Abstract
Neutrophils play a key role in the control of Burkholderia pseudomallei, the pathogen that causes melioidosis. Here, we show that survival of intracellular B. pseudomallei was significantly increased in the presence of 3-methyladenine or lysosomal cathepsin inhibitors. The LC3-flux was increased in B. pseudomallei-infected neutrophils. Concordant with this result, confocal microscopy analyses using anti-LC3 antibodies revealed that B. pseudomallei-containing phagosomes partially overlapped with LC3-positive signal at 3 and 6 h postinfection. Electron microscopic analyses of B. pseudomallei-infected neutrophils at 3 h revealed B. pseudomallei-containing phagosomes that occasionally fused with phagophores or autophagosomes. Following infection with a B. pseudomallei mutant lacking the Burkholderia secretion apparatus Bsa Type III secretion system, neither this characteristic structure nor bacterial escape into the cytosol were observed. These findings indicate that human neutrophils are able to recruit autophagic machinery adjacent to B. pseudomallei-containing phagosomes in a Type III secretion system-dependent manner.
Keywords: autophagy, Burkholderia pseudomallei, LC3-II, melioidosis, neutrophils, T3SS
Abbreviations: 3MA, methyladenine; Bp, Burkholderia pseudomallei; Bsa, Burkholderia secretion apparatus; KM, kanamycin; LAMP1, lysosomal-associated membrane protein 1; LC3-I, unlipidated form of LC3; LC3-II, LC3-phospholipid conjugated and phagophore or autophagosome-associated form of LC3; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; MOI, multiplicity of infection; NET, Neutrophil Extracellular Taps; p.i., postinfection; T3SS, Type III secretion system; WT, wild type
Introduction
Neutrophils are highly specialized effector cells of the innate immune system, which are involved in host inflammatory responses and immune surveillance. They play a key role in controlling bacterial infections, including those caused by Burkholderia pseudomallei, a gram-negative bacterium that causes melioidosis, a serious invasive disease in humans and animals. Melioidosis is endemic in Northern Australia and Southeast Asia, especially in the northeast part of Thailand. The mortality rate from melioidosis can be as high as 50%; when associated with septic shock, it is close to 90%.1,2 Depletion of neutrophils renders mice exquisitely susceptible to experimental B. pseudomallei infection,3 and defects in neutrophil function are believed to underlie the elevated risk of melioidosis in humans with diabetes mellitus.4
B. pseudomallei is a facultative intracellular pathogen that can invade both phagocytic and nonphagocytic cells.5 Following internalization into epithelial cells and macrophages, B. pseudomallei can escape from the phagosomes into the cytosol in a Bsa Type III secretion system (T3SS)-dependent manner. In J774 murine macrophage-like cells, bacterial escape into the cytosolis completed within after 3 h of infection, which allows the bacteria to circumvent oxidative antimicrobial agent and gain replicative niche.6 Once migrated into the cytosol, B. pseudomallei can replicate and form actin comet tail at one pole of the bacterium. Such F-actin polymerization-driven motility is mediated by the bacterial surface exposed BimA protein7 and facilitates intercellular spreading of B. pseudomallei into neighboring cells, leading to formation of multinucleated giant cells, which have been observed in both cultured cells and tissues from infected patients.8
Recently, we reported that neutrophils could kill more than 90% of intracellular B. pseudomallei, and that neutrophils from patients with diabetes mellitus are impaired in phagocytosis, migration, apoptosis,4 and production of neutrophil extracellular traps (NETs) in response to B. pseudomallei infection.9 Elucidation of the intracellular life cycle of B. pseudomallei in neutrophils and the response of host cells to B. pseudomallei infection is therefore essential to our better understanding of the basis of pathogenesis and protection during melioidosis. However, whether B. pseudomallei escapes into the cytoplasm following phagocytosis by primary human neutrophils, and how the bacteria are eliminated, remains unclear.
Autophagy is a cellular activity that acts as an autonomous defense against intracellular bacteria, such as Group A Streptococcus,10 Salmonella,10,11 Shigella12 and Listeria.13 It has been reported that B. pseudomallei can induce autophagy and be engulfed by LC3-associated phagosomes in a mouse macrophage cell line,14 and partially evade killing in these phagosomes by producing BopA, a putative effector protein secreted by the Bsa T3SS.15 The role of autophagy in the clearance of bacterial pathogens by neutrophils has received relatively little study. In 1984, Rikihisa reported that rickettsiae induce the rapid formation of autophagosomes in guinea pig peritoneal neutrophils.16 Mice with ATG5-deficient neutrophils show increased susceptibility to infection with Listeria monocytogenes, Toxoplasma gondii, and uropathogenic Escherichia coli.17 Narni-Mancinelli et al. report that cytolytic memory T lymphocytes can enhance the functional pathogen-killing capacities of neutrophils by inducing autophagy.18 Remijsen et al. report that autophagy plays an essential role in NET formation in neutrophils that can trap and degrade microbes.19 Given the key role of neutrophils in resistance to B. pseudomallei infection, we investigated whether autophagy plays a role in intracellular killing of B. pseudomallei in human neutrophils ex vivo. We found that the autophagic pathway contributes to killing of B. pseudomallei in human neutrophils that possess a characteristic membranous structure associated with phagophore-like structures, in a Bsa T3SS-dependent manner.
Results
Inhibition of autophagy enhances survival of B. pseudomallei in human neutrophils
To investigate whether autophagy plays a role in bacterial killing in human neutrophils, we first analyzed the survivability of the full-genome-sequenced prototype B. pseudomallei strain K96243 in primary human neutrophils ex vivo in the presence or absence of the autophagy inhibitor, 3-methyladenine (3MA). When neutrophils were infected with B. pseudomallei in the absence of 3MA, the majority of intracellular B. pseudomallei were killed in a time-dependent manner, as previously reported.4 Upon treatment with 3MA, the number of intracellular bacteria within neutrophils at 3 and 6 h p.i was about 10-fold higher than that in the absence of 3MA (Fig. 1; P ≤ 0.05). Furthermore, upon treatment with E64d and pepstatin A, inhibitors of lysosomal hydrolases, the number of intracellular bacteria was also increased to the same level of 3MA-treated cells (Fig. 1), suggesting that a 3MA-sensitive(PtdIns3P-dependent) and lysosomal enzymes-dependent pathway contributes to intracellular killing of B. pseudomallei in primary human neutrophils. The viability of neutrophils was not affected by treatment with these inhibitors (Figs. S1 and S2).
Figure 1.
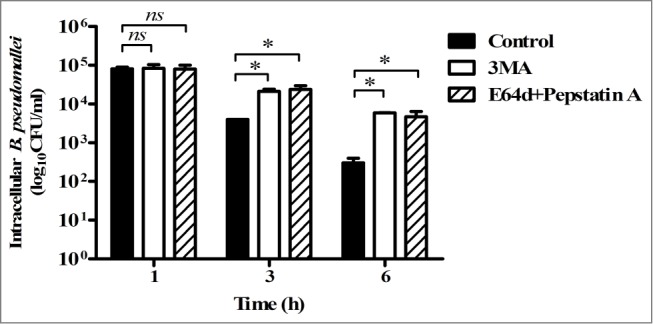
Blocking the induction of autophagy enhances survival of Burkholderia pseudomallei in human PMNs. Neutrophils purified from the blood of healthy subjects (n = 3) were infected with a live B. pseudomallei strain (K96243) at an MOI of 10. After 30 min of incubation to allow uptake, the medium was removed and the cells were incubated with fresh medium in the presence or absence of 5 mM 3MA or 10 μg/ml each of E64d and pepstatin A. After incubation for the indicated periods, the cells were treated with 250 μg/ml of kanamycin at 37°C for 30 min to kill extracellular bacteria. The intracellular survival of B. pseudomallei in neutrophils was determined by bacterial colony counts at 1, 3, and 6 h p.i., as indicated in each graph. All results are shown as the mean ± standard error of the mean (SEM) of duplicate measurements of 3 samples. Statistical significance was determined using an unpaired Student t test. ns denotes not significant, asterisks denote P ≤ 0.05.
Autophagy is activated in response to B. pseudomallei infection of human neutrophils
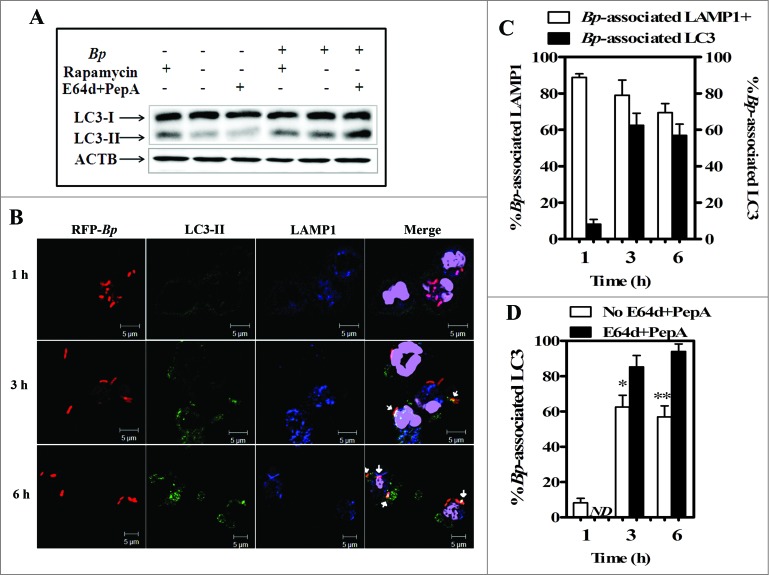
To clarify the contribution of the autophagic activity to B. pseudomallei-elimination in neutrophils, we first investigated the accumulation of LC3-II during infection (Fig. 2A).20,21 LC3-II is an established autophagosome marker specifically localized to autophagosomes and autolysosomes. When uninfected neutrophils were treated with rapamycin, an inducer of autophagy via inactivation of MTORC1, accumulation of LC3-II-positive puncta was observed in the cells, indicating that autophagy can be induced in neutrophils (Fig. 2A). When neutrophils were infected with B. pseudomallei K96243, LC3-II was accumulated, as in rapamycin-treated neutrophils, indicating that an autophagic response was induced in neutrophils during B. pseudomallei infection (Fig. 2A). Treatment of B. pseudomallei-infected neutrophils with rapamycin produced little additional effect on the increase of LC3-II level (Fig. 2A).
Figure 2.
Burkholderia pseudomallei colocalizes with the autophagy marker LC3 and the lysosome marker LAMP1 in human neutrophils. Western blot analysis for LC3-I and LC3-II in lysates of infected or uninfected neutrophils in the presence or absence of rapamycin or cathepsin inhibitor cocktail at 3 h p.i. (A). Representative confocal micrograph images of neutrophils infected with B. pseudomallei K96243 for the indicated periods are shown (B). K96243 bacteria expressing red fluorescent protein (RFP) are shown in red; LC3 is shown in green, LAMP1 in blue, and nuclei in purple. Arrows represent LC3-positive bacteria. Scale bars: 5 μm. Data shown are from a single donor representative of experiments performed with 5 subjects. Quantitative analysis of bacteria colocalized with LC3 and LAMP1 (C). All results are shown as the mean ± SEM of duplicate measurements of all samples. Quantitative analysis of bacteria colocalized with LC3 and LAMP1 in neutrophils treated with and without E64d+pepstatin A at 3 and 6 h postinfection with B. pseudomallei K96243 (D). Statistical significance was determined using an unpaired Student t test. ND denotes not determined, * P ≤ 0.05 and ** P < 0.01. Bp, Burkholderia pseudomallei.
We next focused on autophagic activity during infection, since temporal accumulation of LC3-II is not always observed when autophagy is activated and it is observed either when autophagy is activated or impaired.22 To clarify whether accumulation of LC3-II during B. pseudomallei infection reflects activated autophagy or impaired autophagy, next we performed the autophagy flux assay during bacterial infection using E64d and pepstatin A, inhibitors of lysosomal cathepsins. If autophagy is activated in B. pseudomallei-infected neutrophils, a further accumulation of LC3-II in the infected cells will be observed with inhibition of lysosomal hydrolases. Upon treatment with E64d and pepstatin A for 3 h during infection as previously reported,22 the amount of LC3-II was increased 2-fold compared to untreated infected neutrophils (Fig. 2A and Fig. S3). Our data strongly suggested that B. pseudomallei infection activates autophagy flux in human neutrophils.
If autophagy contributes to the intracellular clearance of B. pseudomallei in neutrophils, LC3 will colocalize with intracellular B. pseudomallei. Next we investigated the colocalization of intracellular B. pseudomallei with LC3 in B. pseudomallei-infected human neutrophils (Fig. 2B and C). At 1 h postinfection (p.i.), LAMP1, but few LC3 signal, was associated with B. pseudomallei. LC3 was associated with intracellular bacteria at 3 h and 6 h p.i. The association of B. pseudomallei with LC3 was not observed when killed bacteria were used, suggesting that some biological activities of B. pseudomallei may be required for the recruitment of LC3 adjacent to the bacterium (Fig. S4).
Upon treatment with E64d and pepstatin A, the population of LC3-associated bacteria was increased in a time-dependent manner (Fig. 2D and Fig. S5), suggesting that LC3 in B. pseudomallei-containing vacuoles was degradated by lysosomal hydrolases. These results were consistent with the results of western blot analysis using anti-LC3 antibody, as shown in Figure 2A, and suggested that the recruitment of LC3 around bacterium and lysosomal hydrolases contributes to the elimination of B. pseudomallei in human neutrophils.
B. pseudomallei-containing phagosomes in human neutrophils are associated with phagophore-like structures containing cytosol and granules
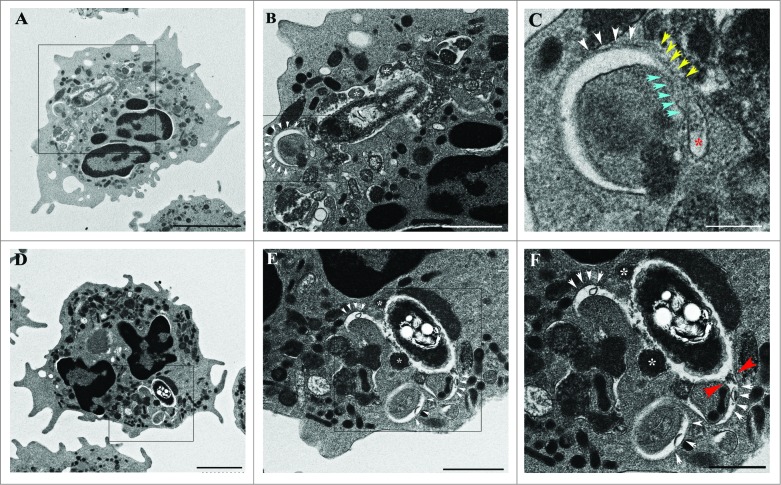
Next, we performed electron microscopic analyses of B. pseudomallei-infected neutrophils at 3 h p.i. to investigate the B. pseudomallei-containing LC3-positive membranous structures. At 3 h p.i., B. pseudomallei was surrounded by characteristic membranous structures composed of partial phagosome-like single membranes and partial elongated membranes. These B. pseudomallei-containing membranous structures also contained granules and cytoplasm. In some cases, the phagophore-like or autophagosome-like structures were partially fused with these B. pseudomallei-containing membranous structures (Fig. 3D to F).
Figure 3.
Electron microscopy analysis of Burkholderia pseudomallei-containing vacuoles in primary human neutrophils. Representative transmission electron micrographs showing the intracellular location of B. pseudomallei in human neutrophils at 3 h p.i. The image shows a B. pseudomallei-containing single-membrane-limited phagosome. The limiting membrane of this phagosome is continuous with a phagophore-like structure, containing a portion of the cytoplasm (white arrowheads) and/or granules (white asterisks) (A to F).Boxed areas are shown as magnified images, as in panels (B and E). The autophagosome-like membranous structure was occasionally fused with a bacteria-containing phagosome; white arrowheads in panels (B, C, E, and F) indicate the outer limiting membrane of the phagophore-like structure. Blue arrowheads in panel (C) indicate the region where the 2 limiting membranes of the phagophore are in close proximity. Yellow arrowheads in panel (C) indicate the single limiting membrane of the phagosome and red asterisks indicated the lumen of a small vesicle or tubule. The 2 red arrowheads in panel (F) indicate continuity of the phagophore and phagosome membranes (F). Scale bars: 2000 nm (A, D), 1000 nm (B, E), and 500 nm (C, F), respectively
Few LAMP1-negative bacteria were observed in neutrophils during the course of infection, suggesting that few bacteria could escape from phagosomes into the cytoplasm. Consistent with this, no bacteria in the cytosol of neutrophils were detected in EM examination (Fig. 3 and Fig. S6) and the actin comet tails formed at one pole of the bacterium could not be detected by fluorescence microscopy (data not shown).
Collectively, the results of confocal and electron microscopy analyses indicate that the B. pseudomallei-containing membranous structures partially associated with LC3 signal could be a novel elongated single-membrane structure, which is fused with autophagosomes or phagophore in part and with granules in part. Our EM results suggest that the B. pseudomallei-containing membranous structures in human neutrophils are likely distinct from those that perform LC3-associated phagocytosis, in which LC3 can be recruited to normal-shaped single-membrane phagosomes in mouse macrophage cell line.
B. pseudomallei induces autophagosome-like structures in a T3SS-dependent manner
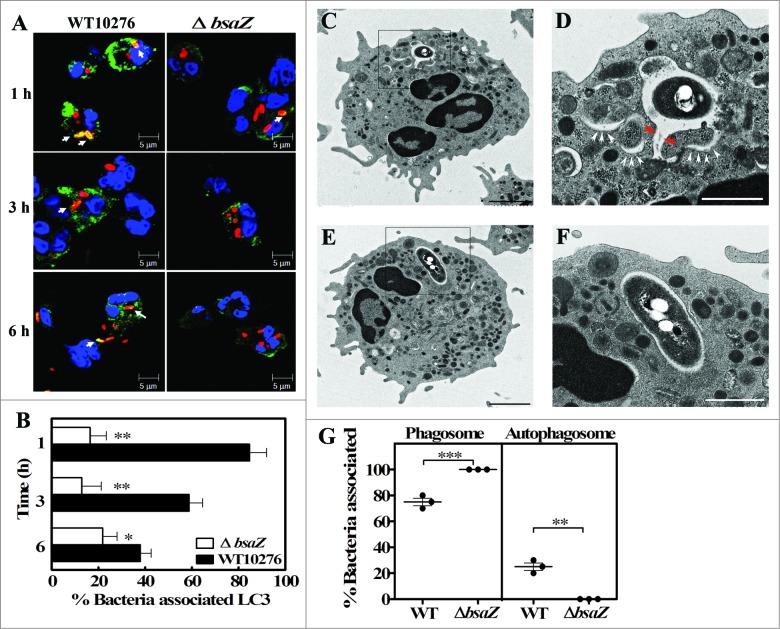
As shown in Figure S4, some biological activities of B. pseudomallei were suggested to be required for autophagy induction after neutrophil infection. Then we assessed the possibility of the autophagy induction via B. pseudomallei Bsa T3SS. It is reported that the Bsa T3SS of B. pseudomallei plays an important role to evade killing in infected macrophages.6 Since the K96243-derived Bsa T3SS fully null mutant strain is unavailable for this study, we employed another B. pseudomallei clinically isolated strain 10276, which was isolated from a severe case of human melioidosis.23 As previously reported,6 10276 exhibits the same infectious activities as K96243, including Bsa T3SS-dependent escape into the cytosol and intracellular replication, as well as actin comet tail-driven motility in macrophages. To investigate whether the Bsa T3SS influences the induction of the LC3-associated B. pseudomallei-containing membranous structures in human neutrophils, we employed a B. pseudomallei 10276 derived bsaZ mutant lacking a functional Bsa T3SS. Confocal microscopy analyses revealed that the population of LC3-positive bacteria is significantly decreased in the bsaZ mutant compared to wild-type (Fig. 4A and B). Electron microscopy analyses revealed that the bsaZ mutant resided in normal-shaped phagosomes (Fig. 4E to G), whereas wild-type bacteria were engulfed in the phagophore-like structure-associated B. pseudomallei-containing membranous structures, similarly observed in K96243 wild-type infected cells (Fig. 4D and G). Taken together, these data suggested that the T3SS machinery or effector proteins secreted via Bsa T3SS play a role in the induction of these membranous structures in B. pseudomallei-infected neutrophils.
Figure 4.
The Burkholderia pseudomallei Bsa machinery is necessary to induce autophagosome-like structures in human neutrophils. Representative confocal micrograph images of neutrophils infected with B. pseudomallei WT10276 or bsaZ mutant for the indicated periods are shown (A). Bacteria are shown in red, LC3 in green, and nuclei in blue. Arrows represent LC3-positive bacteria. Scale bars: 5 μm. Data shown are from a single donor representative of experiments performed with 3 subjects. Quantitative analysis of bacteria colocalized with LC3. All results are shown as the mean ± SEM of duplicate measurements of all samples (B). Transmission electron micrographs show the intracellular location of B. pseudomallei WT10276 (C and D) or bsaZ mutant (E and F) in human neutrophils at 3 h p.i. The image shows a B. pseudomallei-containing single-membrane-limited phagosome. The limiting membrane of this phagosome is continuous with a phagophore-like structure, containing a portion of the cytoplasm (white arrowheads). Putative continuity between the phagosome-limiting membrane and the outer limiting membrane of the autophagosome is indicated by red arrowheads (D). Boxed areas are shown as magnified images, as in panel (D and F). Scale bars: 2000 nm (C and E) and 500 nm (D and F), respectively. Quantitative analysis of bacteria associated with phagosomes or autophagosomes (G). Statistical significance was determined using an unpaired Student t test. ns denotes not significant, * P ≤ 0.05. ** P < 0.01.
Discussion
This is the first report showing that primary human neutrophils can kill intracellular B. pseudomallei through autophagy. We have demonstrated that intracellular survival of the B. pseudomallei K96243 strain is significantly enhanced by inhibition of the autophagic activities. Autophagic flux is increased during B. pseudomallei infection in neutrophils. LC3 could be recruited to the phagosomes via alternative routes: directly recruited to the phagosomal membranes from the cytoplasm, by fusion of phagosomes with LC3-positive autophagosomes or phagophores, or both. Strikingly, electron microscopic analyses revealed the presence of bacteria-containing membranous structures in B. pseudomallei-infected neutrophils. The structures containing B. pseudomallei were variable in shape, composed of a single membrane that was partially fused with phagophore- or autophagosome-like structures. The formation of these membranous structures were dependent on the Bsa T3SS, suggesting that membrane damage by the T3SS apparatus or effector proteins secreted via the T3SS can trigger the formation of these characteristic membranous structures.
It is of interest that the autophagic pathway contributes to the elimination of B. pseudomallei in human neutrophils, as neutrophils contain few acidic compartments, but possess numerous neutral neutrophilic granules in the cytoplasm. Once neutrophils phagocytose bacteria, early phagosomes containing bacteria promptly fuse with granules, followed by killing of bacteria;24 this differs from other types of cells, showing that only late phagosomes that have fused with lysosomes are LAMP1positive. TEM data from neutrophils at 0.5-h p.i. clearly show that B. pseudomallei-containing endosomes and phagosomes are closely associated with high-electron-density granules (shown as asterisks in figures), which may play a role in bacterial killing in advance of autophagy. Autophagy is likely the second line of defense of neutrophils against B. pseudomallei remaining within phagosomes. This kind of fusion between LC3-positive phagophores/autophagosomes and bacteria-containing phagosomes has not previously been described, and differs from the giant autophagosomes that observed at late stage of group A Streptococcus infection.25 Formation of the membranous structure required the presence of live bacteria and the Bsa T3SS. It is noteworthy that a related T3SS from Salmonella is required to trigger autophagy in a manner associated with damage to the membrane of the Salmonella-containing vacuole.26,27 We investigated the colocalization of ubiquitin and SQSTM1/p62 with intracellular bacteria; however, little colocalization was observed (data not shown). It has been reported that B. pseudomallei may use the Bsa system to evade LC3-associated phagocytosis in RAW264.7 macrophage-like cells.15 In macrophage-like cells, B. pseudomallei escapes from phagosomes to the cytoplasm in a Bsa-dependent manner. However, in primary human neutrophils, no escape of B. pseudomallei to the cytoplasm and no actin comet tail could be detected by EM and confocal microscopy observation. Therefore, it is difficult to compare the novel membranous structure observed in B. pseudomallei-infected neutrophils with LC3-associated phagosomes in macrophage-like cells. Strains of B. pseudomallei are genetically diverse and vary widely in infectivity. The differing kinetics of LC3 accumulation between K96423 and 10276 are perhaps due to variation in the clinical isolates of B. pseudomallei. However, the similarity in the membranous structures observed in infected neutrophils suggests that this particular structure is common to B. pseudomallei-infected neutrophils. It is conceivable that B. pseudomallei injects Bsa-secreted effectors into the cytosol to induce the formation of phagophores and autophagosomes, resulting in the fusion of B. pseudomallei-containing phagosomes with phagophores and autophagosomes; however, it will be difficult to distinguish T3SS effector-mediated autophagy from autophagy caused by Bsa-mediated phagosomal membrane damage.
In summary, our study highlights a role for autophagy in the control of intracellular B. pseudomallei by neutrophils, a cell type that plays a vital protective role against melioidosis. We propose a novel type of autophagy, which is induced by live B. pseudomallei in phagosomes of primary neutrophils in a Bsa T3SS-dependent manner. Defects in autophagy may predispose individuals to melioidosis. Current antibiotic therapy is often ineffective against melioidosis; with further research, it may be feasible to potentiate autophagic control of B. pseudomallei.
Materials and Methods
Bacterial strains
The B. pseudomallei wild-type (WT) strain K96243,28 the mStrawberry red fluorescent protein (RFP)-expressing B. pseudomallei strain K96243,29 the WT strain 10276, and the 10276 bsaZ mutant lacking a functional Bsa T3SS apparatus have been described.6 B. pseudomallei was grown in Luria-Bertani (LB) broth for 18 h at 37°C. After washing twice with phosphate-buffered saline (PBS, pH 7.4; Gibco, 10010023), the number of bacteria was estimated by measuring the absorbance of the bacterial suspension at 600 nm. In general, an absorbance of 0.33 to 0.35 was equivalent to approximately 108 CFU/ml of viable bacteria. The number of viable bacteria used in infection studies was determined by retrospective plating of serial 10-fold dilutions of the inoculum to LB agar. Live B. pseudomallei was handled according to the regulations set forth by the US Centers for Disease Control for biosafety containment level 3.
Neutrophil isolation
Human neutrophils were isolated from heparinized venous blood by 3.0 % dextran T-500 sedimentation (Pharmacosmos, 551005004007) and Ficoll-Paque PLUS centrifugation (Sigma-Aldrich, 10771), as previously described by Chanchamroen et al.4 The purity of isolated cells was generally greater than 95%, as determined with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) and FlowJo software (Treestar).
Immunofluorescence microscopy
Purified neutrophils were placed into tissue culture Lab-Tek Chambers (Nunc International, 154534) at a concentration of 2.5 × 106 cells/ml with medium control or B. pseudomallei K96243 was added at a multiplicity of infection (MOI) of 10 and incubated for 30 min. The extracellular bacteria were killed by addition of 250 µg/ml of kanamycin (KM) in complete RPMI 1640 medium (RPMI 1640 containing 10% [vol/vol] heat-inactivated fetal bovine serum) for 30 min and then cells were washed with PBS and maintained with 20 µg/ml of KM in complete RPMI1640 in the presence or absence of 3-methyladenine (3-MA;Sigma-Aldrich, M9281) at 5 mM or a cathepsin inhibitor cocktail (10 µg/ml of E64d [Peptide Inst., 4321-v] and 10 µg/ml of pepstatin A [Peptide Inst., 4397-v]) for the indicated periods. After incubation, the cells were fixed with 4% (wt/vol) paraformaldehyde in PBS and incubated with 50 mM NH4Cl for 10 min. Then, cells were permeabilized with 0.5% (vol/vol) Triton X-100 (Sigma-Aldrich, 9002931) in PBS for 30 min and nonspecific binding was blocked by incubation with 3% (wt/vol) bovine serum albumin (Sigma-Aldrich, 9048) in PBS for 30 min. Cells were stained with rabbit-anti-LC3B (Cell Signaling Technology [CST], 3868), mouse anti-LAMP1 (Abcam, ab25630), or rabbit-anti-CTSD/cathepsin D antibodies (kindly provided by Dr. Takashi Ueno, Juntendo University) for 45 min at 37°C. After washing with PBS, cells were stained with DAPI for staining of nuclei, Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes, A11034), and Alexa Fluor 647-conjugated goat anti-mouse IgG (Molecular Probes, 4410) for 45 min at 37°C. The stained cover slips were mounted using ProLong Antifade (Invitrogen, P7481) and kept in the dark at 4°C. Images were obtained by confocal microscopy using a LSM 510 META microscope (Carl Zeiss, Göttingen, Germany).
Transmission electron microscopy (TEM)
Uninfected neutrophils or neutrophils infected with live B. pseudomallei strains at an MOI of 10 for 1 and 3 h were prefixed with 2.5% (wt/vol) glutaraldehyde in PBS (pH 7.2) at the indicated time points for 2 h at room temperature. Cells were postfixed with 2% OsO4 and embedded in Epon 812 (TAAB Laboratories Equipment, T002) after dehydration, essentially as previously described.30 Ultrathin sections were cut with an ultramicrotome (UC6, Leica, Vienna, Austria), stained with uranium acetate and lead citrate, and examined with a Hitachi HT7700 electron microscope (Hitachi High-Technologies, Tokyo, Japan).
Western blotting
Neutrophils were infected with B. pseudomallei K96243 at an MOI of 10 in the presence or absence of 10 µg/ml of E64d and 10 µg/ml of pepstatin A for 3 h. The cells were lysed with 1× SDS-PAGE sample buffer, subjected to 12.5% SDS-PAGE, and transferred to polyvinylidene fluoride membranes (Pall Corporation, 66543). Membranes were blocked with 5% (wt/vol) bovine serum albumin fraction V (Sigma-Aldrich, 9048) in Tris-buffered saline with 0.1% (vol/vol) Tween-20 (Sigma-Aldrich, P1379) (TBST; 1 ml of Tween-20 and 1000 ml of 1X PBS [Gibco, 10010023]) and incubated with rabbit primary antibodies against LC3B (CST, 7074, 1:1,000 dilution) overnight at 4°C. After washing with TBST 3 times, the membranes were incubated with horseradish-peroxidase-conjugated goat anti-rabbit IgG (1:2,000 dilution; CST, 7074) for 1 h at room temperature. After washing with TBST 3 times, bound antibody was detected by enhanced chemiluminescence (Pierce, 32106), and the intensity of each band was analyzed by densitometry using the software ImageJ.
Assay of bacterial net intracellular survival
Purified neutrophils were cocultured with B. pseudomallei strains at an MOI of 10 in the presence or absence of 3MA(5 mM) or a cathepsin inhibitor cocktail (10 µg/ml of E64d and 10 µg/ml of pepstatin A) at 37°C, 5% CO2 for 30 min. Extracellular bacteria were then killed by the addition of complete RPMI 1640 containing 250 µg/ml KM at 37°C, 5% CO2 for 30 min, and the number of internalized bacteria were enumerated by lysis of the cells with 0.5% (vol/vol) Triton X-100 in PBS and plating of serial 10-fold dilutions of lysates to LB agar plates (1 h postinfection; p.i.). Net intracellular survival over time was then determined by maintaining separate cultures with RPMI containing 20 µg/ml of KM with or without inhibitors for 2 h (3 h p.i.) or 5 h (6 h p.i.), followed by lysis of neutrophils and plating of serial 10-fold dilutions of lysates to LB agar.
Statistical analysis
Statistical analysis (unpaired Student t test) was performed using the software GraphPad Prism version 5 (GraphPad). For quantitative analysis of micrographs, at least 100 bacteria were counted for each condition in each experiment. A P-value of ≤ 0.05 was considered statistically significant.
Acknowledgments
We wish to thank Ms. Keiko Kaneko, Dr. Hideki Hasegawa, and Mr. Mitsutaka Yoshida for assistance with TEM, Dr. Taishi Onodera and Dr. Mayuko Okabe for assistance with confocal microscopy, Dr. Takayuki Matsumura and Mr. Tadasu Tsuchiya (National Institute of Infectious Diseases, NIID, Japan) for assistance with western blot assays, and Ms. Chidchamai Kewcharoenwong for technical assistance. We also wish to thank Dr. Takashi Ueno, Juntendo University, School of Medicine Japan for kindly providing rabbit anti-Cathepsin D.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
10.1080/15548627.2015.1040969 This work was supported in part by the National Institutes of Health: NIAID Cooperative Center for Translational Research on Human Immunology and Biodefense (U01 AI0 82110), Faculty of Associated Medical Sciences, Khon Kaen University (AMS Incubator Postdoctoral Training Program to D.R.), and the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0215/2550 to D.R. and G.L.), and in part by Grants-in-Aid for Scientific Research on Priority Areas “Proteolysis in the Regulation of Biological Processes” from the Ministry of Education, Science, Sports and Culture of Japan (18076005 to I.T. and M.K.) and the Ministry of Health, Labor and Welfare of Japan (I.T., M.O, and M. A.).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Wiersinga WJ, van der Poll T. Immunity to Burkholderia pseudomallei. Curr Opin Infect Dis 2009; 22:102–8; PMID:19276877; http://dx.doi.org/ 10.1097/QCO.0b013e328322e727 [DOI] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol 2006; 4:272–82; PMID:16541135; http://dx.doi.org/ 10.1038/nrmicro1385 [DOI] [PubMed] [Google Scholar]
- 3.Easton A, Haque A, Chu K, Lukaszewski R, Bancroft GJ. A critical role for neutrophils in resistance to experimental infection with Burkholderia pseudomallei. J Infect Dis 2007; 195:99–107; PMID:17152013; http://dx.doi.org/ 10.1086/509810 [DOI] [PubMed] [Google Scholar]
- 4.Chanchamroen S, Kewcharoenwong C, Susaengrat W, Ato M, Lertmemongkolchai G. Human polymorphonuclear neutrophil responses to Burkholderia pseudomallei in healthy and diabetic subjects. Infect Immun 2009; 77:456–63; PMID:18955471; http://dx.doi.org/ 10.1128/IAI.00503-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones AL, Beveridge TJ, Woods DE. Intracellular survival of Burkholderia pseudomallei. Infect Immun 1996; 64:782–90; PMID:8641782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, Wallis TS, Galyov EE. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol 2002; 46:649–59; PMID:12410823; http://dx.doi.org/ 10.1046/j.1365-2958.2002.03190.x [DOI] [PubMed] [Google Scholar]
- 7.Stevens MP, Stevens JM, Jeng RL, Taylor LA, Wood MW, Hawes P, Monaghan P, Welch MD, Galyov EE. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol 2005; 56:40–53; PMID:15773977; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04528.x [DOI] [PubMed] [Google Scholar]
- 8.Suparak S, Kespichayawattana W, Haque A, Easton A, Damnin S, Lertmemongkolchai G, Bancroft GJ, Korbsrisate S. Multinucleated giant cell formation and apoptosis in infected host cells is mediated by Burkholderia pseudomallei type III secretion protein BipB. J Bacteriol 2005; 187:6556–60; PMID:16159789; http://dx.doi.org/ 10.1128/JB.187.18.6556-6560.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riyapa D, Buddhisa S, Korbsrisate S, Cuccui J, Wren BW, Stevens MP, Ato M, Lertmemongkolchai G. Neutrophil extracellular traps exhibit antibacterial activity against Burkholderia pseudomallei and are influenced by bacterial and host factors. Infect Immun 2012; 80:3921–9; PMID:22927051; http://dx.doi.org/ 10.1128/IAI.00806-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215–21; PMID:19820708; http://dx.doi.org/ 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- 11.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 2009; 183:5909–16; PMID:19812211; http://dx.doi.org/ 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- 12.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137–49; PMID:19683680; http://dx.doi.org/ 10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al.. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 2009; 11:1233–40; PMID:19749745; http://dx.doi.org/ 10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- 14.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, Prescott M, Boyce JD, Devenish RJ, Adler B. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy 2008; 4:744–53; PMID:18483470; http://dx.doi.org/ 10.4161/auto.6246 [DOI] [PubMed] [Google Scholar]
- 15.Gong L, Cullinane M, Treerat P, Ramm G, Prescott M, Adler B, Boyce JD, Devenish RJ. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One 2011; 6: e17852; PMID:21412437; http://dx.doi.org/ 10.1371/journal.pone.0017852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat Rec 1984; 208:319–27; PMID:6721227; http://dx.doi.org/ 10.1002/ar.1092080302 [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Mendonsa GR, Symington JW, Zhang Q, Cadwell K, Virgin HW, Mysorekar IU. Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo. Proc Natl Acad Sci U S A 2012; 109:11008–13; PMID:22715292; http://dx.doi.org/ 10.1073/pnas.1203952109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narni-Mancinelli E, Soudja SM, Crozat K, Dalod M, Gounon P, Geissmann F, Lauvau G. Inflammatory monocytes and neutrophils are licensed to kill during memory responses in vivo. PLoS Pathog 2011; 7: e1002457; PMID:22241983; http://dx.doi.org/ 10.1371/journal.ppat.1002457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, Noppen S, Delforge M, Willems J, Vandenabeele P. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res 2011; 21:290–304; PMID:21060338; http://dx.doi.org/ 10.1038/cr.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151–75; PMID:18188003; http://dx.doi.org/ 10.4161/auto.5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy 2007; 3:117–25; PMID:17204850; http://dx.doi.org/ 10.4161/auto.3618 [DOI] [PubMed] [Google Scholar]
- 22.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005; 1:84–91; PMID:16874052; http://dx.doi.org/ 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- 23.Maegraith BG, Leithead CS. Melioidosis: a case-report. Lancet 1964; 1:862–3; PMID:14124069; http://dx.doi.org/ 10.1016/S0140-6736(64)91581-8 [DOI] [PubMed] [Google Scholar]
- 24.Nordenfelt P, Tapper H. Phagosome dynamics during phagocytosis by neutrophils. J Leukoc Biol 2011; 90:271–84; PMID:21504950; http://dx.doi.org/ 10.1189/jlb.0810457 [DOI] [PubMed] [Google Scholar]
- 25.Sakurai A, Maruyama F, Funao J, Nozawa T, Aikawa C, Okahashi N, Shintani S, Hamada S, Ooshima T, Nakagawa I. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J Biol Chem 2010; 285:22666–75; PMID:20472552; http://dx.doi.org/ 10.1074/jbc.M109.100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birmingham CL, Brumell JH. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy 2006; 2:156–8; PMID:16874057; http://dx.doi.org/ 10.4161/auto.2825 [DOI] [PubMed] [Google Scholar]
- 27.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 2006; 281:11374–83; PMID:16495224; http://dx.doi.org/ 10.1074/jbc.M509157200 [DOI] [PubMed] [Google Scholar]
- 28.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, et al.. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 2004; 101:14240–5; PMID:15377794; http://dx.doi.org/ 10.1073/pnas.0403302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.French CT, Toesca IJ, Wu TH, Teslaa T, Beaty SM, Wong W, Liu M, Schröder I, Chiou PY, Teitell MA, et al.. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc Natl Acad Sci U S A 2011; 108:12095–100; PMID:21730143; http://dx.doi.org/ 10.1073/pnas.1107183108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 2008; 9:1728–42; PMID:18637904; http://dx.doi.org/ 10.1111/j.1600-0854.2008.00796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.