579459 | C/T (T) | Dominant | 1.25 (0.575-2.700) | 0.577 | 16 |
| Recessive | 0.95 (0.661-1.371) | 0.791 | 7 | ||
| Allele | 1.00 (0.745-1.349) | 0.989 | 5 | ||
| rs8055236 | G/T (T) | Dominant | 1.70 (1.134-2.561) | 0.01 | 99 |
| Recessive | 0.93 (0.108-7.997) | 1 | 5 | ||
| Allele | 1.56 (1.079-2.260) | 0.01742 | 83 | ||
| 2) Severe coronary artery disease vs control | |||||
| rs17114036 | G/A (A) | Dominant | 1.00 (0.999-1.007) | 1 | NA |
| Recessive | 1.38 (0.574-3.298) | 0.472 | 22 | ||
| Allele | 1.42 (0.602-3.366) | 0.4181 | 17 | ||
| rs9369640 | C/A (A) | Dominant | 0.35 (0.032-3.894) | 0.386 | 34 |
| Recessive | 2.30 (0.814-6.489) | 0.117 | 58 | ||
| Allele | 1.87 (0.737-4.719) | 0.1813 | 32 | ||
| rs515135 | G/A (A) | Dominant | 1.51 (0.956-2.393) | 0.076 | 84 |
| Recessive | 1.01 (1.005-1.023) | 0.37 | NA | ||
| Allele | 1.33 (0.871-2.030) | 0.1852 | 32 | ||
| rs579459 | C/T (T) | Dominant | 2.03 (0.717-5.754) | 0.216 | 50 |
| Recessive | 1.79 (1.160-2.757) | 0.008 | 97 | ||
| Allele | 1.68 (1.154-2.436) | 0.00623 | 90 | ||
| rs8055236 | G/T (T) | Dominant | 1.07 (0.656-1.740) | 0.792 | 7 |
| Recessive | 1.14 (0.132-9.873) | 1 | 4 | ||
| Allele | 1.06 (0.678-1.671) | 0.7871 | 6 | ||
Abbreviations: CI, confidence interval; SNP, single-nucleotidepolymorphism.
Discussion
CHD is the result of a combination of genetic and environmental factors. Until recently, the underlying mechanisms of coronary heart disease formation are only partially understood. More than 200 risk factors have been associated with CHD and, among these, randomized controlled trials have shown that low-density lipoprotein cholesterol (LDL-c) and blood pressure (BP) is causally related to CHD [14-16]. A key factor in reducing the global burden of CAD is early prediction of the disease to target preventive interventions.
Several studies have investigated a number of genes for polymorphisms or mutations that may be associated with coronary heart disease [17]. However, the results genetic studies of coronary heart disease have not been very satisfactory. Recent GWAS studies have identified several genetic variants that are associated with CAD [18-21]. The present study was performed in a Chinese population to determine the positive association signals identified in the GWAS studies with CAD in Caucasians.
We have shown that, there was a significant association of the SNP rs515135 with CHD after age, sex and BMI adjustment in a Chinese population. No association was found betweenSNPs rs17114036, rs9369640, rs579459 and rs8055236 and susceptibility of coronary heart disease.
The SNP rs515135 is located on chromosome 2p23-24 where locates the APOB gene. APOB is the main apolipoprotein component of LDL-C and it plays a central role in lipid metabolism as the major protein component of very-low-density lipoprotein (VLDL) and LDL, and serves as the ligand for the removal of LDL from circulation [22]. The gene encoding for APOB has been cloned and sequenced. It is 43 kb in length with 29 exons [23]. The APOB gene is known to have several sequences that serve as recognition sites for various restriction enzymes. Several polymorphisms have been identified in the APOB gene, which can result in the deletion of amino acids from the signal peptide and have been postulated to affect APOB secretion and assembly with lipids. In our study, polymorphisms of rs515135 in the APOB gene in CHD patients were significantly higher compared to healthy controls, suggesting that these variants increase the risk of CHD.
Stratified analysis of rs579459 revealed that in the recessive mode of inheritance (where C/T+C/C is compared with T/T), the severe coronary artery disease group had significant ORs compared with both the controls and the groupwith mild coronary artery disease. The ORs were maintained after Bonferroni’s correction (OR: 1.79, 95% CI: 1.16-2.76, p-value 0.008, respectively). The ratio of the severe tomild OR for rs579459 (recessive model) is presented in Table 4 (1.79/0.95=1.88). The result indicates that rs579459 is strongly associated with severe coronary artery disease. No association was detected for the other four SNPs regarding the severity of coronary artery disease.
The ABO SNP rs579459 had strong association with severe coronary artery disease in our data. Other GWAS studies have also identified ABO as a locus for low density lipoprotein (LDL-C)[24], type-2 diabetes [25], inflammatory risk biomarkers E-selectin, P-selectin, and sol-ICAM1 [25-27]. These findings suggested that ABO might modulate multiple pathways relating to cardiovascular risk factors, atherosclerosis and thrombosis. The genetic variation(s) in ABO might affect low density lipoprotein and coronary artery disease formation, although further studies are needed to explore its precise role in the development of CHD. There is limitation in the present study, as the cohort is of a limited size, and may not have sufficient power to detect SNPs with a small effect with a true association. Further studies in other ethnic groups and functional studies of the selected SNPs are needed in order to provide more insights into the biological relevance of CHD.
Acknowledgements
We are grateful to all the members who are willingly to participate in this study. This work was funded by the Anhui Provincial Natural Science Foundation (1208085QH155).
Disclosure of conflict of interest
None.
References
- 1.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 2.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. New Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 4.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peden J, Hopewell J, Saleheen D, Chambers J, Hager J, Soranzo N, Collins R, Danesh J, Elliott P, Farrall M, Stirrups K, Zhang W, Hamsten A, Parish S, Lathrop M, Watkins H, Clarke R, Deloukas P, Kooner J, Goel A, Ongen H, Strawbridge R, Heath S, Mälarstig A, Helgadottir A, Öhrvik J, Murtaza M, Potter S, Hunt S, Delepine M, Jalilzadeh S, Axelsson T, Syvanen A, Gwilliam R, Bumpstead S, Gray E, Edkins S, Folkersen L, Kyriakou T, Franco-Cereceda A, Gabrielsen A, Seedorf U, MuTHER C, Eriksson P, Offer A, Bowman L, Sleight P, Armitage J, Peto R, Abecasis G, Ahmed N, Caulfield M, Donnelly P, Froguel P, Kooner A, McCarthy M, Samani N, Scott J, Sehmi J, Silveira A, Hellénius M, van’t Hooft F, Olsson G, Rust S, Assman G, Barlera S, Tognoni G, Franzosi M, Linksted P, Green F, Rasheed A, Zaidi M, Shah N, Samuel M, Mallick N, Azhar M, Zaman K, Samad A, Ishaq M, Gardezi A, Fazal-ur-Rehman M, Frossard P, Spector T, Peltonen L, Nieminen M, Sinisalo J, Salomaa V, Ripatti S, Bennett D, Leander K, Gigante B, de Faire U, Pietri S, Gori F, Marchioli R, Sivapalaratnam S, Kastelein J, Trip M, Theodoraki E, Dedoussis G, Engert J, Yusuf S, Anand S. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 6.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, Silveira A, Malarstig A, Green FR, Lathrop M, Gigante B, Leander K, de Faire U, Seedorf U, Hamsten A, Collins R, Watkins H, Farrall M. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. New Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 7.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. New Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O’Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schafer A, Marz W, Renner W, Bugert P, Kluter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Yee J, Friedlander Y, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Meigs JB, Williams G, Nathan DM, MacRae CA, Havulinna AS, Berglund G, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Daly MJ, Nemesh J, Korn JM, McCarroll SA, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Ouwehand W, Deloukas P, Scholz M, Cambien F, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Scheffold T, Berger K, Huge A, Martinelli N, Olivieri O, Corrocher R, McKeown P, Erdmann E, Konig IR, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Do R, Xie C, Siscovick D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, Willenborg C, Wright B, Chen L, Li M, Salo P, Voight BF, Burns P, Laskowski RA, Xue Y, Menzel S, Altshuler D, Bradley JR, Bumpstead S, Burnett MS, Devaney J, Doring A, Elosua R, Epstein SE, Erber W, Falchi M, Garner SF, Ghori MJ, Goodall AH, Gwilliam R, Hakonarson HH, Hall AS, Hammond N, Hengstenberg C, Illig T, Konig IR, Knouff CW, McPherson R, Melander O, Mooser V, Nauck M, Nieminen MS, O’Donnell CJ, Peltonen L, Potter SC, Prokisch H, Rader DJ, Rice CM, Roberts R, Salomaa V, Sambrook J, Schreiber S, Schunkert H, Schwartz SM, Serbanovic-Canic J, Sinisalo J, Siscovick DS, Stark K, Surakka I, Stephens J, Thompson JR, Volker U, Volzke H, Watkins NA, Wells GA, Wichmann HE, Van Heel DA, Tyler-Smith C, Thein SL, Kathiresan S, Perola M, Reilly MP, Stewart AF, Erdmann J, Samani NJ, Meisinger C, Greinacher A, Deloukas P, Ouwehand WH, Gieger C. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 12.Butterworth A, Braund P, Farrall M, Hardwick R, Saleheen D, Peden J, Soranzo N, Chambers J, Sivapalaratnam S, Kleber M, Keating B, Qasim A, Klopp N, Erdmann J, Assimes T, Ball S, Balmforth A, Barnes T, Basart H, Baumert J, Bezzina C, Boerwinkle E, Boehm B, Brocheton J, Bugert P, Cambien F, Clarke R, Codd V, Collins R, Couper D, Cupples L, de Jong J, Diemert P, Ejebe K, Elbers C, Elliott P, Fornage M, Franzosi M, Frossard P, Garner S, Goel A, Goodall A, Hengstenberg C, Hunt S, Kastelein J, Klungel O, Klüter H, Koch K, König I, Kooner A, Laaksonen R, Lathrop M, Li M, Liu K, McPherson R, Musameh M, Musani S, Nelson C, O’Donnell C, Ongen H, Papanicolaou G, Peters A, Peters B, Potter S, Psaty B, Qu L, Rader D, Rasheed A, Rice C, Scott J, Seedorf U, Sehmi J, Sotoodehnia N, Stark K, Stephens J, van der Schoot C, van der Schouw Y, Thorsteinsdottir U, Tomaszewski M, van der Harst P, Vasan R, Wilde A, Willenborg C, Winkelmann B, Zaidi M, Zhang W, Ziegler A, de Bakker P, Koenig W, Mätz W, Trip M, Reilly M, Kathiresan S, Schunkert H, Hamsten A, Hall A, Kooner J, Thompson S, Thompson J, Deloukas P, Ouwehand W, Watkins H, Danesh J, Samani N. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51:606. doi: 10.1016/s0002-9149(83)80105-2. [DOI] [PubMed] [Google Scholar]
- 14.Kuulasmaa K, Tunstall-Pedoe H, Dobson A, Fortmann S, Sans S, Tolonen H, Evans A, Ferrario M, Tuomilehto J. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 2000;355:675–687. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- 15.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanktree MB, Hegele RA. Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med. 2009;1:28. doi: 10.1186/gm28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 18.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton P, Clayton D, Cardon L, Craddock N, Deloukas P, Duncanson A, Kwiatkowski D, McCarthy M, Ouwehand W, Samani N, Todd J, Donnelly P, Barrett J, Burton P, Davison D, Donnelly P, Easton D, Evans D, Leung H, Marchini J, Morris A, Spencer C, Tobin M, Cardon L, Clayton D, Attwood A, Boorman J, Cant B, Everson U, Hussey J, Jolley J, Knight A, Koch K, Meech E, Nutland S, Prowse C, Stevens H, Taylor N, Walters G, Walker N, Watkins N, Winzer T, Todd J, Ouwehand W, Jones R, McArdle W, Ring S, Strachan D, Pembrey M, Breen G, St C, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green E, Grozeva D, Hamshere M, Holmans P, Jones I, Kirov G, Moskvina V, Nikolov I, O’Donovan M, Owen M, Craddock N, Collier D, Elkin A, Farmer A, Williamson R, McGuffin P, Young A, Ferrier I, Ball S, Balmforth A, Barrett J, Bishop D, Iles M, Maqbool A, Yuldasheva N, Hall A, Braund P, Burton P, Dixon R, Mangino M, Suzanne S, Tobin M, Thompson J, Samani N, Bredin F, Tremelling M, Parkes M, Drummond H, Lees C, Nimmo E, Satsangi J, Fisher S, Forbes A, Lewis C, Onnie C, Prescott N, Sanderson J, Mathew C, Barbour J, Mohiuddin M, Todhunter C, Mansfield J, Ahmad T, Cummings F, Jewell D, Webster J, Brown M, Clayton D, Lathrop G, Connell J, Dominczak A, Samani N, Marcano C, Burke B, Dobson R, Gungadoo J, Lee K, Munroe P, Newhouse S, Onipinla A, Wallace C, Xue M, Caulfield M, Farrall M, Barton A, Bruce I, Donovan H, Eyre S, Gilbert P, Hider S, Hinks A, John S, Potter C, Silman A, Symmmons D, Thomson W, Worthington J, Clayton D, Dunger D, Nutland S, Stevens H, Walker N, Widmer B, Todd J, Frayling T, Freathy R, Lango H, Perry J, Shields B, Weedon M, Hattersley A, Hitman G, Walker M, Elliott K, Groves C, Lindgren C, Rayner N, Timpson N, Zeggini E, McCarthy M, Newport M, Sirugo G, Lyons E, Vannberg F, Hill A, Bradbury L, Farrar C, Pointon J, Wordsworth P, Brown M, Franklyn J, Heward J, Simmonds M, Gough S, Seal S, Stratton M, Rahman N, Ban M, Goris A, Sawcer S, Compston A, Conway D, Jallow M, Newport M, Sirugo G, Rockett K, Kwiatowski D, Bumpstead S, Chaney A, Downes K, Ghori M, Gwilliam R, Hunt S, Inouye M, Keniry A, King E, McGinnis R, Potter S, Ravindrarajah R, Whittaker P, Widden C, Withers D, Deloukas P, Leung H, Nutland S, Stevens H, Walker N, Todd J, Easton D, Clayton D, Burton P, Tobin M, Barrett J, Evans D, Morris A, Cardon L, Cardin N, Davison D, Ferreira T, Pereira-Gale J, Hallgrimsdottir I, Howie B, Marchini J, Spencer C, Su Z, Teo Y, Vukcevic D, Donnelly P, Bentley D, Brown M, Gordon L, Caulfield M, Clayton D, Compston A, Craddock N, Deloukas P, Donnelly P, Farrall M, Gough S, Hall A, Hattersley A, Hill A, Kwiatkowski D, Mathew C, McCarthy M, Ouwehand W, Parkes M, Pembrey M, Rahman N, Samani N, Stratton M, Todd J, Worthington J. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Xu Y, Wang X, Wang Q, Zhang L, Tu Y, Yan J, Wang W, Hui R, Wang CY, Wang DW. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ-Cardiovasc Gene. 2009;2:338–346. doi: 10.1161/CIRCGENETICS.108.810226. [DOI] [PubMed] [Google Scholar]
- 21.Lv X, Zhang Y, Rao S, Qiu J, Wang M, Luo X, Zuo X, Su D, Feng X, Yang Y, Ouyang P, Chen Y, Li X, Xiao Y, Ling W. Joint effects of genetic variants in multiple loci on the risk of coronary artery disease in Chinese Han subjects. Circ J. 2012;76:1987–1992. doi: 10.1253/circj.cj-12-0156. [DOI] [PubMed] [Google Scholar]
- 22.Machado MO, Hirata MH, Bertolami MC, Hirata RD. Apo B gene haplotype is associated with lipid profile of higher risk for coronary heart disease in Caucasian Brazilian men. J Clin Lab Anal. 2001;15:19–24. doi: 10.1002/1098-2825(2001)15:1<19::AID-JCLA4>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig EH, Blackhart BD, Pierotti VR, Caiati L, Fortier C, Knott T, Scott J, Mahley RW, Levy-Wilson B, McCarthy BJ. DNA sequence of the human apolipoprotein B gene. DNA. 1987;6:363–372. doi: 10.1089/dna.1987.6.363. [DOI] [PubMed] [Google Scholar]
- 24.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, Ordovas JM, Ripatti S, Parker AN, Miletich JP, Ridker PM. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, Girman CJ, Laurie CC, Mirel DB, Hunter DJ, Rimm E, Hu FB. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19:1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, Shen E, Mirea L, Bharaj B, Sun L, Bull SB. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscl Throm Vas. 2009;29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, Nambi V, Bretler M, Smith NL, Peters A, Lu C, Tracy RP, Aleksic N, Heeriga J, Keaney JF Jr, Rice K, Lip GY, Vasan RS, Glazer NL, Larson MG, Uitterlinden AG, Yamamoto J, Durda P, Haritunians T, Psaty BM, Boerwinkle E, Hofman A, Koenig W, Jenny NS, Witteman JC, Ballantyne C, Benjamin EJ. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]


