Abstract
The ADP-ribosyltransferase 3 gene (ART3) has been reported to be associated with non-obstructive azoospermia (NOA) in the Japanese population. In this study, we aim to explore the possible association between the four single nucleotide polymorphisms (SNPs) (rs11097230, rs17001385, rs14773 and rs6836703) in ART3 gene and male infertility with spermatogenesis impairment in the Chinese population. The study population included 321 idiopathic infertile males with azoospermia or oligozoospermia and 250 fertile males. Four SNPs of ART3 gene were genotyped using the method of SNaPshot. The results showed that an SNP (rs6836703) in the intron11 region of ART3 gene is significantly associated with male infertility (odds ratio: 0.632, 95% confidence interval: 0.440-0.910). No significant associations were found between any of the other three variants (rs11097230, rs17001385 and rs14773) in ART3 gene and male infertility. SNP rs6836703 in ART3 gene may contribute to male infertility risk in the Chinese population.
Keywords: ART3, Chinese population, gene, genotype, male infertility, single nucleotide polymorphisms
Introduction
Infertility, a reproductive health problem of humans all around the world, has been estimated to affect 10-15% of childbearing couples and approximately 50% of it is due to male infertility [1]. Among those factors known to influence male infertility, genetics which influence a variety of physiological processes is perhaps one of the best known. Ferlin et al suggested that genetic abnormalities account for about 15%-30% of male factor infertility [2]. Many genetic factors have been implicated in spermatogenesis impairment that results in male infertility [2-5]. Spermatogenesis, a major function of mammalian testes, is a complex developmental process regulated by many genes [6]. The mutations of these genes may cause spermatogenesis impairment and male infertility [7]. Despite advances have been made in technologies and diagnostic methods of male infertility, the genetic causalities of most cases of male infertility are not known. Therefore, further research is needed to explore the genetic basis of reproductive failure while manage an infertile couple.
ADP-ribosyltransferase 3 (ART3), a gene located on chromosomes 4p14-p15, encodes an enzyme that influences the function of target proteins via mono-ADP-ribosylation which transfer the ADP-ribose moiety of NAD+ to a specific amino acid residue on the target protein while the nicotinamide moiety is released [8,9]. In the last decade, the roles of ART3 in male infertility have been received much attention. There is increasing evidence that ARTs play critical roles in human cells [10-12]. ART3 was reported to be specifically expressed in testis [9]. As the existence of ART3 protein was detected in testis where it may be involved in the regulation of spermatogenesis, a better understanding of the roles of ART3 in male infertility is necessary [11,13].
Recently, a study of Okada et al. provided evidence that single-nucleotide polymorphisms (SNPs) of the ART3 gene are significantly associated with non-obstructive azoospermia (NOA) in the Japanese population [13]. In that study, rs6836703, rs11097230, rs17001385 and rs14773 SNPs in ART3 gene have been shown association with non-obstructive azoospermia (NOA) in Japanese population [13]. However, the association between these polymorphisms and male infertility in the Chinese population is still unclear. In the present report, we investigated the possible association of the four SNPs (rs11097230, rs17001385, rs14773 and rs6836703) of ART3 gene with male infertility in the Chinese population.
Materials and methods
Subjects
This study was approved by the Medical Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University, and all the participants in our study gave informed consent for the genetic analyses. Blood samples were obtained from three hundred and twenty-one idiopathic non-obstructive infertile male patients with azoospermia or oligozoospermia (sperm concentration <20×106/ml) who were seeking infertility care at The First Affiliated Hospital of Wenzhou Medical University, Zhejiang province, China. Their diagnoses were based on at least two semen samples tests, physical examinations and hormone assays according to the WHO guidelines. Patients with recognizable chromosomal abnormalities and microdeletions in AZF region on Y chromosome of male infertility were excluded. Two hundred and fifty fertile men who had at least one offspring conceived without the use of assistant reproduction technique from the general population in area of East China (Zhejiang province) were selected as controls. Total genomic DNAs were extracted from blood samples using QIAamp DNA Blood Mini Kit (GIAGEN) according to the manufacturer’s instructions.
Genotyping
Four SNPs in a total of 571 samples were genotyped by SNaPshot Multiplex Kit (ABI) as previously described [14], with technical support from Center for Genetic & Genomic Analysis, Genesky Biotechnologies Inc. PCR and the specific single base extension primers are shown in Table 1. Briefly, assay for the four SNPs were performed in PCR amplification, and a product of each PCR-amplification was used as a template to perform single base extension reaction according to the manufacturer’s protocol (ABI Prism SNaPshot multiplex assay; Applied Biosystems). Finally, the SNaPshot products were analyzed by use of ABI 3730×l (96 Capillary) DNA analyzer (Applied Biosystems). The resulting data was analyzed by GeneMapper 4.1 (Applied Biosystems Co., Ltd., USA).
Table 1.
Primers sequence for genotyping of multiple single nucleotide polymorphisms using SNaPshot
| Primer sequence (5’-3’) | Primer for single base extension reaction (5’-3’) |
|---|---|
| rs11097230 | |
| F; TTGGGTGCCAGATGCTGTGA | T20CTTGAGCTTTGTTCTGGTACATAGTTA |
| R; 5’-GGGTACAGCCTCAGCAGTGAGAA-3’ | |
| rs17001385 | |
| F; TGGGAACAGGGAAGAATGTCGT | T38CTCTAATATACCAAACCAGTAAGCTAA |
| R; GTTATGCCCTGCCCAATATCC | |
| rs14773 | |
| F; TCCAGGTCCCAAAAGCCATC | T25ATACATCATTCCTTTTGATCTCCTGTG |
| R; GTGACTTTGGCCAACACGTAAAAA | |
| rs6836703 | |
| F; TGAGAACCACTGGTCTGTGGTAAC | T46AGTAGTGTCCCAGGCCTTCACAT |
| R; AGAAGGAAAATTTGTTATGAATTTAGTGTAGC |
Statistical analyses
The allele and genotype frequencies of the SNPs (rs11097230, rs17001385, rs14773 and rs6836703) in patients and controls were calculated and statistical analyzed using SNPStats software (http://bioinfo.iconcologia.net/SNPStats). The deviation from Hardy-Weinberg equilibrium was examined by the χ2 test. The differences in the distribution of genotypes and alleles between the patients group and the controls were calculated using two-tailed χ2 test. An unconditional logistic regression analysis was used to calculate the ORs (odds ratios) and 95% CIs (confidence intervals). The level of significance was set at P<0.05.
Results
Main effects on the risk of male infertility by individual polymorphisms
The polymorphism distributions of SNPs (rs11097230, rs17001385, rs14773 and rs6836703) in ART3 gene were investigated in 321 infertile and 250 control men. The distributions of allele and genotype of the four SNPs are listed in Table 2. ART3 haplotype frequencies with genotyping data of four tag SNPs were shown in Table 3. The distributions of genotypes of the four SNPs were in accordance with the Hardy-Weinberg equilibrium both in controls and patients.
Table 2.
Genotype and allele distribution of ART gene SNPs in infertile patients and fertile controls
| SNP | Genotype | All subjects (N=571) | Control (N=250) | Patients (N=321) | P Value | OR (95% CI) |
|---|---|---|---|---|---|---|
| rs11097230 | AA | 195 (34.2%) | 90 (36%) | 105(32.7%) | -- | -- |
| AG | 276 (48.3%) | 120 (48%) | 156 (48.6%) | 0.565 | 1.134 (0.888-1.448) | |
| GG | 100 (17.5%) | 40 (16%) | 60 (18.7%) | 0.314 | 1.114 (0.771-1.611) | |
| A allele | 666 (58.3%) | 300 (60%) | 366 (57%) | -- | -- | |
| G allele | 476 (41.7%) | 200 (40%) | 276 (43%) | 0.309 | 1.131 (0.892-1.434) | |
| rs17001385 | CC | 204 (35.7%) | 96 (38.4%) | 108 (33.6%) | -- | -- |
| CG | 275 (48.2%) | 119 (47.6%) | 156 (48.6%) | 0.410 | 1.165 (0.810-1.677) | |
| GG | 92 (16.1%) | 35 (14%) | 57 (17.8%) | 0.148 | 1.203 (0.936-1.547) | |
| C allele | 679 (59.5%) | 307 (61.4%) | 372 (57.9%) | -- | -- | |
| G allele | 463 (40.5%) | 193 (38.6%) | 270 (42.1%) | 0.238 | 1.155 (0.909-1.466) | |
| rs6836703 | GG | 222 (38.9%) | 84 (33.6%) | 138 (42.9%) | -- | -- |
| AG | 253 (43.8%) | 128 (51.2%) | 133 (41.4%) | 0.013 | 0.632 (0.440-0.910) | |
| AA | 88 (15.4%) | 38 (15.2%) | 50 (15.57%) | 0.385 | 0.895 (0.696-1.150) | |
| G allele | 705 (61.8%) | 296 (59.2%) | 409 (63.7%) | -- | -- | |
| A allele | 437 (38.3%) | 204 (40.8%) | 233 (36.3%) | 0.120 | 0.827 (0.650-1.051) | |
| rs14773 | AA | 167 (29.2%) | 69 (27.6%) | 98 (30.5%) | -- | -- |
| AC | 300 (52.5%) | 134 (53.6%) | 166 (51.7%) | 0.484 | 0.872 (0.595-1.279) | |
| CC | 104 (18.2%) | 47 (18.8%) | 57 (17.8%) | 0.531 | 0.924 (0.722-1.183) | |
| A allele | 634 (55.5%) | 272 (54.4%) | 362 (56.4%) | -- | -- | |
| C allele | 508 (44.5%) | 228 (45.6%) | 280 (43.6%) | 0.503 | 0.923 (0.729-1.167) |
Values are frequency (no. of individuals). The number of alleles is based on the genotype. The bold entries emphasize that P value <0.05.
Table 3.
Estimated haplotype frequencies and the association analysis with male infertility
| Haplotype frequencies estimation (n=571) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Haplotype | ART3-SNP# | Frequence | P value | OR (95% CI) | |||||
|
| |||||||||
| 1 | 2 | 3 | 4 | Controls | Patients | Total | |||
| H1 | G | G | G | A | 38.06% | 40.06% | 39.13% | -- | -- |
| H2 | A | C | A | C | 39.60% | 38.25% | 38.88% | 0.503 | 0.918 (0.706-1.193) |
| H3 | A | C | G | A | 15.94% | 15.21% | 15.53% | 0.578 | 0.952 (0.800-1.133) |
| H4 | A | C | G | C | 4.14% | 3.64% | 3.89% | 0.550 | 0.942 (0.766-1.157) |
| H5 | G | G | G | C | 1.03% | 1.06% | 1.05% | 0.927 | 0.994 (0.746-1.326) |
| Rare | * | * | * | * | 1.23% | 1.78% | 1.52% | 0.243 | 1.134 (0.915-1.407) |
The expectation-maximization (EM) algorithm (Excoffier and Slatkin, 1995) was used to infer ART3 haplotype frequencies with genotyping data of four tag SNPs, ART3-SNP1 (rs11097230), SNP2 (rs17001385), SNP3 (rs6836703) and SNP4 (rs14773).
SNP1, SNP2, SNP3 and SNP4.
refers to any haplotype except those listed in this table.
As shown in Table 2, there were no significant differences in the frequencies of allele and genotype of SNPs rs11097230, rs17001385 and rs14773 between patients and controls. However, the frequency of genotype AG (41.4% vs. 51.2%, P=0.013, OR: 0.632, 95% CI: 0.440-0.910) was significantly lower in patients than those in controls at SNP rs6836703 locus.
Pairwise linkage disequilibrium
Pairwise LD (linkage disequilibrium) between the four SNPs in the ART3 gene was calculated for all the participants in this study. The result of linkage disequilibrium analysis which indicated that most of the four SNPs were in nearly complete LD evaluated with D’ statistic (|D’|>0.75) in both patients and controls was shown in Figure 1 (D: the deviation between the observed haplotype frequency and the expected frequency, D’: the proportion of the maximum value of D, R: the correlation coefficient between alleles.). We found strong LD between several pairs of markers in the ART3 gene such as SNP1/SNP2 (D’=1.000), SNP1/SNP3 (D’=0.956), SNP1/SNP4 (D’=0.917), SNP2/SNP3 (D’=0.978), SNP2/SNP4 (D’=0.946), SNP3/SNP4 (D’=0.969).
Figure 1.
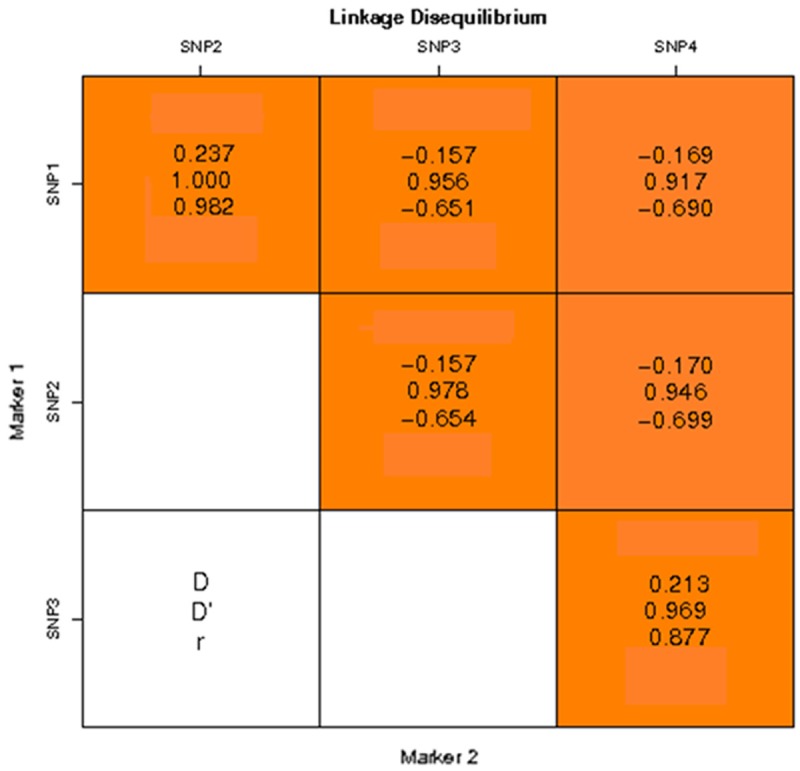
Linkage disequilibrium analysis of SNPs of ART3. D: the deviation between the observed haplotype frequency and the expected frequency, D’: the proportion of the maximum value of D, R: the correlation coefficient between alleles.
Haplotype analyses
For the haplotype-based association study, the four SNPs (rs11097230, rs17001385, rs14773, and rs6836703) as tag SNPs in ART3 were analyzed. Haplotype frequencies were inferred using an expectation-maximization (EM) algorithm. After excluding rare haplotypes (frequency <0.01), association of ART3 haplotypes with male infertility was examined in 321 cases and 250 controls. All of the 6 haplotypes showed no significant association with male infertility in frequencies between patients and controls (Table 3).
Discussion
In recent years, the association of ART3 gene with human diseases has been investigated. Mounting evidence suggests that ADP-ribosyltransferases (ARTs) activity in the extracellular compartment may provide sophisticated regulatory mechanisms for cell communication which imply it as new candidate for drug targeting [15-20]. In other studies, the expression of ARTs were investigated in human and animal tissues at the RNA level [10,12,21,22]. The major function of ARTs is Mono-ADP-ribosylation which transfers the ADP-ribose moiety of NAD+ onto a specific amino acid residue on the target protein. Until nowadays a small group of mammalian ARTs named ART1 to ART5 has been cloned and characterized but the biological role of each human ARTs in contrast to mouse ARTs is poorly defined [12,23]. Though a few studies have found out that the human neutrophil peptide1 (HNP-1) and the platelet-derived growth factor-BB (PDGF-BB) are the target proteins of ART1, the biological function of the remaining human ART family members is almost unknown [21,24]. A study of Friedrich et al. showed that ART3 expression in human cells appears to be governed by a combination of differential splicing and tissue-preferential use of two alternative promoters [20]. Moreover, ART3 was reported to be specifically expressed in testis [11]. And a Japanese study provided the evidence that SNPs of the ART3 gene were significantly associated with non-obstructive azoospermia (NOA) in Japanese population [13]. And in that study the most significant association was observed with SNP (rs6836703) in intron 11 of ART3. Moreover ART3 as a member of the ARTs family genes has conserved exon and intron structures and its protein is specifically expressed in spermatocytes but not in spermatozoa, spermatogonia and spermatids [11]. In view of the roles of ART3 in spermatocytes, it is likely that ART3 gene polymorphisms could be a risk factor for male infertility.
In the present study, the possible associations between SNPs (rs11097230, rs17001385, rs14773 and rs6836703) in ART3 gene and male infertility were investigated in Chinese infertile patients and fertile controls. As a result, analysis of the SNP polymorphisms in ART3 revealed no significant differences in the frequencies of allele and genotype of SNPs (rs11097230, rs17001385 and rs14773) between patients and controls, which indicated that those SNPs in ART3 gene are not associated with spermatogenesis impairment in the Chinese population. However, the frequency of genotype AG was significantly changed in patients compared with controls at SNP ra6836703 locus. The frequency of genotype AG was significantly lower in patients than those in controls, suggesting that genotype AG was associated with male infertility and it may have some protection effect from spermatogenesis impairment (OR=0.632). These findings is not coincide with the study of Okada et al. which provided evidence that rs6836703, rs11097230, rs17001385 and rs14773 SNPs in ART3 gene are significantly associated with non-obstructive azoospermia (NOA) in the Japanese population. Thus we conjecture that the effects of SNPs in ART3 maybe different in different race. In haplotype-based association study of ART3 gene, no significant difference in frequencies were found between patients and controls, which indicated that those haplotypes in ART3 gene are not associated with male infertility. We previously described that SNP (rs6836703) is in the intron11 region of ART3 gene, and as we all know mutations in introns cannot directly affect protein structure. But these mutations can prevent the production of the messenger RNA. So we infer that the polymorphism of SNP rs6836703 in ART3 gene may block the production of the mRNA by inhibiting the splicing together of exons. As a result, we indicated that the polymorphism of SNP rs6836703 in ART3 gene may modify the susceptibility to male infertility in the present study.
In conclusion, the results presented here provide new insights into the role of ART3 in association with male infertility and its possibility to modify the susceptibility to male infertility in the Chinese population. Our newly gained knowledge about the ART3 gene polymorphism and its association with male infertility will guide our attempts in finding out this enigmatic underlying mechanism. However, the ART3 protein expression and its function and molecular epidemiological studies were not included in this study. Considering that the biological function of ART3 is almost unknown, lots of studies are needed in the future.
Acknowledgements
This work was supported by Wenzhou Science and Technology Bureau Program (Y20140728).
Disclosure of conflict of interest
None.
References
- 1.De Kretser DM, Baker HW. Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metab. 1999;84:3443–3450. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 2.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14:734–745. doi: 10.1016/s1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 3.Cram DS, O’Bryan MK, de Kretser DM. Male infertility genetics--the future. J Androl. 2001;22:738–746. [PubMed] [Google Scholar]
- 4.Toshimori K, Ito C, Maekawa M, Toyama Y, Suzuki-Toyota F, Saxena DK. Impairment of spermatogenesis leading to infertility. Anat Sci Int. 2004;79:101–111. doi: 10.1111/j.1447-073x.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 5.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14:40–48. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seshagiri PB. Molecular insights into the causes of male infertility. J Biosci. 2001;26:429–435. doi: 10.1007/BF02704745. [DOI] [PubMed] [Google Scholar]
- 7.Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update. 2009;15:623–637. doi: 10.1093/humupd/dmp023. [DOI] [PubMed] [Google Scholar]
- 8.Bourne HR. ADP-Ribosylating Toxins and G Proteins. Insights into Signal Transduction. Joel Moss and Martha Vaughan, Eds. American Society for Microbiology, Washington, DC, 1990. xviii, 567 pp., illus. $79; to ASM members, $69. Science. 1990;250:841–842. doi: 10.1126/science.250.4982.841-a. [DOI] [PubMed] [Google Scholar]
- 9.Koch-Nolte F, Haag F, Braren R, Kuhl M, Hoovers J, Balasubramanian S, Bazan F, Thiele HG. Two novel human members of an emerging mammalian gene family related to mono-ADP-ribosylating bacterial toxins. Genomics. 1997;39:370–376. doi: 10.1006/geno.1996.4520. [DOI] [PubMed] [Google Scholar]
- 10.Grahnert A, Friedrich M, Pfister M, Haag F, Koch-Nolte F, Hauschildt S. Mono-ADP-ribosyltransferases in human monocytes: regulation by lipopolysaccharide. Biochem J. 2002;362:717–723. doi: 10.1042/0264-6021:3620717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich M, Grahnert A, Paasch U, Tannapfel A, Koch-Nolte F, Hauschildt S. Expression of toxin-related human mono-ADP-ribosyltransferase 3 in human testes. Asian J Androl. 2006;8:281–287. doi: 10.1111/j.1745-7262.2006.00125.x. [DOI] [PubMed] [Google Scholar]
- 12.Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11:1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada H, Tajima A, Shichiri K, Tanaka A, Tanaka K, Inoue I. Genome-wide expression of azoospermia testes demonstrates a specific profile and implicates ART3 in genetic susceptibility. PLoS Genet. 2008;4:e26. doi: 10.1371/journal.pgen.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni W, Li H, Wu A, Zhang P, Yang H, Yang X, Huang X, Jiang L. Lack of association between genetic polymorphisms in three folate-related enzyme genes and male infertility in the Chinese population. J Assist Reprod Genet. 2015;32:369–74. doi: 10.1007/s10815-014-0423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- 16.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly (ADP-ribosyl) ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZX, Yu Y, Dennert G. A cell surface ADP-ribosyltransferase modulates T cell receptor association and signaling. J Biol Chem. 1999;274:17399–17401. doi: 10.1074/jbc.274.25.17399. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto S, Azhipa O, Yu Y, Russo E, Dennert G. Expression of ADP-ribosyltransferase on normal T lymphocytes and effects of nicotinamide adenine dinucleotide on their function. J Immunol. 1998;160:4190–4198. [PubMed] [Google Scholar]
- 19.Liu ZX, Azhipa O, Okamoto S, Govindarajan S, Dennert G. Extracellular nicotinamide adenine dinucleotide induces t cell apoptosis in vivo and in vitro. J Immunol. 2001;167:4942–4947. doi: 10.4049/jimmunol.167.9.4942. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich M, Grahnert A, Klein C, Tschop K, Engeland K, Hauschildt S. Genomic organization and expression of the human mono-ADP-ribosyltransferase ART3 gene. Biochim Biophys Acta. 2006;1759:270–280. doi: 10.1016/j.bbaexp.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci U S A. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glowacki G, Braren R, Cetkovic-Cvrlje M, Leiter EH, Haag F, Koch-Nolte F. Structure, chromosomal localization, and expression of the gene for mouse ecto-mono(ADP-ribosyl)transferase ART5. Gene. 2001;275:267–277. doi: 10.1016/s0378-1119(01)00608-4. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki IJ, Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J Biol Chem. 1998;273:23617–23620. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 24.Saxty BA, Yadollahi-Farsani M, Upton PD, Johnstone SR, MacDermot J. Inactivation of platelet-derived growth factor-BB following modification by ADP-ribosyltransferase. Br J Pharmacol. 2001;133:1219–1226. doi: 10.1038/sj.bjp.0704187. [DOI] [PMC free article] [PubMed] [Google Scholar]


