Abstract
Recent studies in cancer have demonstrated that cancerous tissues have a significantly higher MALAT1 level than in noncancerous tissues. Overexpression of MALAT1 is associated with susceptibility to lymph node metastasis. This meta-analysis collected all relevant articles and explored the association of MALAT1 expression levels with lymph node metastasis in patients with carcinoma. Literature collections were conducted by searching electronic databases PubMed, Cochrane Library, Web of Science (up to January 20, 2015). The odds ratio (OR) and its corresponding 95% confidence interval (CI) were calculated to assess the strength of the association by using RevMan5.1 software. A total of 573 patients from 5 studies were included in this meta-analysis. The results showed lymph node metastasis occurred more frequently in patients with high MALAT1 expression group than in patients with low MALAT1 expression group (OR = 2.64, 95% CI 1.06-6.56, P = 0.04 random-effects model). This meta-analysis demonstrated that overexpression of MALAT1 is significantly associated with lymph node metastasis in carcinoma patients.
Keywords: MALAT1, lncRNA, lymph node metastasis, cancer, meta-analysis
Introduction
Detection of lymph node metastasis is of major prognostic significance in most cancers, and is an important part of the TNM classification system, which dictates the choice of future post-operative therapies and predicts prognosis of cancer patients [1]. In the multistep process of metastasis, invasion into the lymphatic system has generally been believed as a key step of tumor cell dissemination [2]. The exact mechanism of metastasis through lymph nodes is still unclear. Recent studies have attempted to explain the process at the genetic level. Various kinds of genomic signatures have been reported associated with lymph node and distant metastasis [3,4]. However, most of the reports were just talking about a molecule specific to a particular tumor. A putative biomarker for predicting lymph node metastasis at the transcriptional level has not been demonstrated [5].
Long noncoding RNAs (lncRNAs) are generally defined as transcribed RNA molecules with a length greater than 200 nt and lacking an open reading frame of significant length (less than 100 amino acids), and most lack protein coding capability [6]. However, increasing evidences have been presented to suggest that lncRNAs participated in a wide range of biological pathways. LncRNAs could regulate gene expression and function, including dosage compensation, genome rearrangement, chromatin modifications, gene imprinting, alternative splicing, nuclear- cytoplasmic trafficking, cell cycle control, and inactivation of major tumor suppressor genes [7-10]. Moreover, lncRNAs play a critical role in tumor development, progression, and metastasis [11].
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), also known as non-coding nuclear enriched abundant transcript 2 (NEAT2), is a ~ 8000-nt long non-coding RNA, which is highly conserved in human [12]. A number of studies have revealed that the expression level of MALAT1 was up-regulated in cancerous tissues of a variety of cancers, such as esophageal squamous cell carcinoma [13], gastric cancer [14], pancreatic cancer [15], hepatocellular cancer [16], colorectal cancer [17], prostate cancer [18] and cervical cancer [19]. In addition, regulating the expression of MALAT1 in vitro had a significant impact on the viability, proliferation, and invasion of tumor cells. We collected all relevant articles and carried out a meta-analysis to explore the relationship between MALAT1 expression levels with lymph node metastasis and furthermore to determine whether MALAT1 can be used as a putative biomarker for lymph node metastasis.
Methods
Literature search strategies
Articles up to January 20, 2015, which related to the long noncoding RNA MALAT1 serving as a putative biomarker for lymph node metastasis, were searched in several computerized databases, including PubMed, Cochrane Library, Web of Science and Chinese National Knowledge Infrastructure (CNKI). The search terms we used were listed as follows: MALAT1 or MALAT-1 or Metastasis associated lung adenocarcinoma transcript 1 and cancer or carcinoma or neoplasms or tumor and lymph node metastasis. Besides, the reference lists are manually viewed to obtain additional relevant articles.
Inclusion and exclusion criteria
Inclusion criteria are the following: (1) articles investigating the roles of MALAT1 in the development of cancer, (2) the expression levels of MALAT1 in primary cancerous tissues were measured, (3) patients were grouped according to the expression levels of MALAT1, (4) related clinicopathologic parameters were described, (5) studies containing sufficient data for the computation of odds ratios (OR) and corresponding 95% confidence intervals (CI). Exclusion criteria are the following: (1) studies investigating the molecular structure and functions of MALAT1; (2) letters, expert opinions, editorials, reviews and case reports; (3) duplicate publications; (4) studies without usable data.
Date extraction and quality assessment
Two investigators (HZ and QJC) extracted data from the eligible studies independently, according to the inclusion and exclusion criteria above. For disagreements, a consensus was reached by a third investigator (WL). The following information was collected from each eligible study: first author, publication date, country of origin, ethnicity, tumor type, total number of patients, number of high MALAT1 expression group and low MALAT1 expression group, number of patients with lymph node metastasis in each group, and detection method of MALAT1 expression levels. Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) was used to evaluate the quality of included publications.
Statistical methods
All statistical analyses used RevMan5.1 software and Stata SE12.0 (Stata Corporation). To determine the heterogeneity among the included studies, chi-square-based Q test and I2 statistics were used. For the Q test, a P value less than 0.05 indicated significant heterogeneity; for the I2 statistics, an I2 value greater than 50% was considered severe heterogeneity. The potential publication bias was assessed using a “funnel plot” and the Begg’s test. The fixed effects model was adopted in the initial calculation of odds ratio with corresponding 95% CIs. If there was a significant statistical heterogeneity among the studies, the random-effects model was applied for the analysis. By comparing the incidence of lymph node metastasis between high MALAT1 expression group and low MALAT1 expression group, we tried to make a thorough inquiry on the relationship between MALAT1 expression levels with lymph node metastasis.
Results
Studies characteristics
A total of five studies were identified as eligible according to the criteria for selection [20-24] (Figure 1). These studies included a total of 573 patients. The mean patient sample size was 114.6 (range 45 to 150). Four studies came from China and one study from America. All the research methods were qRT-PCR. Among the five studies, two focused on pancreatic cancer, one on clear cell renal cell carcinoma, and one on gastric cancer. All cancerous specimens were well preserved before RNA extraction. No patient received chemotherapy or radiotherapy before surgery. All the diagnoses of lymph node metastasis were based on pathology. QUADAS-2 system was to assess the qualities of included articles, and the assessment results turned out to be from moderate to high.
Figure 1.
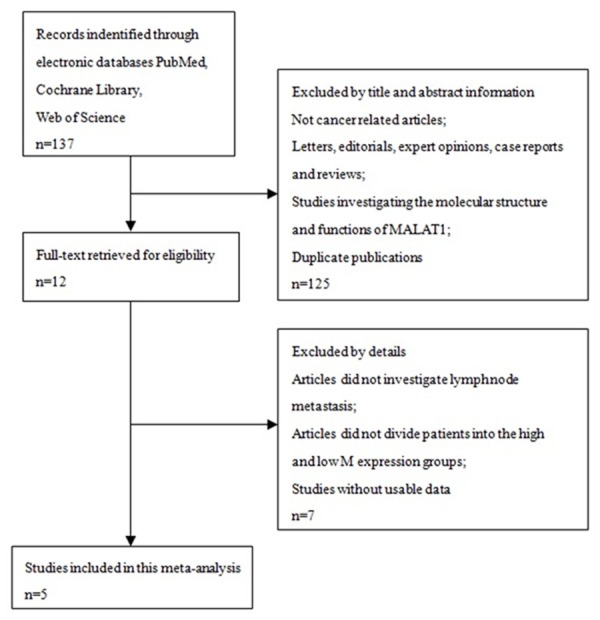
Flowchart presenting the steps of literature search and selection.
In the five studies, a total of four kinds of methods were used to divide high MALAT1 expression group and low MALAT1 expression group: (1) using qRT-PCR (quantitative real-time PCR) to measure the expression levels of MALAT1 in cancerous tissues (high MALAT1 group-MALAT1 expression ratio ≥ median value 6.23, low MALAT1 group-MALAT1 expression ratio < median value 6.23); (2) also using qRT-PCR to measure the expression levels of MALAT1 in cancerous tissues (high MALAT1 group-MALAT1 expression ratio mean value, low MALAT1 group-MALAT1 expression ratio < mean value; (3) using qRT-PCR to measure the expression levels of MALAT1 in cancerous and noncancerous tissues (MALAT1 expression value of 0.985 was used as a cutoff value based on ROC analyses with Youden’s index, and the expression levels of MALAT1 were defined as high (cutoff value ≥ 0.985) or low (cutoff value ≥ 0.985); (4) using qRT-PCR to measure the expression levels of MALAT1 in cancerous and noncancerous tissues (high MALAT1 group-MALAT1 expression level in cancerous tissues ≥ 2.26 fold the level in noncancerous tissues, low MALAT1 group-0MALAT1 expression level in cancerous < 2.26 fold the level in noncancerous tissues). The main characteristics of the eligible studies were summarized in Table 1.
Table 1.
Characteristics of the eligible studies in this meta-analysis
| First author | Year | Country | Cancer type | Total number | MALAT1 expression | MALAT1 Detection method | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| High | High with LNM | Low | Low with LNM | ||||||
| Er-Jun | 2014 | China | PC | 126 | 63 | 46 | 63 | 22 | qRT-PCR |
| Hai-min | 2014 | China | ccRCC | 106 | 22 | 13 | 60 | 6 | qRT-PCR |
| Yoshinaga | 2014 | USA | GC | 150 | 88 | 66 | 62 | 39 | qRT-PCR |
| Hong-Tu | 2014 | China | CC | 146 | 73 | 19 | 73 | 23 | qRT-PCR |
| Jiang-Hua | 2014 | China | PC | 45 | 26 | 15 | 19 | 8 | qRT-PCR |
LNM lymph node metastasis, PC pancreatic cancer, ccRCC clear cell renal cell carcinoma, GC gastric cancer, CC colorectal cancer.
Meta-analysis results
There was a significant heterogeneity among the studies (I2 = 82%, P = 0.0002), and then the random-effects model was adopted (Figure 2). The odds ratios, expressed as high MALAT1 expression group versus low MALAT1 expression group, was 2.64 (CI 95% 1.06-6.56, P = 0.04 random-effects model). Through comparing the incidence of lymph node metastasis between high MALAT1 expression group and low MALAT1 expression group, we found that there was a significant difference in the incidence of lymph node metastasis between the two groups. This result demonstrated that patients detected with high MALAT1 expression in cancerous tissues were more prone to lymph node metastasis.
Figure 2.
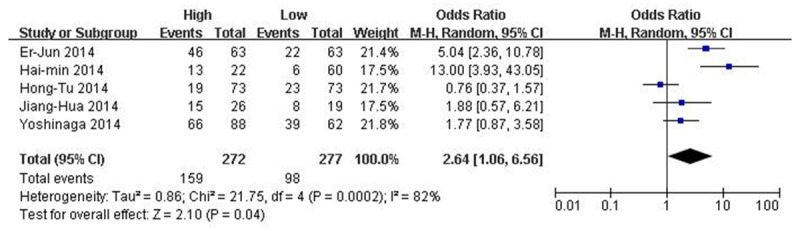
Forest plot for the association between MALAT1 expression levels with lymph node metastasis. Forest plot of OR was assessed for the difference in the incidence of lymph node metastasis between high MALAT1 expression group and low MALAT1 expression group (OR = 2.64, 95% CI = 1.06-6.56, P = 0.04 random-effects model).
Publication bias
We used a funnel plot to test for publication bias (Figure 3). No significant publication bias was observed. Bias was assessed statistically using the Begg’s test; still, the results of Begg’s test (Pr > |z| = 0.462) revealed no publication bias (P > 0.05).
Figure 3.
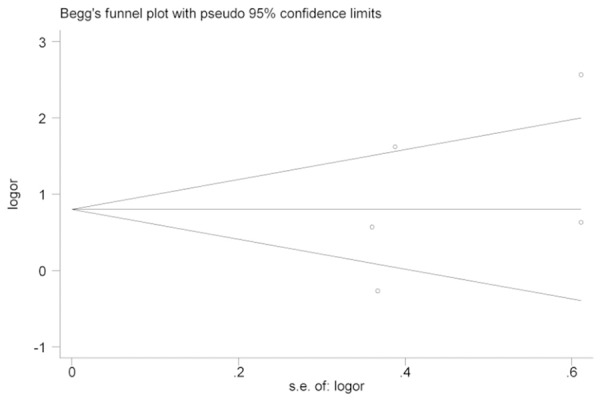
Funnel plot analysis of potential publication bias.
Discussion
LncRNAs, which are non-coding RNAs greater than 200 nt in length, were first described by Brockdorff et al [25]. Since then, especially in recent years, an increasing number of studies have reported that lncRNAs are involved in the progression of diverse diseases, especially cancers [26].
Emerging studies have proved that unique lncRNAs are associated with the development and progression of cancer through diverse pathways, including regulation of the cell cycle [27], metastasis, apoptosis, chemotherapy resistance, autophagy, and epigenetic regulation [28-32] in tumor tissues or malignant cell lines. A great number of lncRNAs can be commonly detected in various tumors, such as HOTAIR, CCAT1, ANRIL, SRHC, and MALAT1 [33-37]. Thus, lncRNAs have opened a new field of cancer genomics.
MALAT1 is a ~8000-nt long lncRNA, which is located on chromosome 11q13.1. It regulates the expression of metastasis-associated genes [38]. It also positively regulates cell motility via the transcriptional and/or post-transcriptional regulation of motility-related genes [39]. Research report indicated that MALAT1 regulated gene expression and also post-transcriptionally modified primary transcripts [40]. In addition, functional domain study also showed that 3’ end of MALAT1 played a crucial role in cell proliferation, invasion and migration [41]. MALAT1 was highly expressed in lung, pancreas and multiple types of cancers [42].
To date, there have been many studies focusing on the expression levels of MALAT1 in paired primary cancerous tissues and adjacent noncancerous tissues. Overexpression of MALAT1 was significantly associated with high-risk grade, metastasis and survival of cancer patients. In addition, knocking down MALAT1 obviously inhibited cell proliferation, migration, and invasion, and regulated MMPs, stemness, and EMT-associated genes expression [43]. However, the precise mechanism of how MALAT1 promotes tumor cell invasion and migration is unclear. Ren S et al. [44] found that silencing MALAT1 resulted in the inhibition of CRPC cell growth and metastasis in vivo. Previous studies have shown that the up-regulation of the expression levels of matrix metalloproteinases (MMPs) is a key step for promoting cell invasion [45]. Knockdown of MALAT1 inhibited the proliferation and invasion of human osteosarcoma cell and suppressed its metastasis in vitro and vivo, and the expression of matrix metallopeptidase 9 (MMP-9) was significantly inhibited in MALAT1-deleted cells. Moreover, Dong et al. [46] found that that MALAT1 might suppress the tumor growth and metastasis via PI3K/AKT signaling pathway [46].
This meta-analysis explored the relationship between MALAT1 expression levels with lymph node metastasis. The meta-analysis results showed that the incidence of lymph node metastasis in patients detected with high MALAT1 expression was higher than that in patients with low MALAT1 expression. Nevertheless, it is still necessary to conduct larger-size and better design studies to confirm our results. In addition, the major limitation of this meta-analysis was that patients included in our study were most of Asians, only one study was from America. Because of this, our finding may just represent patients from Asia. Another limitation was that there was a high heterogeneity among the studies (I2 = 82%, P = 0.0002), it may be due to the different methods used to divide high MALAT1 expression group and low MALAT1 expression group. Therefore, we suggest that setting up a unified criterion for the division of MALAT1 expression groups is important for further researches.
In conclusion, as a novel minimally invasive biomarker for lymph node metastasis, MALAT1 shows great potential in lymph node metastasis for cancer and warrants further study to explore its clinical application.
Acknowledgements
This article is supported by the National Natural Science Foundation of China (grants No. 81460069). The Science and Technology for Xinjiang Autonomous Region Project Plan (grant No. 201491176).
Disclosure of conflict of interest
None.
References
- 1.Datta K, Muders M, Zhang H, Tindall DJ. Mechanism of lymph node metastasis in prostate cancer. Future Oncol. 2010;6:823–836. doi: 10.2217/fon.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Nam H, Lee D. Exploring molecular links between lymph node invasion and cancer prognosis in human breast cancer. BMC Syst Biol. 2011;5(Suppl 2):S4. doi: 10.1186/1752-0509-5-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter CR, Sansom OJ, Evans TR, McKay CJ, Oien KA. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18:534–45. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 4.Ellsworth RE, Field LA, Love B, Kane JL, Hooke JA, Shriver CD. Differential gene expression in primary breast tumors associated with lymph node metastasis. Int J Breast Cancer. 2011;2011:142763. doi: 10.4061/2011/142763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt B, Wessels LF, Bosma AJ, Glas AM, Nuyten DS, He YD, Dai H, Peterse JL, van’t Veer LJ. No common denominator for breast cancer lymph node metastasis. Br J Cancer. 2005;93:924–32. doi: 10.1038/sj.bjc.6602794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 7.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T, Hammerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217inhibits proliferation, migration and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2014;290:3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2014;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 16.Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL, Huang JF. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314–21. doi: 10.3748/wjg.v20.i20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–74. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. Whole Transcriptome Sequencing Reveals Extensive Unspliced mRNA in Metastatic Castration-Resistant Prostate Cancer. Mol Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2014;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2014;36:2947–55. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 22.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–81. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–7. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 25.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 26.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–96. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–11. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 32.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long Noncoding RNA ANRIL Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol Cancer Ther. 2015;14:268–77. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 36.Zheng H, Yang S, Yang Y, Yuan SX, Wu FQ, Wang LL, Yan HL, Sun SH, Zhou WP. Epigenetically silenced long noncoding-SRHC promotes proliferation of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2014;141:1195–203. doi: 10.1007/s00432-014-1871-4. [DOI] [PubMed] [Google Scholar]
- 37.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–92. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 38.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility- related genes. FEBS Lett. 2010;584:4575–80. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–6. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in earlystage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 42.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long noncoding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 43.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–92. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 44.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, Xu C, Huang J, Zhao Y, Sun Y. Long Noncoding RNA MALAT-1 is a New Potential Therapeutic Target for Castration Resistant Prostate Cancer. J Urol. 2013;190:2278–87. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 46.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–86. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]


