Abstract
Background: A large body of studies has investigated the potential role of ABCB1 polymorphism in ALL susceptibility. However, the results are conflicting. The aim of the present meta-analysis was to define the effect of ABCB1 polymorphism on ALL risk. Methods: We identified 8 eligible studies involving 1,308 cases and 1,427 controls through searching PubMed and Enbase databases. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to access the strength of the association with both fixed effects and random effect models. Results: We found ABCB1 polymorphism was associated with an increased risk of ALL under the homozygote genotypes (TT vs. CC: OR, 1.29, 95% CI, 1.08-1.54), the recessive model (TT vs. CT + CC: OR, 1.47, 95% CI, 1.02-2.13) and the allele model (T vs. C: OR, 1.14, 95% CI, 1.04-1.25). Similar results were indicated in Asian populations (TT vs. CC: OR, 1.79, 95% CI, 1.32-2.43; TT vs. CT + CC: OR, 2.55, 95% CI, 1.47-4.43; T vs. C: OR, 1.38, 95% CI, 1.18-1.62), but not in Caucasian populations. Conclusions: These findings indicate that ABCB1 polymorphism may play a critical role in the development of ALL in Asians.
Keywords: ABCB1, polymorphism, ALL
Introduction
Acute lymphoblastic leukemia (ALL) is a malignant neoplasm of hematopoietic stem cells, highly occurring among children with a proportion of 30% of all pediatric malignancies [1,2]. The initial peak incidence of this hematologic malignancy is at 2 to 5 years of age, followed by a second peak over age 50. Although the etiology is currently unknown, environmental factors interacting with genetic components may jointly contribute to leukemogenesis [3,4].
ABCB1 gene is mapped to chromosome 7 and encodes for P-glycoprotein (P-gp), a 170-kDa member belonging to ATP-binding cassette (ABC) superfamily of membrane transporters [5]. P-gp responsible for transporting phospholipids across the cell membrane may act as an anti-apoptotic molecule. Dysregulation of P-gp expression in cancer cells induces accumulation of carcinogens and decreases drug accumulation, thereby regulating cellular resistance to various anti-cancer agents [6]. P-gp mainly expressed in liver, kidney, testis, placenta, gastrointestinal tract, and blood-tissue barrier [7]. A large body of evidence also showed that P-gp is involved in the release of interleukin-2, interleukin-4, and IFN-γ from lymphocytes [8-10]. In addition, over-expression of ABCB1 has been indicated in peripheral leukocytes and bone marrow [11-14].
ABCB1 is a highly polymorphic gene, in which more than 28 single nucleotide polymorphisms (SNPs) have been identified in genetic studies [15-17]. The SNP located in exon 26 at position 3435 led to over 2-fold lower P-gp expression in duodenum and higher plasma concentration of P-gp substrate digoxin TT genotype carriers in comparison with carriers with CC genotype [15]. A case-control study conducted in Poland demonstrated a significant association between the silent ABCB1 polymorphism (rs1045642) and ALL risk [18]. Later, a subsequent report addressing four ABCB1 SNPs (T-129C, C1236T, G2677T/A, C3435T) did not show evidence of a statistically significant association with ALL in non-Hispanic White or Hispanic children [19]. Possible reasons for these discrepancies are differences in small or non-homogeneous populations, and the subjects of diverse ethnicities.
In this study, we undertook a meta-analysis based on all available data, in an attempt to determine the relationship between ABCB1 polymorphism and susceptibility to ALL.
Methods
Identification and eligibility of relevant studies
We performed an exhaustive search in PubMed and Embase to identify studies concerning the association between ABCB1 polymorphism and susceptibility to ALL until February 2013. The keywords used in the search strategy were “multidrug resistance 1” or “ABCB1” or “rs1045642”, “polymorphism” or “variant”, and “acute lymphoblastic leukemia” or “ALL”. All references of the retrieved articles were then reviewed to identify additional relevant works. The inclusion criteria for eligible studies required that the study had to investigate the association of ABCB1 polymorphism and susceptibility to ALL; that each study must be based on a case-control design; and that the original article provided adequate data of genotype frequency. The article was not considered if required information was not supplied or a case-only design was used.
Data extraction
Data extraction was conducted independently by two reviewers. The information gathered from each study included first author’s name, year of publication, study country, race/ethnicity, total cases and controls, gender distribution, mean age, genotyping method, and genotype frequency in cases and controls. A third reviewer was consulted to reach a consensus when disagreements were encountered.
Statistical analysis
Using the genetic data extracted from each study and meta orders, we calculated pooled odds ratios (ORs) along with 95% confidence intervals (CIs) with Stata software, to evaluate the strength of association between ABCB1 polymorphism and ALL risk. Statistical significance of the pooled ORs was examined by Z-test, and P < 0.10 was deemed significant. x2-based Q-test and the I 2 statistic were performed to measure heterogeneity across the studies. A P value more than 0.10 and I 2 <50% represented absence of heterogeneity. In such a case, the fixed-effects model based on Mantel-Haenszel method that assumes the same homogeneity of effect size among studies was used to pool the summary ORs [20]. Otherwise, the random-effects model based on DerSimonian and Laird method was performed in order to provide wider 95% CIs for the studies with different findings [21]. We created funnel plots where the standard error of log (OR) of each study is plotted against its log (OR) to assess publication bias. The funnel plot asymmetry was further examined using Egger’s liner regression test and P < 0.10 was considered statistically significant [22]. Deviation of genotype distributions from Hardy-Weinberg equilibrium (HWE) was checked in controls by the chi-square test (P < 0.10 indicated significant HWE violation). Sensitivity analysis by sequentially excluding the independent studies was performed to assess the influence on pooled ORs.
Results
Eligible studies and studies’ characteristics
We identified 203 records in the initial search of PubMed and Embase. After eliminating the obviously irrelevant records, 137 articles were left for further review. Of these, 121 were discarded after reviewing the key words and abstracts. The full texts of the remaining 16 articles were examined in detail. Among them, 8 studies [6,18,19,23-27] satisfied the pre-described inclusion criteria and were finally included in the meta-analysis. The genotype distributions among the controls of all studies were in agreement with HWE except for two studies [18,26]. The genotyping for ABCB1 polymorphism was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and Taqman PCR on the genomic DNA from the human blood samples. Moreover, of the 8 studies, five were for the population of Caucasian ancestry, and three were for the subjects of Asian ancestry. The summary characteristics of the eligible studies are described in Table 1.
Table 1.
Characteristics of literatures included in the meta-analysis
| First author (year) | Country | Ethnicity | Control source | Genotyping method | Gender (F/M) | Mean age | Cases | Controls | HWE |
|---|---|---|---|---|---|---|---|---|---|
| Jamroziak (2004) | Poland | Caucasian | Population | PCR-RFLP | 96/79 | 21.4 | 113 | 175 | 0.043 |
| Urayama (2007) | USA | Caucasian | Population | TaqMan | NR | NR | 294 | 369 | 0.506 |
| Hattori (2007) | Japan | Asian | Population | TaqMan | 67/90 | 6.1 | 118 | 96 | 0.543 |
| Semsei (2008) | HunCTry | Caucasian | Population | TaqMan | 180/126 | 6.1 ± 3.9 | 378 | 189 | 0.127 |
| Leal-Ugarte (2008) | Mexico | Caucasian | Population | PCR-RFLP | 44/63 | 7.0 | 107 | 111 | 0.298 |
| Rao (2010) | India | Asian | Population | PCR-RFLP | 47/100 | 16.5 | 147 | 249 | 0.560 |
| Bektas-Kayhan (2012) | Turkey | Caucasian | Population | PCR-RFLP | 13/34 | 8.7 ± 4.9 | 47 | 68 | 0.015 |
| Lv (2012) | China | Asian | Hospital | PCR-RFLP | 68/108 | 5.7 ± 3.3 | 176 | 170 | 0.781 |
PCR-RFLP: polymerase chain reaction- restriction fragment length polymorphism; NR: not report; HWE: Hardy-Weinberg equilibrium.
Meta-analysis
Table 2 shows the detailed results of the meta-analysis. By combining all available studies in one dataset, the homozygote genotypes of ABCB1 polymorphism was found to be associated with an elevated risk of ALL (TT vs. CC: OR, 1.29, 95% CI, 1.08-1.54) (Figure 1). Meanwhile, the recessive model (TT vs. CT + CC: OR, 1.47, 95% CI, 1.02-2.13) and the allele model (T vs. C: OR, 1.14, 95% CI, 1.04-1.25) also showed a significant tend to increase ALL risk (Figure 2).
Table 2.
Meta-analysis of ABCB1 polymorphism and AMD risk
| Variables | TT vs. CC (FM) | TT + CT vs. CC (FM) | TT vs. CT + CC (RM) | T vs. C (FM) | CT vs. CC (FM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| OR (95% CI) | P h | I 2 | OR (95% CI) | P h | I 2 | OR (95% CI) | P h | I 2 | OR (95% CI) | P h | I 2 | OR (95% CI) | P h | I 2 | |
| Ethnicity | |||||||||||||||
| Total | 1.29 (1.08, 1.54) | 0.109 | 40.5% | 1.04 (0.93, 1.17) | 0.997 | 0 | 1.47 (1.02, 2.13) | < 0.10 | 76.7% | 1.14 (1.04, 1.25) | 0.101 | 41.6% | 0.95 (0.82, 1.10) | 0.427 | 0.2% |
| Caucasian | 1.07 (0.86, 1.34) | 0.778 | 0 | 1.02 (0.88, 1.17) | 0.979 | 0 | 1.07 (0.86, 1.32) | 0.375 | 5.5% | 1.03 (0.92, 1.15) | 0.757 | 0 | 1.00 (0.84, 1.19) | 0.954 | 0 |
| Asian | 1.79 (1.32, 2.43) | 0.183 | 41.1% | 1.09 (0.89, 1.34) | 0.964 | 0 | 2.55 (1.47, 4.43) | 0.044 | 68.0% | 1.38 (1.18, 1.62) | 0.503 | 0 | 0.84 (0.64, 1.11) | 0.067 | 63.0% |
| Source of control | |||||||||||||||
| Population | 1.17 (0.97, 1.42) | 0.546 | 0 | 1.04 (0.92, 1.18) | 0.991 | 0 | 1.26 (0.96, 1.65) | 0.052 | 52.0% | 1.09 (0.99, 1.20) | 0.441 | 0 | 1.01 (0.87, 1.18) | 0.994 | 0 |
| Hospital | 2.54 (1.46, 4.42) | - | - | 1.06 (0.75, 1.50) | - | - | 4.11 (2.41, 6.98) | - | - | 1.57 (1.20, 2.07) | - | - | 0.50 (0.30, 0.85) | - | - |
P h: P value of heterogeneity test; CI: confidence interval; OR: odds ratio; FM: fixed-effects model; RM: random-effects model.
Figure 1.
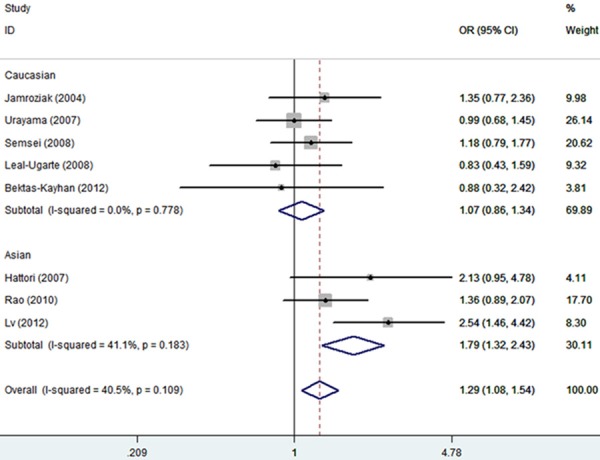
Forest plot of estimates of the odds ratios (ORs) for ABCB1 polymorphism in acute lymphoblastic leukemia (ALL) under TT vs. CC.
Figure 2.
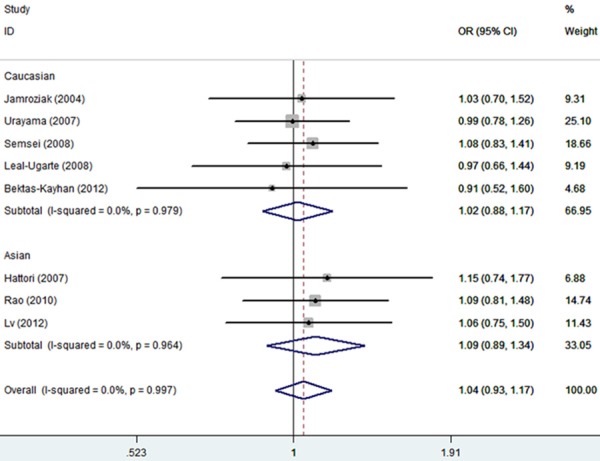
Forest plot of estimates of the odds ratios (ORs) for ABCB1 polymorphism in ALL under TT + CT vs. CC.
In the stratified analysis based on ethnicity, an obviously increased risk of ALL risk was indicated among Asians under the homozygote genotypes, the recessive model, and the allele model (TT vs. CC: OR, 1.79, 95% CI, 1.32-2.43; TT vs. CT + CC: OR, 2.55, 95% CI, 1.47-4.43; T vs. C: OR, 1.38, 95% CI, 1.18-1.62, respectively) (Table 2). However, there was no indication of significant association under any genetic model among Caucasian populations.
When stratifying the general population according to control source, we found that the OR of the homozygote genotypes was 2.54 (TT vs. CC: 95% CI, 1.46-4.42), and that of the recessive model was 4.11 (TT vs. CT + CC: 95% CI, 2.41-6.98) in the studies based on hospital controls. Interestingly, we observed a reverse association under the heterozygote genotypes (CT vs. CC: OR, 0.50, 95% CI, 0.30-0.85).
Significant heterogeneity was observed in the recessive model and the random-effects model was selected to pool the OR. Sensitivity analysis was subsequently performed and identified Lv et al. [27] was the main source of the between-study heterogeneity. Exclusion of this study increased the homogeneity across the studies. No significant publication bias was tested by performing Begg’s funnel plot and Egger’s test (P = 0.223 for TT vs. CC) (Figure 3).
Figure 3.
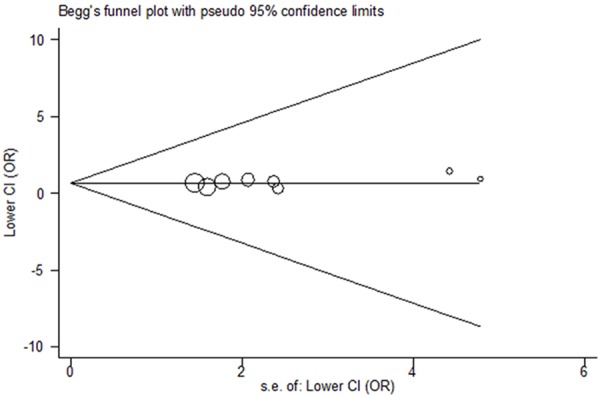
Funnel plot for publication bias test (TT vs. CC).
Discussion
ALL is the primary subtype of childhood leukemia characterized by the uncontrolled proliferation of hematopoietic cells in the bone marrow. Approximately 80% of children with newly diagnosed ALL and 50% of them with myeloid neoplasm can be cured, due to contemporary improvements in risk assessment, chemotherapy, hematopoietic stem cell transplantation and supportive care [28]. However, the resistance to chemotherapeutic drugs remains a major problem that results in poor treatment outcome of this disease [29,30]. The human ABCB1 is located on chromosome 7q21, encoding a 170 KD integral membrane protein product Pgp, which functions as a protective role against harmful chemicals and active metabolites. Available data reported that an increased risk of ALL is attributable to polymorphisms in genes that participate in transport and metabolism of xenobiotics [31]. The widely studied ABCB1 gene contains a number of polymorphisms that may confer susceptibility to ALL [18].
The ABCB1 polymorphism has been consistently investigated in the epidemiological studies regarding the risk of ALL. Nevertheless, the results are controversial [24,27]. Therefore, it is necessary to perform this meta-analysis to identify the association between ABCB1 polymorphism and ALL risk by combining the relevant studies published to date. After pooling available data from all included studies, we found a significantly elevated risk of ALL. When stratified according to ethnic origin, an obvious correlation between ABCB1 polymorphism and ALL risk was observed in Asian populations, but not in Caucasian populations. Interestingly, the subgroup analysis by source of controls showed ABCB1 polymorphism was likely to increase or decrease the risk of suffering ALL in the hospital-based studies.
A previous meta-analysis of the association between this polymorphism and cancer risk (five studies for ALL) demonstrated no significant association in ALL under all analyzed genetic models, either in Asians or in Caucasians [32]. While the current study expanded the sample size by adding additional three subsequently published articles showed very different results. The explanation may be that the ABCB1 is a low-penetrant polymorphism that needs a sufficiently large study to detect the susceptibility to cancer. Besides, the use of typical control populations is vitally important, especially for the genetic association studies. The failure to reach a statistical significance in population-based studies implies that the selection of representative controls may reduce bias of the results.
Several strong points need to be addressed. First, this is the first study on the association between ABCB1 polymorphism and susceptibility to ALL to date, which supplies reference information to future studies. Second, our meta-analysis based on all available data may provide new insights into the role of ABCB1 polymorphism in ALL risk. Three are also some limitations. Significant between-study heterogeneity indicated across the studies may have potentially influenced the results. Furthermore, we failed to carry out further stratification analysis by other confounders such as gender, because only one study supplied genotype frequency of this polymorphism in man and women [6].
In summary, we conclude that there is statistical evidence to support ABCB1 polymorphism is associated with an increased risk of ALL in Asians. However, larger studies taking gender into consideration is required to validate the current findings.
Acknowledgements
This work was supported by the “863 Projects” of Ministry of Science and Technology of PR China (No. 2011AA020114), Military Clinical High-Tech Key Program (No. 2010gxjs100) and Clinical feature and Application Research of Capital (No. Z111107058811107).
Disclosure of conflict of interest
None.
References
- 1.Koppen IJ, Hermans FJ, Kaspers GJ. Folate related gene polymorphisms and susceptibility to develop childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;148:3–14. doi: 10.1111/j.1365-2141.2009.07898.x. [DOI] [PubMed] [Google Scholar]
- 2.Johnston WT, Lightfoot TJ, Simpson J, Roman E. Childhood cancer survival: a report from the United Kingdom Childhood Cancer Study. Cancer Epidemiol. 2010;34:659–666. doi: 10.1016/j.canep.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Sinnett D, Krajinovic M, Labuda D. Genetic susceptibility to childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2000;38:447–462. doi: 10.3109/10428190009059264. [DOI] [PubMed] [Google Scholar]
- 4.Kim YI. Methylenetetrahydrofolate reductase polymorphisms, folate, and cancer risk: a paradigm of gene-nutrient interactions in carcinogenesis. Nutr Rev. 2000;58:205–209. doi: 10.1111/j.1753-4887.2000.tb01863.x. [DOI] [PubMed] [Google Scholar]
- 5.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 6.Rao DN, Anuradha C, Vishnupriya S, Sailaja K, Surekha D, Raghunadharao D, Rajappa S. Association of an MDR1 gene (C3435T) polymorphism with acute leukemia in India. Asian Pac J Cancer Prev. 2010;11:1063–1066. [PubMed] [Google Scholar]
- 7.Schinkel AH. The physiological function of drug-transporting P-glycoproteins. Semin Cancer Biol. 1997;8:161–170. doi: 10.1006/scbi.1997.0068. [DOI] [PubMed] [Google Scholar]
- 8.Drach J, Gsur A, Hamilton G, Zhao S, Angerler J, Fiegl M, Zojer N, Raderer M, Haberl I, Andreeff M, Huber H. Involvement of P-glycoprotein in the transmembrane transport of interleukin-2 (IL-2), IL-4, and interferon-gamma in normal human T lymphocytes. Blood. 1996;88:1747–1754. [PubMed] [Google Scholar]
- 9.Pawlik A, Baskiewicz-Masiuk M, Machalinski B, Kurzawski M, Gawronska-Szklarz B. Involvement of C3435T and G2677T multidrug resistance gene polymorphisms in release of cytokines from peripheral blood mononuclear cells treated with methotrexate and dexamethasone. Eur J Pharmacol. 2005;528:27–36. doi: 10.1016/j.ejphar.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 10.Raghu G, Park SW, Roninson IB, Mechetner EB. Monoclonal antibodies against P-glycoprotein, an MDR1 gene product, inhibit interleukin-2 release from PHA-activated lymphocytes. Exp Hematol. 1996;24:1258–1264. [PubMed] [Google Scholar]
- 11.Chaudhary PM, Roninson IB. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell. 1991;66:85–94. doi: 10.1016/0092-8674(91)90141-k. [DOI] [PubMed] [Google Scholar]
- 12.Elliott JI, Raguz S, Higgins CF. Multidrug transporter activity in lymphocytes. Br J Pharmacol. 2004;143:899–907. doi: 10.1038/sj.bjp.0705940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitzl M, Drescher S, van der Kuip H, Schaffeler E, Fischer J, Schwab M, Eichelbaum M, Fromm MF. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics. 2001;11:293–298. doi: 10.1097/00008571-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Sakaeda T, Nakamura T, Okumura K. MDR1 genotype-related pharmacokinetics and pharmacodynamics. Biol Pharm Bull. 2002;25:1391–1400. doi: 10.1248/bpb.25.1391. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, Ieiri I, Tanabe M, Suzuki A, Higuchi S, Otsubo K. Polymorphism of the ABC transporter genes, MDR1, MRP1 and MRP2/cMOAT, in healthy Japanese subjects. Pharmacogenetics. 2001;11:175–184. doi: 10.1097/00008571-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Kim RB, Leake BF, Choo EF, Dresser GK, Kubba SV, Schwarz UI, Taylor A, Xie HG, McKinsey J, Zhou S, Lan LB, Schuetz JD, Schuetz EG, Wilkinson GR. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70:189–199. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 18.Jamroziak K, Mlynarski W, Balcerczak E, Mistygacz M, Trelinska J, Mirowski M, Bodalski J, Robak T. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72:314–321. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 19.Urayama KY, Wiencke JK, Buffler PA, Chokkalingam AP, Metayer C, Wiemels JL. MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev. 2007;16:1172–1177. doi: 10.1158/1055-9965.EPI-07-0007. [DOI] [PubMed] [Google Scholar]
- 20.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattori H, Suminoe A, Wada M, Koga Y, Kohno K, Okamura J, Hara T, Matsuzaki A. Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia. Leuk Res. 2007;31:1633–1640. doi: 10.1016/j.leukres.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Leal-Ugarte E, Gutierrez-Angulo M, Macias-Gomez NM, Peralta-Leal V, Duran-Gonzalez J, De La Luz Ayala-Madrigal M, Partida-Perez M, Barros-Nunez P, Ruiz-Diaz D, Moreno-Ortiz JM, Peregrina-Sandoval J, Meza-Espinoza JP. MDR1 C3435T polymorphism in Mexican children with acute lymphoblastic leukemia and in healthy individuals. Hum Biol. 2008;80:449–455. doi: 10.3378/1534-6617-80.4.449. [DOI] [PubMed] [Google Scholar]
- 25.Semsei AF, Erdelyi DJ, Ungvari I, Kamory E, Csokay B, Andrikovics H, Tordai A, Csagoly E, Falus A, Kovacs GT, Szalai C. Association of some rare haplotypes and genotype combinations in the MDR1 gene with childhood acute lymphoblastic leukaemia. Leuk Res. 2008;32:1214–1220. doi: 10.1016/j.leukres.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Bektas-Kayhan K, Kucukhuseyin O, Karagoz G, Unur M, Ozturk O, Unuvar A, Devecioglu O, Yilmaz-Aydogan H. Is the MDR1 C3435T polymorphism responsible for oral mucositis in children with acute lymphoblastic leukemia? Asian Pac J Cancer Prev. 2012;13:5251–5255. doi: 10.7314/apjcp.2012.13.10.5251. [DOI] [PubMed] [Google Scholar]
- 27.Lu H, Du ZZ, Wang W, Zhao WL, Wang Y, Hu SY, Chai YH. [Relationship between genetic polymorphism of multidrug resistance 1 gene and the risk of childhood acute lymphocytic leukemia] . Zhonghua Er Ke Za Zhi. 2012;50:692–696. [PubMed] [Google Scholar]
- 28.Pui CH, Schrappe M, Ribeiro RC, Niemeyer CM. Childhood and adolescent lymphoid and myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2004:118–145. doi: 10.1182/asheducation-2004.1.118. [DOI] [PubMed] [Google Scholar]
- 29.Chessells JM, Veys P, Kempski H, Henley P, Leiper A, Webb D, Hann IM. Long-term follow-up of relapsed childhood acute lymphoblastic leukaemia. Br J Haematol. 2003;123:396–405. doi: 10.1046/j.1365-2141.2003.04584.x. [DOI] [PubMed] [Google Scholar]
- 30.Aladjidi N, Auvrignon A, Leblanc T, Perel Y, Benard A, Bordigoni P, Gandemer V, Thuret I, Dalle JH, Piguet C, Pautard B, Baruchel A, Leverger G. Outcome in children with relapsed acute myeloid leukemia after initial treatment with the French Leucemie Aique Myeloide Enfant (LAME) 89/91 protocol of the French Society of Pediatric Hematology and Immunology. J. Clin. Oncol. 2003;21:4377–4385. doi: 10.1200/JCO.2003.11.066. [DOI] [PubMed] [Google Scholar]
- 31.Belson M, Kingsley B, Holmes A. Risk factors for acute leukemia in children: a review. Environ Health Perspect. 2007;115:138–145. doi: 10.1289/ehp.9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Wang B, Bi J, Li K, Di J. MDR1 gene C3435T polymorphism and cancer risk: a meta-analysis of 34 case-control studies. J Cancer Res Clin Oncol. 2012;138:979–989. doi: 10.1007/s00432-012-1171-9. [DOI] [PubMed] [Google Scholar]


