Abstract
Asthma is a complex airways disease resulting from the input of both biological and environmental factors. Previous studies of single-nucleotide polymorphisms in toll-like receptor 4 (TLR4), which produces a protein involved in regulating T cell populations, have presented conflicting results regarding its role in asthma severity. In the current study, individuals with asthma were genotyped for variants of TLR4, and the genotypes were compared with asthma severity and T cell subpopulations. TLR4 rs11536879 (A>G) and rs1927907 (G>A) genotypes were determined in 350 asthma patients using TaqMan. Asthma severity was graded by clinical symptoms, and blood markers and lung function measures were also collected. T cell subpopulations were identified from peripheral blood by flow cytometry. No significant correlations were observed between genotypes at TLR4 rs11536879 or rs1927907 and eosinophil counts, total serum IgE, serum hypersensitive C-reactive protein, forced expiratory volume in 1 second (FEV1%), or FEV1/forced vital capacity (FVC) in asthma patients (P > 0.05). However, the GG genotype of rs1927907 was correlated with higher asthma severity (P < 0.05). No associations were detected between genotypes at rs11536879 or rs1927907 and CD4+CD25high regulatory T cell counts in peripheral blood from asthmatic patients (P > 0.05), but the rs1927907 genotype was associated with TLR4 expression on the surface of CD4+CD25high regulatory T cells (P < 0.05). Therefore, the TLR4 variant rs1927907 appears to be related to asthma severity and TLR4 expression on the surface of CD4+CD25high regulatory T cells, suggesting the potential influence of TLR4 on T cell population balances.
Keywords: Asthma, polymorphism, CD4+CD25high regulatory T cell, toll-like receptor 4
Introduction
Bronchial asthma (hereafter, asthma) is a common chronic inflammatory disorder of the airways that results from both genetic and environmental factors [1]. Importantly, asthma is also an autoimmune disease, wherein T lymphocytes control inflammatory responses. Imbalances in the Th1 and Th2 types of T helper cells (Th) contribute to the etiology of asthma [2]. The Th1/Th2 balance, as well as the functions of regulatory T cells, are regulated by toll-like receptors (TLRs), which recognize the microbial pathogen-associated molecular patterns in the environment and bridge the innate and acquired immune systems [3].
In particular, one family member, TLR4, is of interest in understanding the pathogenesis of asthma. TLR4 recognizes lipopolysaccharide (LPS) in the bacterial outer membrane; LPS plays an important role in regulating allergic inflammatory diseases of the airways, including asthma [6,7]. TLR4 fights bacterial infections by triggering innate and acquired immunity; however, suppressed TLR4 responses may lead to inflammatory responses [8]. TLR4 is activated following binding of LPS, and a series of downstream phosphorylation and dephosphorylation events eventually leads to the activation of transcription factors that regulate inflammatory factors including interferon, tumor necrosis factor, and interleukin; it also induces antigen-presenting cell maturation and promotes a Th0 to Th1 shift [9]. Indeed, activated TLR4 can directly or indirectly affect the function of regulatory T cells, thus influencing the Th1/Th2 imbalance [10] and reducing inflammatory responses [11].
Thus, TLR4 is believed to play an important role in the pathogenesis of asthma. However, investigations into whether polymorphisms in the TLR4 gene influence susceptibility to asthma have produced inconsistent findings [12-14]. To better understand the contribution of TLR4 to asthma pathogenesis, the current case-only study was designed to assess the correlation between TLR4 gene polymorphism and the severity and clinical indices of morbidity in asthma patients. Here, two single-nucleotide polymorphism (SNP) sites in TLR4 were genotyped in the patient population: rs11536879 and rs1927907.
Methods
Subjects
The study recruited 205 patients with clinically confirmed asthma from the Division of Respiratory Diseases of the First Affiliated Hospital of Zhengzhou University. There were 93 males and 112 females, mean age (38.1 ± 13.6) years. Of them, 176 cases were smokers and the remaining 29 cases were non-smokers. For asthma symptoms, 42 cases had intermittent episodes (Grade 1), 31 cases had mild persistence (Grade 2), 54 cases had moderate persistence (Grade 3), and the remaining 78 cases had severe persistence (Grade 4). A number of clinical indices were measured (see methods below): eosinophil count, (0.48 ± 0.25) × 106/mL; plasma log10IgE, 1.85 ± 0.56 IU/mL; hsCRP, 2.60 ± 1.17 mg/L; FEV1% 75.0 ± 11.7%; and FEV1/FVC 75.3 ± 10.6%, which occurred after inhalation of hormones. Diagnostic criteria met the 2003 Chinese guidelines for the prevention and management of bronchial asthma formulated by the Asthma Workgroup of the Chinese Society of Respiratory Diseases (CSRD), Chinese Medical Association [15]: 1) Patient complained of recurrent wheezing, shortness of breath, chest tightness and coughs, which were frequently associated with exposure to allergens, cold air, physical or chemical triggers, and with upper airway viral infections or physical exertion; 2) Scattered or diffuse wheezing, typically during the expiratory phase, heard over both lungs; 3) These signs and symptoms may resolve spontaneously or with treatment; 4) In patients with atypical clinical presentations (e.g., no symptoms or signs of wheezing), the diagnosis of asthma was made based on any of the following: (i) peak expiratory flow (PEF) with a diurnal variation ≥ 20% as determined by a mini peak flow meter; or (ii) bronchial provocation test with a change of ≥ 12% and ≥ 200 mL in forced expiratory volume in 1 second (FEV1); 5) Other causes of wheezing, shortness of breath, chest tightness, and coughs were excluded. Any subject who met criteria items 1 through 4 or criteria items 4 plus 5 was diagnosed with bronchial asthma. Patients were excluded from the study if affected by the following: other respiratory diseases, upper and lower respiratory tract infections in recent history, or chronic obstructive pulmonary disease; pregnancy; heart, liver, and kidney diseases; diabetes; cancer; recent surgery; or systemic inflammatory diseases. All subjects provided informed consent. This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, Henan Province, P. R. China).
Clinical measurements
Peripheral blood (5 mL) was drawn from each subject. Three mL of peripheral blood were centrifuged at 1300 × g for 10 minutes at 4°C. The plasma in the upper layer was collected and stored at -70°C; the middle layer of white blood cells was collected and stored at -20°C for genomic DNA extraction. The eosinophil count was measured with a fully-automatic hematology analyzer. The plasma IgE level was detected using Human IgG ELISA Quantitation Kit (Bethyl Laboratories, Montgomery, TX) according to manufacturer’s instructions; the minimum detection level was 6.51 IU/mL. The hypersensitive C-reactive protein (hs-CRP) was measured using methodology from a previous report [16].
The lung function was measured for each subject using a portable spirometer (Micro Medical, Chatham, Kent). Measurements included forced vital capacity (FVC) and forced expiratory volume in one second (FEV1); FEV1%, FVC%, and FEV1/FVC ratio were calculated; tests were performed 3 times to obtain the mean values.
Genotyping
Genomic DNA was extracted using QIAamp DNA Blood Mini kit (QIAGEN, Shanghai, China) as per the kit instructions. The concentration and purity of the extracted DNA were measured with a UV spectrophotometer. TaqMan SNP genotyping was used for genotyping two SNPs in TLR4, rs11536879 and rs1927907. For validation, 10% of the samples were randomly chosen to determine the call accuracy and consistency rates; the success rate was more than 96%, and the consistency rate was 100%.
T cell analysis
A flow cytometer (Becton Dickinson and Company, Franklin Lakes, NJ, USA) was used to detect T cells. A gate for CD4+CD25-cells was set according to the isotype control, while the one for CD25high cells was set according to the low-intensity CD25 on non-CD4+T cells. When the expression of CD25 cells was higher than that of their isotype control but lower than that of CD25high cells, then the cells were defined as CD25low. The specific methods were based on a previous report [17].
Statistical analysis
SPSS17.0 was used to analyze data. A chi-square goodness-of-fit test was applied to allele frequency to determine fit with Hardy-Weinberg equilibrium and to analyze asthma severity by genotype. Univariate analysis of variance was used to compare the clinical indices in patients with different genotypes; the SNK method was used to perform a pairwise comparison between groups. Measurement data are expressed as mean ± standard deviation (X̅ ± s). Tests were two-tailed, α was equal to 0.05, and P < 0.05 was accepted as statistically significant.
Results
Allele distribution in the asthma population
Genotyping results identified the genotypes and allele frequencies of all 205 asthma patients for both targeted SNPs of TLR4. For rs11536879 (A>G), 153 cases (74.5%) were homozygous for the A allele, 50 cases (24.4%) were heterozygous AG, and 2 cases (0.1%) were respectively homozygous for the G allele. For rs1927907 (G>A), 114 cases (55.6%) were homozygous for the G allele, 68 cases (33.2%) were heterozygous GA, and 23 cases (11.2%) were homozygous for the A allele. Allele frequencies for both rs11536879 and rs1927907 were consistent with Hardy-Weinberg equilibrium (P > 0.05); the minimum allelotype frequencies (MAFs) were 13.2% and 27.8%, respectively.
Genotypes, asthma severity, and clinical indices
The severity (by grade) of asthma symptoms was assessed by genotype for each SNP independently (Table 1). For SNP rs1927907 (G>A), patients with the GG genotype had more severe asthma than those with GA and AA genotypes (P < 0.05); in contrast, there was no significant correlation between genotype at rs11536879 and asthma severity (P > 0.05).
Table 1.
Associations of rs11536879 and rs1927907 genotype with asthma severity
| SNP genotype | Asthma severity, n (%) | |||
|---|---|---|---|---|
|
| ||||
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | |
| rs11536879* | ||||
| AA | 28 (18.3) | 25 (16.3) | 40 (26.1) | 60 (39.2) |
| AG | 11 (22.0) | 9 (18.0) | 14 (28.0) | 16 (32.0) |
| GG | 0 | 0 | 0 | 2 (100.0) |
| rs1927907# | ||||
| GG | 15 (12.9) | 20 (17.2) | 34 (29.3) | 45 (40.5) |
| GA | 16 (23.2) | 9 (13.0) | 16 (23.2) | 27 (40.6) |
| AA | 8 (40.0) | 5 (25.0) | 4 (20.0) | 6 (15.0) |
χ 2 = 4.187, P = 0.651;
χ 2 = 12.972, P = 0.043.
Other clinical measures of asthma were assessed for correlation with genotype (Table 2). No correlations were found between genotype at rs11536879 or rs1927907 and eosinophil count, total plasma IgE level, or hsCRP in peripheral blood, nor were there correlations between genotype and lung function (FEV1% and FEV1/FVC) (P > 0.05).
Table 2.
Associations of rs11536879 and rs1927907 genotype with asthma clinical indices
| SNP genotype | Eos (106/mL) | hsCRP (mg/L) | log10IgE (IU/mL) | FEV1% | FEV1/FVC |
|---|---|---|---|---|---|
| rs11536879 | |||||
| AA | 0.48 ± 0.25 | 2.59 ± 1.19 | 1.81 ± 0.57 | 75.3 ± 11.5 | 74.8 ± 10.5 |
| AG | 0.45 ± 0.25 | 2.64 ± 1.13 | 1.97 ± 0.50 | 74.3 ± 12.0 | 77.0 ± 10.8 |
| GG | 0.63 ± 0.27 | 3.00 ± 1.44 | 1.74 ± 1.05 | 67.6 ± 24.2 | 68.5 ± 3.9 |
| F | 0.699 | 0.146 | 1.627 | 0.525 | 1.248 |
| P | 0.498 | 0.864 | 0.199 | 0.592 | 0.289 |
| rs1927907 | |||||
| GG | 0.50 ± 0.25 | 2.56 ± 1.23 | 1.82 ± 0.58 | 74.7 ± 11.9 | 74.2 ± 10.6 |
| GA | 0.43 ± 0.26 | 2.59 ± 1.07 | 1.95 ± 0.52 | 75.6 ± 11.5 | 75.9 ± 10.8 |
| AA | 0.50 ± 0.22 | 2.88 ± 1.21 | 1.67 ± 0.52 | 74.3 ± 11.6 | 79.6 ± 8.2 |
| F | 2.095 | 0.621 | 2.319 | 0.176 | 2.435 |
| P | 0.126 | 0.538 | 0.101 | 0.839 | 0.090 |
Genotypes and CD4+CD25high regulatory T cells
The genotype at either rs11536879 or rs1927907 did not statistically affect the ratios of CD4+CD25high regulatory T cells to CD4+T cells in the peripheral blood (P > 0.05; Figure 1). However, for rs1927907, cases with the GG genotype had a significantly reduced ratio of TLR4+CD4+CD25high regulatory T cells to CD4+CD25high regulatory T cells than cases with GA and AA genotypes (P=0.001; Figure 2, right). For rs11536879, no statistically significant differences were observed for the ratio of TLR4+CD4+CD25high regulatory T cells to CD4+CD25high regulatory T cells (P > 0.05).
Figure 1.
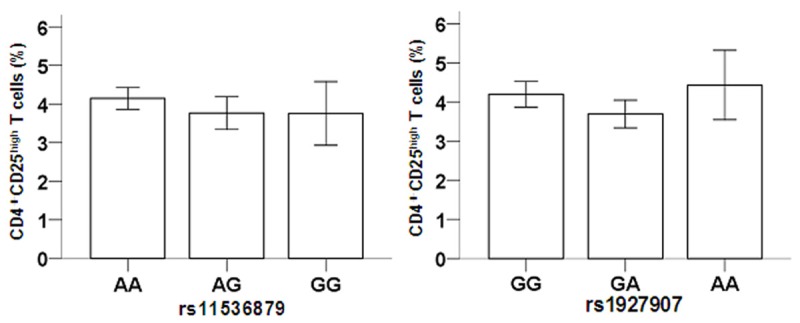
Association of TLR4 genotype with CD4+CD25high regulatory T cells. After T cell analysis, the ratios of CD4+CD25high regulatory T cells to CD4+T cells in the peripheral blood were computed with no statistical difference among genotypes at either rs11536879 or rs1927907.
Figure 2.
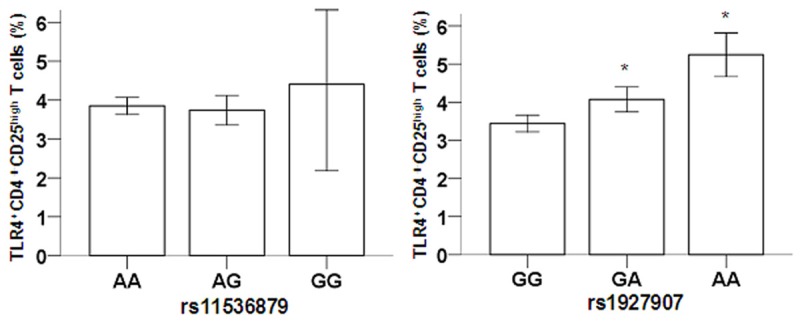
Association of TLR4 genotype with TLR4+CD4+CD25high regulatory T cells. After T cell analysis, the ratio of TLR4+CD4+CD25high regulatory T cells to CD4+CD25high regulatory T cells was computed, which was significantly reduced in cases with GG genotype than those with GA and AA genotypes in rs1927907 (*P < 0.05), but no statistically significant differences were observed among different genotypes for rs11536879.
Discussion
Previous studies of allelic variants of TLR4 reported conflicting results [12-14]. In 2011, Zhang et al. reported that the polymorphisms in TLR4 gene are associated with asthma severity but not susceptibility in a Chinese Han population, but their results revealed that the TT homozygote of rs1927914 was associated with lower forced expiratory volume in the first second (percent predicted) in asthmatic patients, and an evidently positive association was found between asthma severity and both the TT genotype of rs1927914 and the GG genotype of rs10983755 and rs1927907 (P < 0.05), indicating that the C allele of rs1927914 and the A allele of rs10983755 and rs1927907 have a protective effect on asthma severity [18]. Recently, the role of TLR4 Asp299Gly polymorphisms in the asthma susceptibility, progress, control levels and lung functions was investigated in Iranian patients, and the data demonstrated that TLR2 Arg753Gln and TLR4 Asp299Gly gene variants were not risk factors for asthma or its features in Iranian patients. Genetic complexity, ethnicity, influence of other genes or polymorphisms may overcome these polymorphisms in our asthmatics [19].
In the present study, the genotype at one of two SNPs analyzed, rs1927907, was significantly correlated with asthma severity as well as T cell subtypes present in the peripheral blood. To be more exact, patients with the GG genotype had more severe asthma than those with GA and AA genotypes. Neither SNP at rs11536879 nor rs1927907 was significantly associated with eosinophil count, total plasma IgE level, hsCRP, FEV1%, or FEV1/FVC ratio. That rs1927907 genotype was correlated with TLR4 expression on the surface of CD4+CD25high regulatory T cells suggests that the TLR4 polymorphism may influence the severity of asthma by changing the TLR4 expression on the surface of CD4+CD25high regulatory T cells. Meanwhile, the genetic variant of TLR4 may have an altered response to LPS, which could aggravate existing asthma symptoms, thus affecting the severity of asthma.
Disclosure of conflict of interest
None.
References
- 1.Martinez FD. Gene-Environment Interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc. 2007;4:26–31. doi: 10.1513/pats.200607-144JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson DS. The role of the T cell in asthma. J Allergy Clin Immunol. 2010;126:1081–1091. doi: 10.1016/j.jaci.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Drexler SK, Foxwell BM. The role of toll-like receptors in chronic inflammation. Int J Biochem Cell Biol. 2010;42:506–518. doi: 10.1016/j.biocel.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Chen K, Xiang Y, Yao X, Liu Y, Gong W, Yoshimura T, Wang JM. The active contribution of Toll-like receptors to allergic airway inflammation. Int Immunopharmacol. 2011;11:1391–1398. doi: 10.1016/j.intimp.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwamura C, Nakayama T. Toll-like receptors in the respiratory system: their roles in inflammation. Curr Allergy Asthma Rep. 2008;8:7–13. doi: 10.1007/s11882-008-0003-0. [DOI] [PubMed] [Google Scholar]
- 6.Dong L, Li H, Wang S, Li Y. Different doses of lipopolysaccharides regulate the lung inflammation of asthmatic mice via TLR4 pathway in alveolar macrophages. J Asthma. 2009;46:229–233. doi: 10.1080/02770900802610050. [DOI] [PubMed] [Google Scholar]
- 7.Peters M, Dudziak K, Stiehm M, Bufe A. T-cell polarization depends on concentration of the danger signal used to activate dendritic cells. Immunol Cell Biol. 2010;88:537–544. doi: 10.1038/icb.2010.3. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Takeda K. Toll-like receptor signaling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 10.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 11.Khatri SB, Holguin FC, Ryan PB, Mannino D, Erzurum SC, Teague WG. Association of ambient ozone exposure with airway inflammation and allergy in adults with asthma. J Asthma. 2009;46:777–785. [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh YY, Wan L, Chang CC, Tsai CH, Tsai FJ. STAT 2*C related genotypes and allele but not TLR4 and CD40 gene polymorphisms are associated with higher susceptibility for asthma. Int J Biol Sci. 2009;5:74–81. doi: 10.7150/ijbs.5.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reijmerink NE, Bottema RW, Kerkhof M, Gerritsen J, Stelma FF, Thijs C, van Schayck CP, Smit HA, Brunekreef B, Koppelman GH, Postma DS. TLR-related pathway analysis: novel gene-gene interactions in the development of asthma and atopy. Allergy. 2010;65:199–207. doi: 10.1111/j.1398-9995.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- 14.Lachheb J, Dhifallah IB, Chelbi H, Hamzaoui K, Hamzaoui A. Toll-like receptors and CD14 genes polymorphisms and susceptibility to asthma in Tunisian children. Tissue Antigens. 2008;71:417–425. doi: 10.1111/j.1399-0039.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 15.Asthma Workgroup of Chinese Society of Respiratory Diseases (CSRD), Chinese Medical Association. Chinese guideline for the prevention and management of bronchial asthma (the definition, diagnosis, treatment as well as education and management programs of bronchial asthma) Chinese Journal of Tuberculosis and Respiratory Diseases. 2003;26:132–138. [Google Scholar]
- 16.Qian FH, Zhang Q, Zhou LF, Liu H, Huang M, Zhang XL, Yin KS. High sensitivity C reactive protein: a predicative marker in severe asthma. Respirology. 2008;13:664–669. doi: 10.1111/j.1440-1843.2008.01314.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Qian FH, Liu H, Zhou LF, Huang M, Zhang XL, Yin KS. Expression of surface markers on peripheral CD4+CD25high T cells in patients with atopic asthma: role of inhaled corticosteroid. Chin Med J. 2008;121:205–212. [PubMed] [Google Scholar]
- 18.Zhang Q, Qian FH, Zhou LF, Wei GZ, Jin GF, Bai JL, Yin KS. Polymorphisms in toll-like receptor 4 gene are associated with asthma severity but not susceptibility in a Chinese Han population. J Investig Allergol Clin Immunol. 2011;21:370–377. [PubMed] [Google Scholar]
- 19.Bahrami H, Daneshmandi S, Heidarnazhad H, Pourfathollah AA. Lack of association between Toll like receptor-2 and Toll like receptor-4 gene polymorphisms and other feature in Iranian asthmatics patients. Iran J Allergy Asthma Immunol. 2015;14:48–54. [PubMed] [Google Scholar]


