Abstract
Objective: This study aimed to evaluate the relationships between members of APOBEC3 in tumor tissues and hepatocellular carcinoma (HCC) aggressiveness and prognosis. Methods: Using the expression profile GSE36376 from Gene Expression Omnibus (GEO), we compared APOBEC3 expression between tumor and non-tumor tissues, and correlated this with clinico-pathological features and outcomes of HCC patients. Results: A3B, A3D, A3F and A3H were overexpressed in HCC tumor tissues compared to non-tumor tissues (all P≤0.001). Cox regression shown that A3G was negatively associated with overall survival of HCC patients (HR=2.277, 95% CI=1.324-3.915, P=0.033), in contrast, A3C level in tumor tissues might play a positive role in HCC overall survival (HR=0.364, 95% CI=0.182-0.727, P=0.004). Interestingly, A3F contributed to a poor disease-free survival of HCC (HR=3.383, 95% CI=1.249-9.715, P=0.017), while A3H may be a positive factor associated with HCC disease-free survival (HR=0.25, 95% CI=0.098-0.636, P=0.004). Cirrhosis, tumor size and intrahepatic metastasis were associated with HCC poor disease-free survival (HR=1.838, 95% CI=1.308-2.583, P<0.001; HR= 1.095, 95% CI=1.042-1.15, P<0.001 and HR=3.669, 95% CI=2.447-5.5, P<0.001; respectively). Logistic regression analysis indicated that up-regulation of A3F in tumor tissues promoted HCC vascular invasion, intrahepatic metastasis and AFP elevation (all P<0.05). In contrast, A3H might decrease these risks (all P<0.05). Conclusions: APOBEC3G and APOBEC3F might be risk factors for HCC development and survival, while APOBEC3C and APOBEC3H should play positive roles in HCC aggressiveness and prognosis. Further investigation for APOBEC3 mechanisms are needed in the future.
Keywords: APOBEC3, APOBEC3F, APOBEC3H, APOBEC3G, APOBEC3C, liver cancer, hepatocellular carcinoma, survival, prognosis
Introduction
Among APOBEC family, APOBEC3 proteins play an important role in innate cellular immunity inhibiting retroviral infection, hepatitis B virus propagation, and the retrotransposition of endogenous elements. Members of this gene family like A3B, A3D, A3F, and A3G contain two conserved cytidine deaminase domains, instead of one in A3A, A3C, and A3H [1-4]. APOBEC3 proteins defend against retroviruses by deaminating cytosine residues to uracil, resulting in hypermutation and degradation of the viral genome. Moreover, the APOBEC3 family has been subjected to strong and continuing selective pressures at the amino acid level [5,6], which are associated with an increased risk of HCC [7,8].
The worldwide incidence of HCC has increased; presently it constitutes the fifth most common cancer, representing around 5% of all cancers [9]. HBV replication and genome mutations were both contributed to HCC development. Previous reports have reported gene expression patterns from the tumor tissues that can predict the recurrence of HCC, and these gene signatures were shown to predict HCC recurrence [10,11]. Recent studies indicate that a subclass of APOBEC cytidine deaminases may induce mutation clusters in human tumors. Two studies [12,13] independently reached the same conclusion that the A3B mutation signature is specifically enriched in six types of cancers, including those of the cervix, bladder, lung, head and neck, and breast, leading to a conclusion that A3B catalyzed genomic uracil lesions are responsible for a large proportion of both dispersed and clustered mutations in multiple distinct cancers. In another side, APOBEC3-mediated HBx mutants, especially the C-terminally truncated mutants, cause a gain of function that enhances the colony forming ability and proliferative capacity of neoplastic cells. Some of the APOBEC3 deaminases play a role in the carcinogenesis of HCC through the generation of HBx mutants, providing preneoplastic and neoplastic hepatocytes with a selective clonal growth advantage [14].
Considering APOBEC3 proteins’ critical roles in viral genome hypermutation and degradation and HCC carcinogenesis, a further analysis clarifying relationships between APOBEC3 and HCC prognosis is urgently needed. This study sets out to define relationships between APOBEC3 and outcomes in HCC patients after curative hepatectomy, in the hope that these data may provide novel biomarker candidates as well as useful insights into the pathogenesis and progression of HCC.
Methods
Ethics statement
Informed consent was obtained from each patient included in the study, and this study was approved by the institutional review board of Samsung Medical Center, Seoul, Korea, as reported by Lim HY et al [15].
Patients
240 tumor tissues containing no necrosis or hemorrhage were available from primary HCC patients who were treated with surgical resection or liver transplantation at Samsung Medical Center, Seoul, Korea, from July 2000 to May 2006. None of the patients received preoperative chemotherapy.
Source of data
Tumor tissues of HCC patients after curative hepatectomy in this study were profiled using Illumina HumanHT-12 V4.0 expression beadchip (Illumina Inc., San Diego, CA). The expression data was retrieved from Gene Expression Omnibus (GSE36376, http://www.ncbi.nlm.nih.gov/geo/) [15]. We restricted our search to genes within the APOBEC3 family, and seven APOBEC3s were included in our analysis.
End points
The overall survival was defined as time from surgery to the date of death or last follow-up. The disease-free survival was defined as time from surgery to the date of tumor recurrence or death. The censoring time was defined as the final documented date of no evidence of tumor recurrence by imaging.
Clinico-pathological features of HCC patients including vascular invasion, major portal vein invasion, intrahepatic metastasis, multicentric occurrence, American Joint Committee on Cancer (AJCC) stage, and nontumor liver pathology were all considered. As presented by Lim HY et al [15], vascular invasion was considered present when a neoplastic cell group was surrounded by at least one or more endothelial cells or the tunica media of the vessel. Intrahepatic metastasis and multicentric occurrence were defined according to the criteria of the Liver Cancer Study Group of Japan. Patient serum α-fetoprotein levels were evaluated and three phase dynamic computed tomography scans were performed at least once every 3 months after surgery until December 31, 2010. When tumor recurrence was suspected, precise diagnostic imaging was performed by magnetic resonance imaging.
Statistical analysis
Parametric data were expressed as mean ± standard deviation (SD) or median values. The Kolmogorov-Smirnov test was used as a test of normality. Student’s t-test was used to compare means for normally distributed continuous data, Mann-Whitney U-test was used for non-normally distributed continuous data, and the Chi-squared test or Fisher test was used for categorical variables. Factors associated with the outcomes and clinic-pathological features were assessed by univariate analysis and multivariate analysis separately using Cox and logistic regression. Only covariates significantly associated with outcomes at univariate analysis (two-sided P value<0.10) are shown and included in the multivariate model. Results were reported as hazard ratios (HR) or odd ratios (OR) with 95% confidence intervals (CI). The Kaplan-Meier method was used to compare overall survival between different groups, and the log-rank test was used to estimate the difference in survival. Statistical analyses were performed using PASW Statistics software version 18.0 from SPSS Inc. (Chicago, IL, USA). All statistical tests were two-tailed, and differences with P <0.05 were considered statistically significant.
Results
Patient characteristics
Among 240 HCC, Survival patients had smaller tumor size (P<0.001), lower risk of vascular invasion (P<0.001), major portal vein invasion (P=0.011), intrahepatic metastasis (P<0.001) and high level of body mass index (BMI, P=0.009). On the other side, HCC recurrence patients were younger (P=0.031) and more frequently infected by HBV (84%). Moreover, they also had bigger tumor size (P=0.029), more frequently coexisted with cirrhosis (P=0.01), vascular invasion (P<0.001), intrahepatic metastasis (P<0.001). And, more HCC patients had increased AFP over 200 ng/ml compared to those without recurrence (P=0.017). The AJCC stage and BCLC stage distribution were also presented in Table 1.
Table 1.
Baseline characteristics of hepatocellular carcinoma patients after curative hepatectomy
| Characteristics | Total (n=240) | Survival group (n=135) | P valueΔ | Recurrence group (n=150) | P value▲ |
|---|---|---|---|---|---|
| Male, n (%) | 199 (82.9) | 110 (81.5) | 0.503 | 121 (80.7) | 0.232 |
| Age, median (IQR), years | 53 (45-61) | 53 (45-60) | 0.698 | 52 (44-60) | 0.031 |
| Etiology, n (%) | 0.302 | 0.002 | |||
| HBV infection | 186 (77.5) | 108 (80.0) | 126 (84.0) | ||
| HCV infection | 20 (8.3) | 8 (5.9) | 12 (8.0) | ||
| Other | 34 (14.2) | 19 (14.1) | 12 (8.0) | ||
| Tumor size, median (IQR), cm | 3.7 (2.5-6.15) | 3.2 (2.2-5.0) | <0.001 | 3.95 (2.7-7.05) | 0.029 |
| Liver histology of non-tumor | 0.352 | 0.01 | |||
| Cirrhosis | 115 (47.9) | 57 (42.2) | 84 (56.0) | ||
| Chronic active hepatitis | 58 (24.2) | 36 (26.7) | 37 (24.7) | ||
| Chronic persistent hepatitis | 36 (15.0) | 23 (17.0) | 20 (13.3) | ||
| Reactive hepatitis | 11 (4.6) | 5 (3.7) | 5 (3.3) | ||
| Alcoholic hepatitis | 11 (4.6) | 7 (5.2) | 3 (2.0) | ||
| Vascular invasion, n (%) | 133 (55.4) | 60 (44.4) | <0.001 | 96 (64.0) | 0.001 |
| Major portal vein invasion, n (%) | 9 (3.8) | 1 (0.7) | 0.011 | 8 (5.3) | 0.159 |
| Intrahepatic metastasis, n (%) | 55 (22.9) | 13 (9.6) | <0.001 | 50 (33.3) | <0.001 |
| Multicentric occurrence, n (%) | 13 (5.4) | 7 (5.2) | 0.857 | 8 (5.3) | 0.941 |
| Direct invasion of adjacent organ, n (%) | 5 (2.1) | 2 (1.5) | 0.656 | 4 (2.7) | 0.653 |
| AJCC stage, n (%) | <0.001 | <0.001 | |||
| I | 102 (42.5) | 73 (54.1) | 52 (34.7) | ||
| II | 100 (41.7) | 53 (39.3) | 64 (42.7) | ||
| III | 33 (13.8) | 7 (5.2) | 30 (20.0) | ||
| IV | 5 (2.1) | 2 (1.5) | 4 (2.7) | ||
| BCLC stage, n (%) | <0.001 | 0.012 | |||
| A | 139 (57.9) | 96 (71.1) | 76 (50.7) | ||
| B | 91 (37.9) | 37 (27.4) | 66 (44.0) | ||
| C | 10 (4.2) | 2 (1.5) | 8 (5.3) | ||
| Alpha-fetoprotein >200 ng/ml, n (%) | 87 (36.3) | 45 (33.3) | 0.287 | 63 (42.0) | 0.017 |
| BMI, mean ± SD, kg/m2 | 24.2±2.8 | 24.6±2.9 | 0.009 | 24.0±2.7 | 0.234 |
compared to death patients;
compared to patient without recurrence.
APOBEC3 expression levels
As shown in Table 2, A3B, A3D, A3F and A3H were overexpressed in HCC tumor tissues compared to non-tumor tissues (all P≤0.001). No difference was found in A3A, A3C and A3G levels between tumor tissues and non-tumor tissues of HCC patients.
Table 2.
APOBEC3 expression levels between tumor tissues and non-tumor tissues of HCC patients (unit)
| APOBEC3 | Tumor tissues | Non-tumor tissues | P value |
|---|---|---|---|
| A3A, mean ± SD | 6.3±0.17 | 6.3±0.15 | 0.813 |
| A3B, median (IQR) | 5.95 (5.74-6.21) | 5.95 (5.64-5.85) | <0.001 |
| A3C, median (IQR) | 6.93 (6.84-7.14) | 6.91 (6.83-7.04) | 0.081 |
| A3D, median (IQR) | 6.34 (6.23-6.46) | 6.29 (6.2-6.37) | 0.001 |
| A3F, median (IQR) | 6.72 (6.63-6.87) | 6.65 (6.58-6.72) | <0.001 |
| A3G, median (IQR) | 6.66 (6.53-6.83) | 6.66 (6.54-6.81) | 0.508 |
| A3H, median (IQR) | 5.93 (5.83-6.05) | 5.88 (5.81-5.97) | <0.001 |
Factors associated with HCC overall survival
All characteristics and APOBEC3 included in the analysis are summarized in Table 3. Univariate analysis showed that A3B, A3C, A3D, A3F and A3G expression levels and patient gender were all factors associated with overall survival in HCC patients (all P<0.10). When these factors were evaluated by a multivariate model using forward selection, A3G was negatively associated with overall survival of HCC patients (HR=2.277, 95% CI=1.324-3.915, P=0.033), in contrast, A3C level in tumor tissues might play a positive role in HCC overall survival (HR=0.364, 95% CI=0.182-0.727, P=0.004).
Table 3.
Cox regression analysis of risk factors associated with overall survival of HCC patients
| Covariates | Univariate analysis, HR (95% CI) | P value | Multivariate Analysis, HR (95% CI) | P value |
|---|---|---|---|---|
| A3B, per increase of 1 unit | 1.508 (1.061-2.141) | 0.022 | ||
| A3C, per increase of 1 unit | 0.398 (0.205-0.773) | 0.007 | 0.364 (0.182-0.727) | 0.004 |
| A3D, per increase of 1 unit | 0.411 (0.214-0.788) | 0.007 | ||
| A3F, per increase of 1 unit | 0.403 (0.157-1.036) | 0.059 | ||
| A3G, per increase of 1 unit | 2.077 (1.241-3.476) | 0.005 | 2.277 (1.324-3.915) | 0.003 |
| Gender (male vs. female) | 1.557 (0.989-2.452) | 0.056 |
Despite a significant correlation between A3 (including A3C and A3G) expression and HCC overall survival both in univariate and multivariate regression analyses, the extent of the association by Kaplan-Meier event analysis with log rank method is relatively small (both P>0.05, Figure 1).
Figure 1.
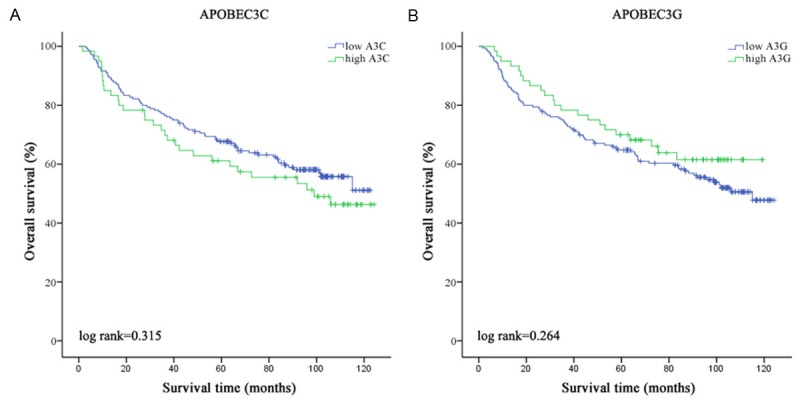
Analysis of overall survival of HCC patients. Overall survival analysis of (A) A3C and (B) A3G by Kaplan-Meier survival method.
Factors associated with HCC disease-free survival
Table 4 summarizes results from univariate and multivariate regression analyses. HBV infection, cirrhosis, age, tumor size, AJCC stage, BCLC stage, vascular invasion, intrahepatic metastasis, AFP and APOBEC3s, including A3F, A3G and A3H, were all associated with disease-free survival of HCC (all P<0.10). Furthermore, cirrhosis, tumor size and intrahepatic metastasis were clinico-pathological features associated with HCC poor disease-free survival (HR=1.838, 95% CI=1.308-2.583, P<0.001; HR= 1.095, 9% CI=1.042-1.15, P<0.001 and HR=3.669, 95% CI=2.447-5.5, P<0.001; respectively). Interestingly, A3F contributed to a poor disease-free survival of HCC (HR=3.383, 95% CI=1.249-9.715, P=0.017), while A3H may be a positive factor associated with HCC disease-free survival (HR=0.25, 95% CI=0.098-0.636, P=0.004).
Table 4.
Cox regression analysis of risk factors associated with disease free survival of HCC patients
| Covariates | Univariate analysis, HR (95% CI) | P value | Multivariate Analysis, HR (95% CI) | P value |
|---|---|---|---|---|
| A3F, per increase of 1 unit | 2.168 (0.868-5.417) | 0.098 | 3.383 (1.249-9.715) | 0.017 |
| A3G, per increase of 1 unit | 0.455 (0.237-0.874) | 0.018 | ||
| A3H, per increase of 1 unit | 0.273 (0.106-0.704) | 0.007 | 0.25 (0.098-0.636) | 0.004 |
| HBV infection (yes vs. no) | 1.782 (1.159-2.739) | 0.008 | ||
| Cirrhosis (yes vs. no) | 1.691 (1.224-2.336) | 0.001 | 1.838 (1.308-2.583) | <0.001 |
| Age, per increase of 1 year | 0.981 (0.966-0.996) | 0.016 | ||
| Tumor size, per increase of 1 cm | 1.129 (1.082-1.177) | <0.001 | 1.095 (1.042-1.15) | <0.001 |
| AJCC stage, per increase 1 stage | 1.944 (1.601-2.36) | <0.001 | ||
| BCLC stage, per increase 1 stage | 1.907 (1.467-2.479) | <0.001 | ||
| Vascular invasion (yes vs. no) | 2.324 (1.662-3.251) | <0.001 | ||
| Intrahepatic metastasis (yes vs. no) | 5.059 (3.528-7.254) | <0.001 | 3.669 (2.447-5.5) | <0.001 |
| AFP (>200 ng/ml vs. <200 ng/ml) | 1.66 (1.198-2.298) | 0.002 |
We performed a Kaplan-Meier event analysis grouping by A3F and A3H median expression identified to be significantly associated with disease-free survival presented above. As shown in Figure 2, A3F in tumor tissues might be a potential risk factor for HCC disease-free survival (P=0.065, Figure 2A), while A3H should play a protective role in HCC disease-free survival (P=0.008, Figure 2B).
Figure 2.
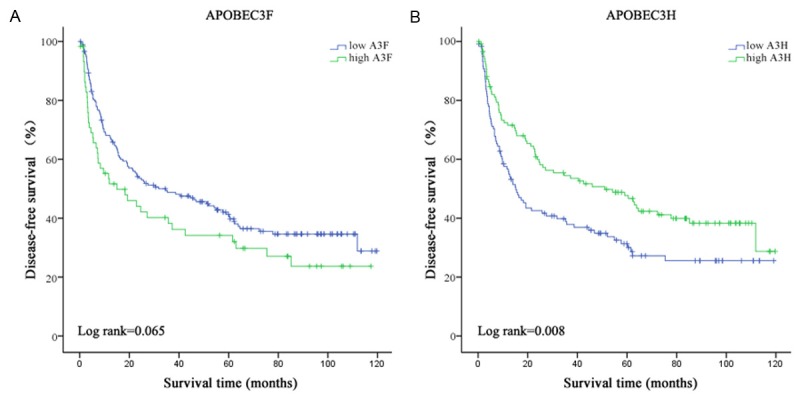
Analysis of disease-free survival of HCC patients. Disease-free survival analysis of (A) A3F and (B) A3H by Kaplan-Meier survival method.
Relationship between APOBEC3 and HCC clinico-pathological features
APOBEC3 in tumor tissues were related to vascular invasion, intrahepatic metastasis and AFP level over 200 ng/ml, which was shown in Table 5. A3F and A3H expression were all associated with HCC clinico-pathological features mentioned above in univariate analysis model (all P<0.10). When all APOBEC3 members were evaluated by a multivariate model using forward selection, overexpression of A3F was negatively related with the vascular invasion, intrahepatic metastasis and AFP level over 200 ng/ml (HR=5.379, 95% CI=1.111-26.039, P=0.037; HR=6.741, 95% CI=1.111-40.885, P=0.038 and HR=7.685, 95% CI=1.483-39.823, P=0.015, respectively). In contrast, A3H plays a positive role in the HCC clinico-pathological features above as shown in Table 5.
Table 5.
Relationship between APOBEC3 and HCC clinico-pathological characteristics by logistic regression analysis
| APOBEC3 | Vascular invasion | |||
|
| ||||
| Univariate analysis, OR (95% CI) | P value | Multivariate Analysis, OR (95% CI) | P value | |
|
| ||||
| A3F, per increase of 1 unit | 3.776 (0.818-17.435) | 0.089 | 5.379 (1.111-26.039) | 0.037 |
| A3H, per increase of 1 unit | 0.263 (0.069-1.009) | 0.051 | 0.197 (0.049-0.798) | 0.023 |
|
| ||||
| APOBEC3 | Intrahepatic metastasis | |||
|
| ||||
| Univariate analysis, OR (95% CI) | P value | Multivariate Analysis, OR (95% CI) | P value | |
|
| ||||
| A3F, per increase of 1 unit | 4.263 (0.758-23.989) | 0.1 | 6.741 (1.111-40.885) | 0.038 |
| A3H, per increase of 1 unit | 0.14 (0.022-0.885) | 0.037 | 0.101 (0.015-0.66) | 0.017 |
|
| ||||
| APOBEC3 | AFP over 200 ng/ml | |||
|
| ||||
| Univariate analysis, OR (95% CI) | P value | Multivariate Analysis, OR (95% CI) | P value | |
|
| ||||
| A3B, per increase of 1 unit | 0.473 (0.246-0.911) | 0.025 | 0.53 (0.277-1.014) | 0.055 |
| A3F, per increase of 1 unit | 5.897 (1.233-28.191) | 0.026 | 7.685 (1.483-39.823) | 0.015 |
| A3H, per increase of 1 unit | 0.226 (0.05-1.015) | 0.052 | 0.166 (0.034-0.81) | 0.026 |
Discussion
It is postulated that APOBEC-mediated mutagenesis of viral DNA may result in an increase in viral mutation load to a level that exceeds the threshold for viral viability [16,17]. Accordingly, induction of APOBEC family members in certain human tumors with an existing high mutation load may similarly increase mutation numbers to a point that exceeds tumor viability [18,19]. Given this hypothesis, it is of great importance to evaluate the relationship between APOBEC3 proteins and clinico-pathological features and prognosis of HCC patients.
As a member of the mammalian apolipoprotein B mRNA-editing enzyme catalytic polypeptide like family, A3G exhibits deoxycytidine deaminase activity to protect against viral infection [20,21]. However, a report by Noguchi et al [22] revealed that overexpression of A3G reduced the number of replicative intermediate of HBV and increased hypermutated genomes, suggesting that A3G might have a dual effect on HBV: induction of hypermutation and reduction of virus synthesis. Furthermore, Ding et al [23] elucidate that A3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. In our study, no A3G expression difference was found between HCC tumor tissues and non-tumor tissues, but, up-regulation of A3G in tumor tissues should be a risk factor contributing to poor overall survival of HCC. To our surprise, in contrast to the results obtained for A3G, A3C showed positive effect on HCC overall survival. To our known, the A3C protein produces hypermutated DNA molecules at high frequency in transfected HepG2 cells and the Huh7 hepatoma cell line [24]. Inducing mutations at a frequency several-fold lower than A3G, A3C is still capable of inhibiting viral infection and blocking viral DNA persistence in the target cells [25]. Unfortunately, Kaplan-Meier analysis did not show significant difference on HCC overall survival base on the median expression of both A3G and A3C in this study. Thus, whether A3G and A3C are truly associated with HCC overall survival still need further investigated. It has been somewhat unclear as to the precise mechanisms of A3G and A3C in HCC overall survival.
A report by Ying et al [26] revealed that interferon-α significantly up-regulated A3F in the HepG2 cell lines but less significantly in Huh7 cells at the same time point. In HIV infection, A3F is often coexpressed [27]. Also, A3F and A3G expressions are coordinately regulated in multiple liver cell lines, primary hepatocytes, and macrophages [26]. On the other hand, A3F mostly generates G-to-A mutations in HBV genome [16], which is an increased risk for persistent HBV infection and HCC development. Our analysis showed that A3F overexpression in tumor tissues is a risk factor for HCC disease-free survival, consistent with our previous result of A3G in HCC overall survival. On the contrary, A3H should be a protective factor in HCC disease-free survival. Kaplan-Meier test also indicated that A3H might more valuable than A3F in HCC disease-free survival. Similar to A3F and A3G, A3H had strong anti-viral activities, including retroviruses and retrotransposons. Previous studies [5,24] demonstrated that A3H mRNA has been detected in several human tissues, like peripheral blood mononuclear cells, liver. But, few data was available for A3H induced HBV genome mutations and HCC carcinogenesis. Compared to rhesus macaque, human A3H shows little activity against retroviruses and LINE-1 elements [5,28,29]. The lack of antiviral activity of human A3H correlates with its low steady-state expression at the protein level although mRNA levels [5]. However, our result showed that A3H was well expressed in HCC tumor tissues. Taken previous research together [30], A3H is very active against exogenous and endogenous retroviruses and the prolific primate non-LTR retroelement, LINE-1 based on its up-regulated expression. In this case, we assumed that antiviral activities of A3H in tumor tissues played more valuable role than genome mutations. Further investigation is essential for this point.
Consistent with previous reports [9,31,32], our analysis also indicated that clinico-pathological characteristics including cirrhosis, tumor size and intrahepatic metastasis contributed to poor disease-free survival of HCC. Therefore, we performed a further analysis on relationships between APOBEC3 proteins and HCC clinico-pathological features. In our logistic regression analysis, up-regulation of A3F in tumor tissues promoted HCC vascular invasion, intrahepatic metastasis and AFP elevation. In contrast, A3H might decrease the risk of vascular invasion, intrahepatic metastasis and AFP elevation in HCC patients. The precise mechanisms of A3F and A3H in HCC clinico-pathological features needed future research.
Taken together, APOBEC3 members played dual roles in HCC clinico-pathological characteristics and survival. Although this study based on national data bank, no direct first-hand data were available, our results presented clues about APOBEC3 in HCC carcinogenesis and tumor progression. Further research should be considered for APOBEC3 mechanisms in this population.
Acknowledgements
This work was supported by the National Science and Technology Major Projects of the Twelfth Five-year Plan (2012ZX10004301004). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;3:285–96. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 2.LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;6944:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 4.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;4:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;8:3853–62. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;9:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Gao CF. Hepatocellular carcinoma risk and hepatitis B virus mutations. Zhonghua Gan Zang Bing Za Zhi. 2013;10:788–92. [PubMed] [Google Scholar]
- 8.Liu S, Zhang H, Gu C, Yin J, He Y, Xie J. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;15:1066–82. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–85. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Ho MC, Lin JJ, Chen CN, Chen CC, Lee H, Yang CY. A gene expression profile for vascular invasion can predict the recurrence after resection of hepatocellular carcinoma: a microarray approach. Ann Surg Oncol. 2006;11:1474–84. doi: 10.1245/s10434-006-9057-1. [DOI] [PubMed] [Google Scholar]
- 11.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;5:1501–12. e2. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;9:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;9:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;6:1810–20. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- 15.Lim HY, Sohn I, Deng S, Lee J, Jung SH, Mao M. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol. 2013;12:3747–53. doi: 10.1245/s10434-013-3070-y. [DOI] [PubMed] [Google Scholar]
- 16.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;11:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 17.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;15:1392–6. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 18.Prindle MJ, Fox EJ, Loeb LA. The mutator phenotype in cancer: molecular mechanisms and targeting strategies. Curr Drug Targets. 2010;10:1296–303. doi: 10.2174/1389450111007011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuong KJ, Loeb LA. APOBEC3B mutagenesis in cancer. Nat Genet. 2013;9:964–5. doi: 10.1038/ng.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu YL, Greene WC. APOBEC3G: an intracellular centurion. Philos Trans R Soc Lond B Biol Sci. 2009;1517:689–703. doi: 10.1098/rstb.2008.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S. Dual effect of APOBEC3G on Hepatitis B virus. J Gen Virol. 2007;88:432–40. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- 23.Ding Q, Chang CJ, Xie X, Xia W, Yang JY, Wang SC. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J Clin Invest. 2011;11:4526–36. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köck J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol. 2008;89:1184–91. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- 25.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;6:1913–23. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying S, Zhang X, Sarkis PT, Xu R, Yu X. Cell-specific regulation of APOBEC3F by interferons. Acta Biochim Biophys Sin (Shanghai) 2007;4:297–304. doi: 10.1111/j.1745-7270.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- 27.Miyagi E, Brown CR, Opi S, Khan M, Goila-Gaur R, Kao S. Stably expressed APOBEC3F has negligible antiviral activity. J Virol. 2010;21:11067–75. doi: 10.1128/JVI.01249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;9:2955–64. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;24:13932–7. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;3:249–59. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truant S, Boleslawski E, Duhamel A, Bouras AF, Louvet A, Febvay C. Tumor size of hepatocellular carcinoma in noncirrhotic liver: a controversial predictive factor for outcome after resection. Eur J Surg Oncol. 2012;12:1189–96. doi: 10.1016/j.ejso.2012.07.112. [DOI] [PubMed] [Google Scholar]
- 32.Li SL, Su M, Peng T, Xiao KY, Shang LM, Xu BH. Clinicopathologic characteristics and prognoses for multicentric occurrence and intrahepatic metastasis in synchronous multinodular hepatocellular carcinoma patients. Asian Pac J Cancer Prev. 2013;1:217–23. doi: 10.7314/apjcp.2013.14.1.217. [DOI] [PubMed] [Google Scholar]


