Abstract
CAV1 (caveolin 1, caveolae protein, 22kDa) is well known as a principal scaffolding protein of caveolae, a specialized plasma membrane structure. Relatively, the caveolae-independent function of CAV1 is less studied. Autophagy is a process known to involve various membrane structures, including autophagosomes, lysosomes, and autolysosomes for degradation of intracellular proteins and organelles. Currently, the function of CAV1 in autophagy remains largely elusive. In this study, we demonstrate for the first time that CAV1 deficiency promotes both basal and inducible autophagy. Interestingly, the promoting effect was found mainly in the late stage of autophagy via enhancing lysosomal function and autophagosome-lysosome fusion. Notably, the regulatory function of CAV1 in lysosome and autophagy was found to be caveolae-independent, and acts through lipid rafts. Furthermore, the elevated autophagy level induced by CAV1 deficiency serves as a cell survival mechanism under starvation. Importantly, downregulation of CAV1 and enhanced autophagy level were observed in human breast cancer cells and tissues. Taken together, our data reveal a novel function of CAV1 and lipid rafts in breast cancer development via modulation of lysosomal function and autophagy.
Keywords: autophagy, breast cancer, caveolin 1, lipid rafts, lysosome
Abbreviations: Baf, bafilomycin A1; CAV1, caveolin 1; CHO, water-soluble cholesterol; CQ, choloroquine; CTxB, cholera toxin subunit B; CTSL, cathepsin L; DRF, detergent-resistant fraction; DSF, detergent-soluble fraction; EGF, epidermal growth factor; KO, knockout; LAMP1, lysosomal-associated membrane protein 1; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; MBCD, methyl-β-cyclodextrin; MEF, mouse embryonic fibroblasts; MTOR, mechanistic target of rapamycin; PBS, phosphate-buffered saline; PI, propidium iodide; PLA, proximity ligation assay; PTRF, polymerase I and transcript release factor; tfLC3B, mRFP-GFP tandem fluorescent-tagged LC3B; TFRC, transferrin receptor; TSC, tuberous sclerosis complex; ATP6V0D1, ATPase H+ transporting lysosomal 38kDa, V0 subunit d1; WT, wild type.
Introduction
CAV1 is the principal scaffolding protein of caveolae, which is one of the major types of lipid rafts that are present at the plasma membrane.1 Lipid rafts are liquid-ordered microdomains enriched with a characteristic structural composition (sphingolipids, cholesterol, and saturated phospholipids) in plasma membrane and intracellular membranes of various organelles, including Golgi, endoplasmic reticulum, mitochondria, and the endosome/lysosome.2 The caveolae (flask-shaped structures) and the planar lipid rafts (also known as noncaveolar rafts, referred to as lipid rafts hereafter) are the 2 main types of lipid rafts.3 At present, the critical role of CAV1 in caveolae is well studied, while there is accumulating evidence suggesting the importance of CAV1 in lipid rafts. For instance, CAV1 plays an important role in cholesterol trafficking and homeostasis, a key component in lipid rafts.4,5 In addition, compared to the limited distribution on the plasma membrane of caveolae,6 CAV1 has a more extensive membrane distribution including mitochondria and endoplasmic reticulum, as well as the late endosome/lysosome, indicating the existence of caveolae-independent function of CAV1 in the intracellular membrane system.7-9
Emerging evidence demonstrates the tumor-suppression functions of CAV1 in several types of cancer, especially in breast cancer.10 For instance, expression of oncogenes (such as ABL1/v-abl and HRAS/H-ras) leads to a significant decrease in CAV1 protein and mRNA levels.11 Loss of CAV1 induces cellular transformation through the activation of the MAPK1/ERK2/p41-MAPK3/ERK1/p44 cascade and subsequent development of an unusual growth pattern with increased phospho-MAPK1/3 independent of EGF (epidermal growth factor).12,13 Consistently, overexpression of CAV1 is sufficient to decrease cell proliferation and tumor size in vivo.11 In breast cancer, a large percentage of patients are deficient in CAV1 expression.10 Several human breast cancer cell lines display a decreased CAV1 expression level compared to benign mammary epithelial cells.14 Moreover, about 35% of breast cancer cases contain mutant CAV1.15 For example, a dominant negative mutant CAV1P132L has been identified in ER-positive patients with well-differentiated breast cancer.16 Although the studies discussed above indicate a tumor suppressive role for CAV1, there is also conflicting evidence showing an opposite role of CAV1. For instance, CAV1 expression in breast tumor stroma increases tumor invasion and metastasis via biomechanical remodeling.17 Therefore, the exact biological function of CAV1 in breast cancer development and the molecular mechanisms remain to be further investigated.
Macroautophagy (referred to as autophagy hereafter) is an evolutionarily well-conserved self-eating process in eukaryotic cells that results in degradation of long-lived proteins and organelles via the lysosomal pathway, which serves as a powerful booster of metabolic homeostasis.18 One key feature of autophagy is that it involves various intracellular membrane structures, including autophagosomes, lysosomes, and autolysosomes. At present, several studies have implicated CAV1 and lipid rafts in the regulation of autophagy. For instance, CAV1 deficiency induces autophagy in adipocytes via suppression of insulin and lipolytic responses.19 Loss of CAV1 promotes autophagy under hypoxia and oxidative stress in adipocytes and fibroblasts.20 These studies indicate a suppressive role for CAV1 in autophagy. Furthermore, there are several clues indicating the potential role of lipid rafts in autophagy. For instance, lipid rafts promote the AKT-MTOR (mechanistic target of rapamycin) pathway,21,22 a key negative regulator of autophagy.23 In contrast, there is conflicting evidence indicating that some components of lipid rafts, such as ceramides and GD3 ganglioside, play a positive role in autophagy regulation.24-26
In this study, we aimed to evaluate the involvement of CAV1 in autophagy. Our data clearly demonstrate that deficiency of CAV1 promotes autophagy and lysosomal function via the disruption of lipid rafts, independent of caveolae. Moreover, the elevated autophagy level induced by CAV1 deficiency serves as a cell survival mechanism under nutritional stress. Importantly, downregulation of CAV1 and enhanced autophagy level were observed in human breast cancer cell lines and cancerous tissues. Thus, our data reveal a novel function of CAV1 in cell stress responses and possibly breast cancer development via modulation of lysosomal function and autophagy.
Results
CAV1 deficiency promotes autophagy via disruption of lipid rafts
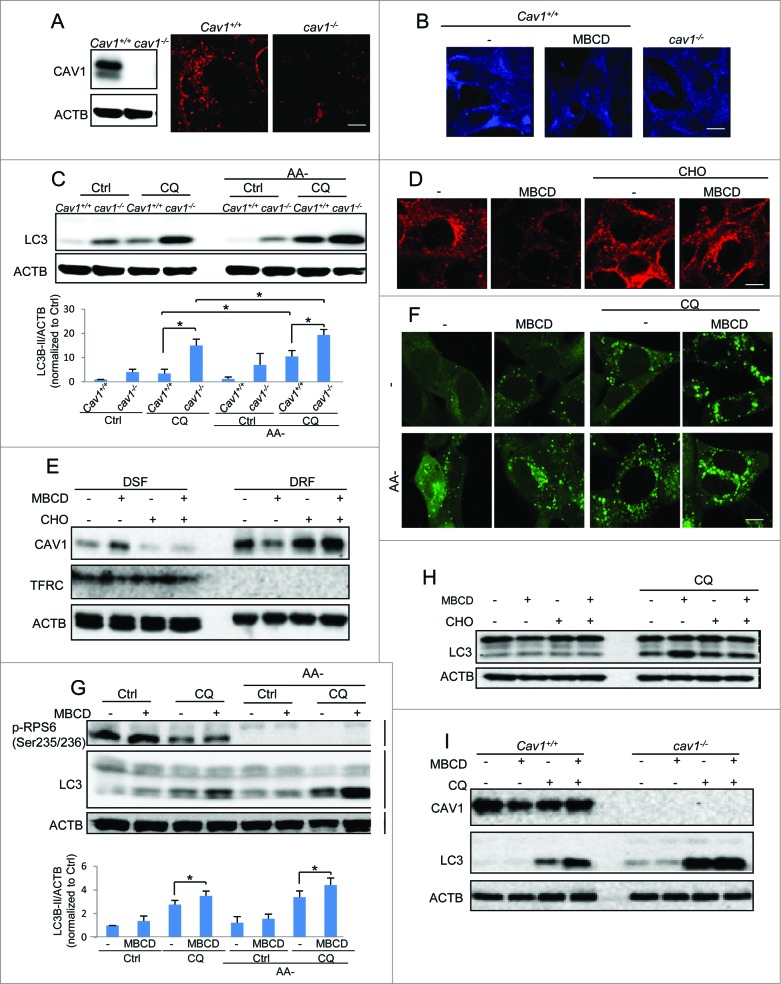
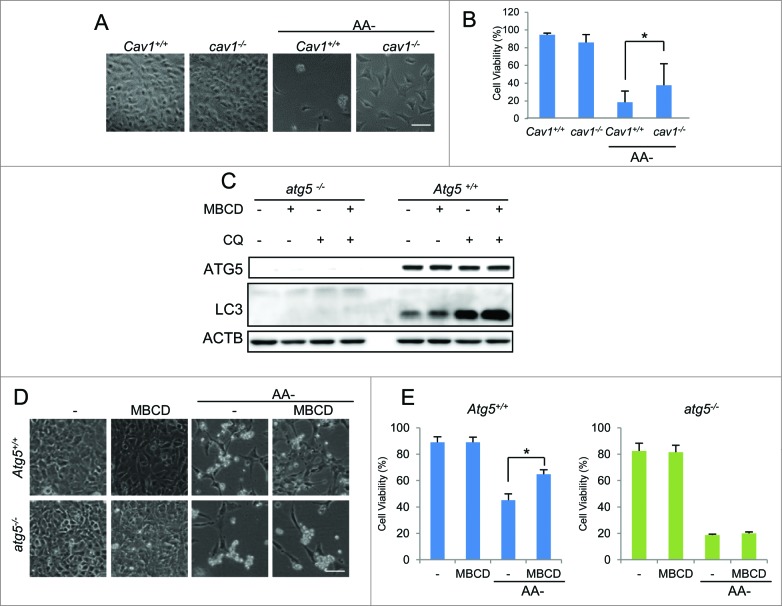
We first tested the effect of CAV1 deficiency on autophagy by using Cav1 wild-type (WT) and knockout (KO) mouse embryonic fibroblasts (MEFs) as reported previously.27 The absence of the CAV1 protein was confirmed in western blots (Fig. 1A). We assessed the lipid raft level using Alexa Fluor 594–conjugated cholera toxin subunit B (CTxB) staining in both Cav1+/+ and cav1−/− cell lines.28 As shown in Figure 1A, cav1−/− cells showed markedly reduced CTxB signal in both intracellular and plasma membranes in comparison to the Cav1+/+ MEFs, indicating a lower level of lipid rafts in cav1−/− cells. These results were confirmed by labeling the cholesterol directly using filipin (Fig. 1B). Next, we compared the autophagy levels by evaluating the well-established autophagy marker MAP1LC3B-II/LC3B-II in normal and amino acid starvation conditions. We found that in both conditions, LC3B-II levels were higher in cav1−/− cells (Fig. 1C). Moreover, to measure the autophagic flux, we inhibited the autophagosomal degradation by addition of the lysosomal inhibitor chloroquine (CQ), and observed an increase in LC3B-II protein levels in cav1−/− cells, excluding the possibility that the increased LC3B-II was caused by a blockage of autophagosomal degradation (Fig. 1C). These data suggest that CAV1 deficiency promotes both basal and inducible autophagy levels.
Figure 1.
For figure legend, see page 764. Figure 1 (See previous page). CAV1 deficiency promotes autophagy via disruption of lipid rafts. (A) Lipid raft marker CTxB (Red) staining in Cav1+/+ and cav1−/− MEFs. Cell identity was shown by western blots. (B) Cholesterol indicator filipin (blue) staining of both CAV1 deficiency and MBCD pretreatment (5 mM, 1 h). Scale bar: 10 μm. (C) CAV1 deficiency increases both basal and amino acid starvation-induced autophagy (AA-, 2 h). Autophagic flux was detected by the treatments with or without chloroquine diphosphate (CQ, 20 μM) for 2 h. Quantification of the LC3B-II/ACTB ratios is presented (*P <0.05). (D) Cholesterol replenishment rescues the lowering effect of MBCD on CTxB staining. MEFs were pretreated with or without MBCD (5 mM) for 1 h, then incubated in the presence or absence of cholesterol (CHO, 30 μg/ml) and then stained with CTxB. (E) CAV1 redistribution in detergent-resistant fractions (DRF, proteins rich in membrane rafts) and detergent soluble fractions (DSF) after the same treatments as indicated in (D). Cell lysates were separated and immunoblotted with the indicated markers. (F) MBCD increases the GFP-LC3B puncta formation in both normal and starvation condition. MEFs with stable expression of GFP-LC3B were pretreated with or without MBCD (5 mM) for 1 h, then incubated in both normal DMEM and AA- medium with or without CQ (20 μM) for 2 h. The GFP-LC3B punctation and lipid raft marker CTxB were observed under a confocal microscope (×600). Scale bar: 10 μm. (G) MBCD increases autophagic flux in both normal and amino acid starvation-induced autophagy. Cells were exposed to the same treatments indicated in panel. Quantification of the LC3B-II/ACTB ratios is presented (*P < 0.05). (F) then cell lysates were collected and subjected to western blot analysis for the indicated markers. Quantification of the LC3B-II/ACTB is presented. (H) Cholesterol replenishment overcomes the effect of MBCD disruption on autophagy. MEFs were pretreated with or without MBCD (5 mM) for 1 h, then incubated in the presence or absence of CHO (30 μg/ml) and CQ (20 μM). (I) MBCD disruption does not further enhance autophagy flux in cav1−/− MEFs. Cav1+/+ and cav1−/− MEFs were pretreated with or without MBCD (5 mM) for 1 h, then incubated in the presence or absence of CQ (20 μM).
Since CAV1 has been established as a critical scaffolding protein in caveolae, we attempted to test whether the role of CAV1 in autophagy is caveolae-dependent. To do this, we utilized cells deficient in PTRF (polymerase I and transcript release factor), another key component for caveolae formation.1 Deficiency of PTRF blocks caveolae formation and promotes CAV1 degradation.29 Consistently, we found lower CAV1 levels in ptrf−/− cells, and the CQ increased CAV1 level via blockage of lysosomal degradation (Fig. S1A). Interestingly, we found that both CTxB and filipin staining did not differ significantly between Ptrf+/+ and ptrf−/− MEFs (Fig. S1B and S1C). The autophagy level was also comparable in these 2 cell lines in basal and amino acid starvation conditions (Fig. S1A). Moreover, disruption of lipid rafts by methyl-β-cyclodextrin (MBCD) had a similar effect on both Ptrf+/+ and ptrf−/− cells (Fig. S1A). Taken together, these data indicate the possibility that the implication of CAV1 in autophagy is mainly via noncaveolar lipid rafts, but not PTRF-related caveolae.
To further examine whether CAV1 is implicated in autophagy via lipid rafts, we tested the effect of MBCD on autophagy. MBCD depletes cholesterol from the plasma membrane and disrupts lipid rafts.30 The disruptive effects of MBCD on lipid rafts were confirmed by using 3 different approaches: (i) reduction of Alexa Fluor 594–CTxB staining, (ii) reduction of filipin staining and (iii) loss of CAV1 in the isolated raft fraction and the accumulation in the nonraft fraction.31 MBCD treatment significantly decreased the intensity of both CTxB staining (Fig. 1D) and filipin staining (Fig. 1B), and caused CAV1 and the other lipid raft protein FLOT1 (flotillin 1) to redistribute from the detergent-resistant lipid raft fraction (DRF, representing the lipid raft components) to the detergent-soluble fraction (DSF, representing the nonlipid raft components, using TFRC [transferrin receptor] as the marker) (Fig. 1E and S2), indicating that MBCD is able to disrupt lipid rafts effectively. As expected, MBCD enhanced both the basal and amino acid starvation-induced autophagy level, based on the observations of GFP-LC3B puncta formation (Fig. 1F) and increased LC3B-II level (Fig. 1G) in both normal and amino acid starvation conditions, with or without the presence of CQ. These observations demonstrate that disruption of lipid rafts by MBCD increases autophagic flux.
Moreover, to exclude the possibility that MBCD-mediated autophagy observed above was caused by mechanisms independent of lipid rafts, we used cholesterol (CHO) replenishment to restore the lipid rafts. As shown in Figure 1D and E, CHO replenishment recovered the decreased intensity of CTxB staining and the redistribution of CAV1 caused by MBCD. Notably, CHO loading decreases LC3B-II flux, as noticed by the reduction in the CQ-induced accumulation of LC3B-II (Fig. 1H). To further confirm the notion that the regulatory role of CAV1 in autophagy is associated with cholesterol, we performed MBCD treatment in cav1-KO cells. As shown in Figure 1I, compared to Cav1 WT cells, MBCD treatment was less effective in inducing autophagy in cav1-KO cells, suggesting that the regulatory role of CAV1 in autophagy is associated with cholesterol and lipid rafts.
In addition, lipid rafts are implicated in the AKT-MTOR signaling pathway, which is a key negative regulator of autophagy.21,22 We next tested the involvement of MTORC1 in autophagy induced by lipid raft disruption. As shown in Figure S3A, MBCD disruption did not show a significant inhibitory effect on MTOR downstream target protein RPS6/S6 (Ser235/Ser236). Consistently no difference was found in the phosphorylation level of between CAV1 WT and KO MEFs (Fig. S3B). Moreover, we disrupted lipid rafts by MBCD treatment in tsc2 (tuberous sclerosis 2)-KO MEFs. TSC2 is an essential component in the formation of a functional heterodimer to suppress MTORC1 activity, and loss of TSC2 leads to the constitutive MTOR activation and impaired autophagy.32 The effects of MBCD disruption on the autophagic flux were similar in both Tsc2+/+ and tsc2−/− MEFs (Fig. S3A), suggesting that MTORC1 is unlikely to play a role in autophagy-mediated by CAV1 deficiency and lipid raft disruption.
CAV1 deficiency promotes lysosomal function
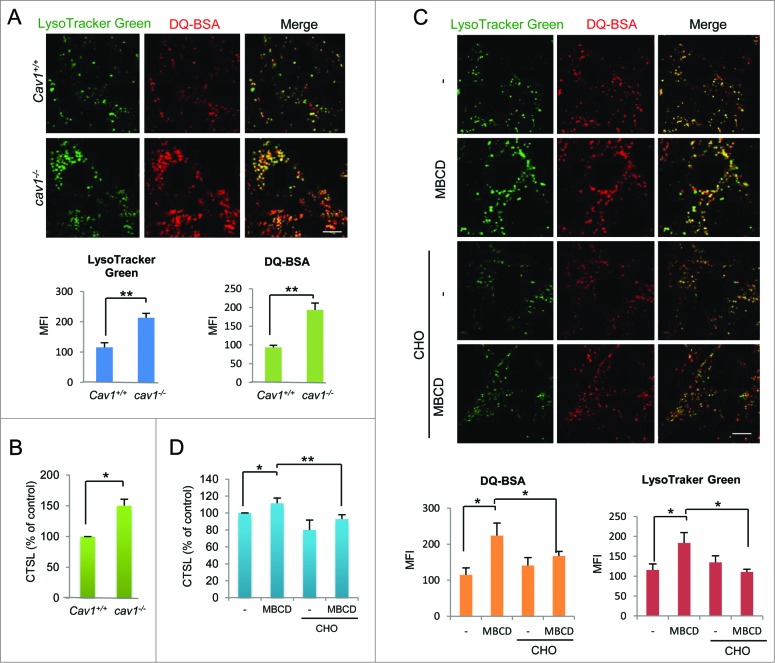
Lysosomes are the key organelles responsible for autophagy degradation.33 Recently we have demonstrated upregulation of lysosomal function in the course of autophagy induced by starvation and MTOR inhibition.23 In this study, we examined whether CAV1 deficiency affects lysosomal function, measured by the following assays: (i) lysosomal pH evaluation by LysoTracker and LysoSensor, (ii) lysosomal proteolysis by DQ-BSA, and (iii) detection of cathepsin enzyme activity coupled with flow cytometry and confocal microscopy. First, there was a significant increase in LysoTracker Green, LysoSensor and DQ-BSA signal in cav1−/− MEFs when comparing to WT cells (Fig. 2A and, S4A and B), indicating enhanced acidification of lysosomes and proteolysis activity, respectively. The lysosomal pH was also significantly decreased in cav1−/− cells (Fig. S4C). Second, a significant increase in lysosomal CTSL (cathepsin L) enzyme activity in cav1−/− cells was detected by flow cytometry (Fig. 2B) and confocal microscopy (Fig. S4D).
Figure 2.
CAV1 deficiency promotes lysosomal function. (A) cav1−/− MEFs has a higher DQTM Red BSA (red) and LysoTracker® Green DND-26 (green) staining intensity when comparing to Cav1+/+ cells. Cav1+/+ and cav1−/− MEFs were preloaded with DQTM Red BSA (red) and costained with LysoTracker® Green DND-26 (green). Then examined by confocal microscopy (×600). Scale bar: 10 μm. The mean florescence intensity were analyzed by ImageJ, means±SD were presented. (B) CTSL enzyme activity is increased in cav1−/− MEFs. Cav1+/+ and cav1−/− MEFs were loaded with Magic RedTM CTSL reagent for 15 min. Fluorescence intensity of 10,000 cells per sample was determined by flow cytometry using the BD FACS cytometer. Three independent experiments were analyzed, means ±SD were presented. (C). MBCD increases the DQTM Red BSA and LysoTracker Green DND-26 staining intensity, while cholesterol replenishment overcomes these effects. MEFs were costained with DQTM Red BSA and LysoTracker Green DND-26, then pretreated with MBCD (5 mM) for 1 h, and then treated with or without CHO (30 μg/ml) and examined by confocal microscopy (×600). Scale bar: 10 μm. (D) MBCD increases the CTSL enzyme activity, while cholesterol replenishment overcomes this effect. MEFs were pretreated with MBCD (5 mM) for 1 h. Then CTSL enzyme activities were measured by FACS cytometer. Three independent experiments were analyzed, means ±SD were presented (*P < 0.05, ** P < 0.01).
To further understand the role of lipid rafts in regulation of lysosomal function, we also assessed the effect of MBCD and cholesterol replenishment on lysosomal function. Consistently, a higher level of LysoTracker Green staining, LysoSensor staining, DQ-BSA intensity, and CTSL enzyme activity were found in cells treated with MBCD (Fig. 2C, D, S4A and B). MBCD treatment also led to a significant decrease of the lysosomal pH (Fig. S4C). Notably, the same cholesterol replenishment which effectively recovered lipid rafts (Figs. 1D and E) reversed the promoting effect of MBCD on lysosomal function (Figs. 2C and D).
To confirm whether the increase of these markers used for lysosomal function was due to increased lysosomal membrane permeability or even increased lysosome numbers, we examined the distribution of CTSL and the LAMP1 (lysosomal-associated membrane protein 1) level. In this study, most of the CTSL magic red staining colocalized with LysoTracker Green in both Cav1+/+ and cav1−/− cell lines before and after raft disruption by MBCD (Fig. S4D and E), indicating that increased CTSL by lipid raft disruption was in the lysosome but not due to its leakage into the cytosol. Moreover, in our data, the LAMP1 protein level was not affected by CAV1 deficiency or MBCD treatment (Fig. S5A and B), thus excluding the possibility that the observed changes of lysosomal functions are due to increased lysosome numbers.
CAV1 deficiency promotes V-ATPase assembly
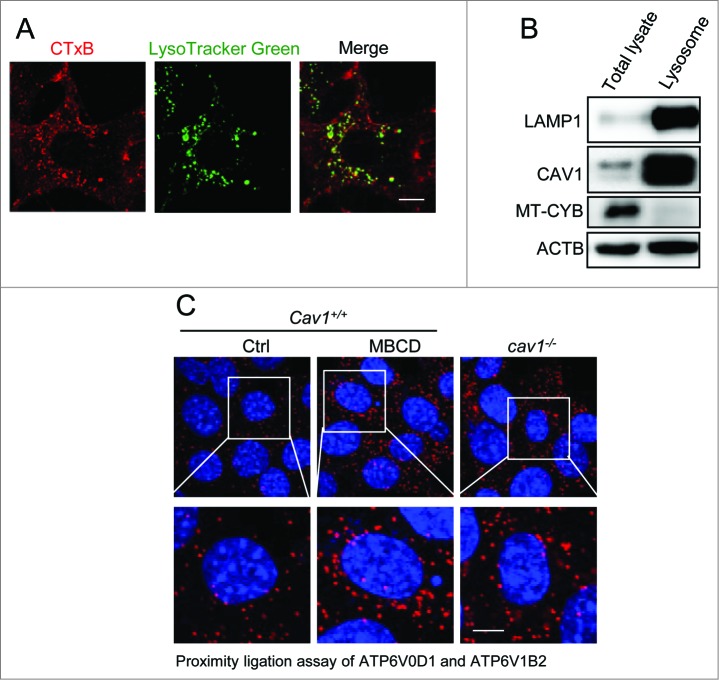
In order to understand the molecular mechanisms underlying lysosomal activation mediated by CAV1 deficiency, we examined changes in lysosomal V-ATPase assembly. V-ATPase is known as the crucial protein complex that regulates the acidification process of the lysosomal lumen and lysosomal degradation.34 The V-ATPase, a proton-pumping membrane protein complex, is controlled by the assembly of 2 domains (the cytosolic V1 domain and the integral membrane V0 domain).34 Since there are studies showing that lipid rafts contain V-ATPase subunits,35 we thus determined whether CAV1 deficiency regulates the V-ATPase assembly via lipid raft disruption. First, more than 50% of lysosomes (labeled by LysoTracker Green) were colocalized with lipid rafts (visualized by CTxB) (Fig. 3A), suggesting the existence of lipid rafts on lysosomes. Such a finding was further supported by the evident colocalization between LAMP1 and CAV1 (Fig. S6). Second, isolated lysosome membranes contained a significant amount of CAV1 (Fig. 3B), suggesting the possible existence of CAV1-dependent lipid rafts on the lysosome membrane. And finally, we used the proximity ligation assay (PLA) to assess the colocalization of the V0 and V1 subunits. Indeed, a higher level of signal was detected in CAV1-deficient cells and after MBCD treatment (Fig. 3C), indicating enhanced V1 translocation to lysosomes and V-ATPase assembly in cells after lipid raft disruption. In addition, we found that the colocalization between CAV1 and ATP6V0D1 (ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1) was decreased after lipid raft disruption by MBCD (Fig. S7). Interestingly, the amino acid-free starvation, which promotes lysosomal function,23 also decreased the colocalization between CAV1 and ATP6V0D1 (Fig. S7). These observations support the notion that the CAV1-dependent lipid rafts localize at lysosome membranes and disruption of these lipid rafts increases the V-ATPase V0 and V1 assembly.
Figure 3.
CAV1 deficiency promotes V-ATPase assembly. (A) Lipid raft marker CTxB (red) and lysosome marker LysoTracker Green DND-26 (green) are colocalized. MEFs were costained with CTxB (red) and LysoTracker Green DND-26 (green), then observed by confocal microscopy (×600). Scale bar: 10 μm. (B) CAV1 is accumulated in lysosome fractions. Lysosome fractions were collected as described in Materials and Methods. LAMP1 was used as a marker for the lysosome fraction, and MT-CYB was used as a marker for mitochondria. Lysosome fractions and total cell lysates were blotted for indicated proteins. (C) V-ATPase assembly is increased in CAV1-deficient cells or after MBCD treatment. Cav1+/+ MEFs, which were treated with or without MBCD (5 mM) for 1 h, and cav1−/− MEFs were fixed and followed by proximity ligation assay (PLA). The nuclei were counterstained with DAPI. Representative PLA images were selected and shown. Scale bar: 10 μm.
Disruption of lipid rafts promotes autophagosome-lysosome fusion
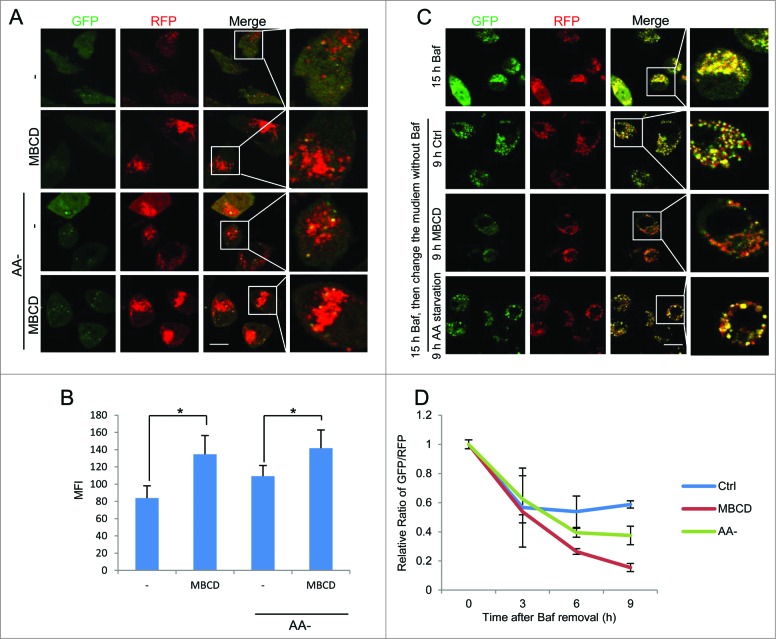
It is known that activation of lysosomal function in the course of autophagy is also dependent on autophagosome-lysosome fusion.23 Here, we assessed the effect of lipid rafts on this fusion process by using the L929 cells with stable transfection of tfLC3B (mRFP-GFP tandem fluorescent-tagged LC3B) as a tool. First, in these cells, the RFP is more resistant than GFP to the acidic lysosome environment. Therefore, RFP-positive/GFP-negative puncta can be used to evaluate the fusion of autophagosomes and lysosomes.36 We observed increased RFP puncta after MBCD pretreatment and Cav1 knockdown, indicating a positive effect of raft disruption on autophagosome-lysosome fusion (Figs. 4A, B and S8). Second, as shown in Figure 4C and D, in cells treated with bafilomycin A1 (Baf) for 15 h, all of the puncta were double positive for GFP and RFP, indicating a complete blockage of autophagosome-lysosome fusion. Removal of Baf from the cell culture medium would eliminate the blockage and resume the autophagosome-lysosome fusion process.37 After Baf was removed, the nonfused autophagosomes (both GFP and RFP positive) would proceed with lysosome fusion to form autolysosomes, which are detectable with RFP due to the quenching of GFP in the acidic autolysosome.36 Therefore, by comparing the ratio of GFP/RFP signals after Baf removal we are able to evaluate the autophagosome-lysosome fusion process. In comparison to control cells, MBCD treatment markedly reduced the relative ratio of GFP/RFP, indicating a higher rate of autophagosome-lysosome fusion and autolysosome formation in MBCD-treated cells (Figs. 4C and D). Notably, the effect of MBCD was even more evident than those observed in the positive control with amino acid starvation (Figs. 4C and D).
Figure 4.
Disruption of lipid rafts promotes autophagosome-lysosome fusion. (A) MBCD pretreatment increases the RFP signal in the tfLC3B-L929 cells. The tfLC3B-L929 cells were pretreated with or without MBCD (5 mM) for 1 h, then incubated in both normal DMEM and amino acid-free medium for 2 h. Cells were examined using a confocal microscope (×600). Scale bar: 10 μm. (B) Statistical analysis of the figures in panel (A) by the average RFP fluorescence per cell. Values were represented as means ±SD of 3 experiments (*P < 0.05). (C) MBCD pretreatment increases autophagosome-lysososme fusion. The tfLC3B-L929 cells were treated with bafilomycin A1 (Baf, 50 nM) for 15 h. Then medium was changed to the indicated treatments and cells were observed under confocal microscopy at each time (3, 6, and 9 h). Representative data from 9-h treatment were presented. Scale bar: 10 μm. (D) Statistical analysis of GFP/RFP ratio. The rate of autophagosome maturation was measured by the green/red fluorescence ratio (Relative Ratio of GFP/RFP) of indicated treatments at each time (3, 6, and 9 h) after Baf removal (0). The ratio of GFP/RFP was normalized to the value at T0. Values were represented as means ± SD of 3 experiments.
Autophagy induced by CAV1 deficiency promotes cell survival under starvation
One important physiological role of autophagy is to prolong cell survival under starvation by recycling cytoplasmic components to provide the essential nutrients needed.38-40 Here, we examined the cell viability by morphological changes and propidium iodide (PtdIns) exclusion assay to test if CAV1 deficiency-induced autophagy would have any impact on the cell death induced by amino acid starvation. As expected, we observed that cav1−/− MEFs were more resistant to amino acid starvation than WT MEFs (Figs. 5A and B).
Figure 5.
Autophagy induced by CAV1 deficiency promotes cell survival under starvation. (A and B) cav1−/− MEFs are more resistant to amino acid starvation compared to Cav1+/+ cells. Cav1+/+ and cav1−/− MEFs were treated with amino acid-free medium for 48 h. Then the cells were observed under phase-contrast microscopy (A) Scale bar: 200 μm. The percentage of dead cells was measured with the propidium iodide (PtdIns) uptake assay coupled with flow cytometry (B) Values were represented as means ±SD of 3 experiments (*P <0.05). (C) MBCD pretreatment induces autophagic flux in Atg5+/+ cells, and autophagy is deficient in atg5−/− cells. WT and atg5−/− MEFs were pretreated with or without MBCD (5 mM) for 1 h, then incubated in the presence or absence of CQ (20 μM). (D and E) MBCD protects against cell death in ATG5 WT cells after amino acid starvation (AA-). Atg5+/+ and atg5−/− MEFs were pretreated with MBCD (5 mM) for 1 h, followed by amino acid starvation. Then the cells were observed under phase-contrast microscopy. Scale bar: 200 μm. The percentage of dead cells was measured with the PtdIns uptake assay coupled with flow cytometry.
Next, to confirm whether this survival advantage in CAV1-deficient cells is autophagy dependent, we compared the effect of MBCD treatment, which mimics the lipid raft disruption effect of CAV1 deficiency, on the cell viability in both Atg5+/+ and atg5−/− MEFs after starvation. First, we found that treatment with MBCD enhanced autophagic flux level only in Atg5+/+ MEFs (Fig. 5C). Second, MBCD alone did not affect cell viability in normal growth medium in both Atg5+/+ and atg5−/− cells, while pretreatment with MBCD offered significant protection against starvation-mediated cell death in Atg5+/+ cells, but not in atg5−/− cells (Figs. 5D and E). As expected, the atg5−/− MEFs were much more susceptible to cell death under starvation (Figs. 5D and E). These results thus indicate that the protective effect by lipid raft disruption against starvation-induced cell death is autophagy or Atg5-dependent.
Altogether, we provide evidence to demonstrate that the higher level of autophagy induced by CAV1 deficiency via lipid raft disruption promotes cell survival under starvation conditions.
Reduced CAV1 expression level in human breast cancer cells and re-expression of CAV1 in MCF7 cells recovers lipid rafts and suppresses autophagy and lysosome function
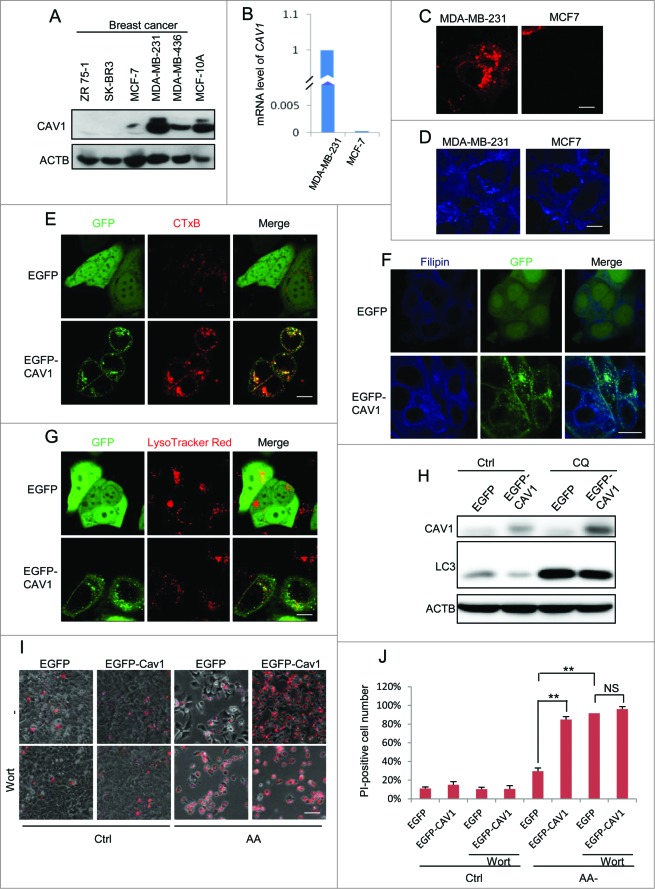
Data from an earlier part of our study demonstrate that CAV1 deficiency promotes autophagy. Since autophagy is widely considered to be a tumor-suppressive mechanism,38,39,41 we hypothesized that CAV1 expression is associated with cancer development via modulation of autophagy. We first measured the CAV1 protein level in a set of different breast cancer cell lines and a nontumorigenic MCF10A. Among them, ZR 75–1, SK-BR3 and MCF7 had low levels of CAV1 expression, while the CAV1 protein levels in MDA-MB-231, MDA-MB-436 were found to be higher, similar to MCF10A cells (Fig. 6A). Such results are generally consistent with the earlier reports with regard to CAV1 expression in breast cancer cells.14 We next chose 2 breast cancer cell lines MDA-MB-231 and MCF7 to elucidate the function of CAV1 in autophagy and cell survival. The mRNA level of CAV1 was found to be consistent with the protein level in these 2 cell lines (Fig. 6B). We then compared CTxB and filipin staining intensity as direct evidence for lipid raft levels. Consistent with CAV1 expression differences, MDA-MB-231 cells with high CAV1 expression had a relatively higher CTxB and filipin level when comparing to MCF7 cells (Figs. 6C and D). These data thus suggest that CAV1 expression level is associated with lipid rafts in breast cancer cell lines.
Figure 6.
Reduced CAV1 expression level in human breast cancer cells and re-expression of CAV1 in MCF7 cells recovers lipid rafts and suppresses autophagy and lysosome function. (A) The CAV1 level in different breast cancer cell lines. The lysates of MCF-10A and 5 breast cancer cell lines were immunoblotted with the indicated markers. (B) mRNA level of CAV1 in MCF7 and MDA-MB-231 cells. Quantitative real-time RT-PCR analysis of mRNA levels of CAV1 in the MDA-MB-231 and MCF7 cells. The amount of CAV1 expression in MDA-MB-231 cells was set to 100% (C and D). Lipid raft marker in MDA-MB-231 and MCF7 cells. MDA-MB-231 and MCF7 cells were stained by raft marker CTxB (C) and filipin (D) and observed by confocal microscopy (X600). (E and F) Re-expression of CAV1 in MCF7 recovers lipid raft levels. MCF7 cells with stable expression of EGFP and EGFP-CAV1 were stained by the raft marker CTxB (E) and filipin (F). (G) Re-expression of CAV1 decreases lysosome function. MCF7 cells with stable expression of EGFP and EGFP-CAV1 were incubated with LysoTracker Red and assessed by confocal microscopy (×600). To avoid signal saturation, the power of the green laser was reduced. (H) Re-expression of CAV1 decreases the autophagy level in MCF7 cells. MCF7 cells with stable expression of EGFP and EGFP-CAV1 were treated with CQ (20 μM) for 2 h, then the cell lysates were blotted for the indicated markers. (I) Re-expression of CAV1 decreases cell survival after amino acid starvation. EGFP and EGFP-CAV1 MCF7 cells were treated with or without wortmannin (Wort, 50 nM) under both normal and amino acids starvation conditions after 72 h, then stained with PI. The PtdIns staining was observed through fluorescence microscopy. Scale bar: 200 μm. (J) The percentage of PI-positive cells were counted and analyzed. Three independent experiments were analyzed, Means ±SD are presented (** P < 0.01).
In order to evaluate the role of CAV1 in autophagy regulation, we established a stable cell line in which the MCF7 cells were reconstituted with CAV1. As expected, the lipid raft markers CTxB and filipin staining were recovered in MCF7 cells with CAV1 expression (Figs. 6E and F). Moreover, expression of CAV1 led to reduced LysoTracker Red staining (Fig. 6G). More importantly, the autophagy marker LC3B-II level was decreased in MCF7 cells with exogenous expression of CAV1 with or without CQ treatment (Fig. 6H), consistent with the earlier notion that CAV1 negatively regulates autophagy. Finally, the expression of CAV1 sensitized the MCF7 cells to cell death induced by amino acid starvation (Figs. 6I and J), indicating that deficiency of CAV1 in MCF7 cells promotes cell survival under starvation conditions. In addition, the inhibition of autophagy by wortmannin (Wort) almost totally diminished the prosurvival advantages in CAV1-deficient cells (Figs. 6I and J). These results further confirm that the protective effect of CAV1 deficiency against starvation-induced cell death is autophagy-dependent.
Downregulation of CAV1 with enhanced autophagy in human breast cancer tissues
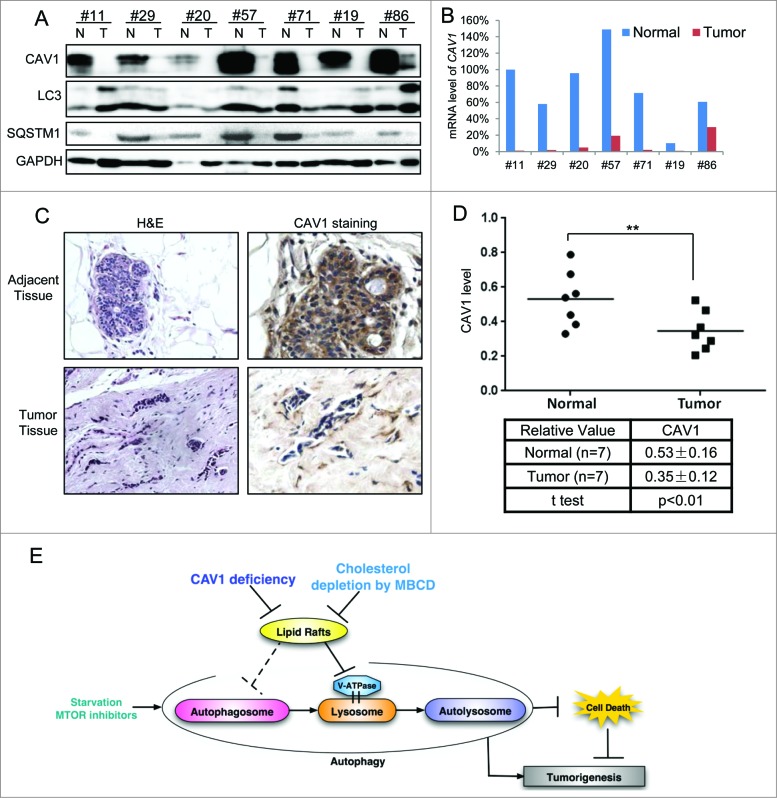
In order to understand the pathological implication of CAV1 in human breast cancer, we analyzed and compared the CAV1 mRNA and CAV1 protein levels in 7 pairs of breast cancer and their adjacent normal tissues. As shown in Figure 7A, we found marked downregulation for CAV1 protein levels in the cancerous tissues. Similarly, the mRNA level of CAV1 was significantly lower or undetectable in the cancerous tissues (Fig. 7B). Consistently, in all of the samples, we observed a lower level of CAV1 in breast tumor tissues (one representative pair was shown in Fig. 7C and the statistical analysis of the semiquantification of the CAV1 level in 7 pairs of tissue samples were shown in Fig. 7D). Since CAV1 is associated with autophagy as shown earlier, we further analyzed and compared the autophagic markers in those human breast cancer and adjacent tissue samples. As shown in Figure 7A, among the 7 pairs tested, LC3B-II level was increased in cancerous tissues in 4 pairs of the samples (#11, #20, #19 and #86), with corresponding reduction of the level of the autophagy substrate SQSTM1/p62 in all of the 7 samples. Taken together, these data suggest that downregulation of CAV1 is associated with upregulated autophagy in human breast cancer tissues.
Figure 7.
Downregulation of CAV1 with enhanced autophagy in human breast cancer tissues. (A) Levels of CAV1 and autophagy markers in breast tumor and adjacent normal tissues. The lysates of 7 pairs of tumors and their adjacent normal tissue samples were immunoblotted with the indicated markers. (B) mRNA level of CAV1 in breast tumor and their adjacent normal tissue samples. Quantitative real-time RT-PCR analysis of mRNA levels of CAV1 in the 7 pairs of tumor and their adjacent normal tissue samples. The amount of CAV1 expression in the normal tissue of the first pair was set to 100%. (C) Immunostaining of CAV1 in paraffin-embedded sections from breast tumor and their adjacent normal samples. Samples were immunostained with anti-CAV1, then counterstained with hematoxylin (400X). The tissue morphology was confirmed by H&E staining (200X). (D) The statistical analysis of 7 pairs of breast tumor and adjacent normal samples. The intensities of CAV1 proteins were quantified using the Motic Images Advanced software, followed by statistical analysis. Means ±SD are presented (** P < 0.01). (E) Model of the regulatory role of lipid rafts and CAV1 in autophagy and the cellular stress response. Loss of CAV1 and the associated disruption of lipid rafts promote autophagy by promoting lysosome function and possibly increasing of autophagosome formation. Inducible autophagy serves as a cell survival mechanism under stress conditions such as starvation.
Discussion
In this study, we report a novel function of CAV1 in modulation of lysosomal function and autophagy. This regulatory function of CAV1 is believed to act via lipid rafts independent of caveolae. As summarized in Figure 7E, CAV1 deficiency and lipid raft disruption promote lysosomal function, increase autophagic flux and promote cell survival under starvation. Moreover, in breast cancer cells and tissues, downregulated CAV1 was associated with an enhanced autophagy level. Thus our data reveal a novel function of CAV1 in cell stress responses and possibly cancer development via modulation of lysosomal function and autophagy.
Caveolae and lipid rafts are structurally and functionally distinct, while CAV1 has been well established to play a critical role in the structure and function of caveolae.6 At present, there are 2 lines of evidence demonstrating that CAV1 is also involved in lipid rafts: (i) CAV1 plays an important role in the regulation of cholesterol homeostasis in various membrane structures,4,6 and (ii) distribution of CAV1 in cellular organelles such as mitochondria, endoplasmic reticulum, and the late endosome/lysosome, has also been well reported.7-9 In this study, we provide convincing evidence demonstrating that the function of CAV1 in autophagy is most probably lipid raft-dependent but caveolae-independent. First, depletion of another critical caveolae regulator PTRF did not affect autophagy (Fig. S1). Second, depletion of cholesterol by MBCD treatment had a similar effect as Cav1 deficiency on autophagy, and moreover, MBCD was unable to further promote autophagy in Cav1-KO cells (Fig. 1I). We acknowledge that treatment with MBCD would have a different impact on lipid rafts than CAV1 deficiency, the fact that MBCD is ineffective in Cav1-KO cells to promote autophagy, indicating the proautophagy function of MBCD and CAV1 deficiency are most likely lipid raft-related. Third, there was partially colocalization of lipid rafts with lysosomal markers, and a large quantity of CAV1 accumulation in the lysosomal fraction (Figs. 3A, B). Based on the fact that caveolae primarily localize at the plasma membrane and PTRF stabilizes CAV1 protein at the plasma membrane,29 it is thus believed that CAV1 regulates lysosomal function and autophagy in a caveolae-independent and lipid raft-dependent manner. Therefore, data from this study demonstrate a novel function of CAV1 in regulation of lysosomal functions and autophagy.
Our results are generally consistent with several earlier studies in which CAV1 and lipid rafts are able to negatively regulate autophagy, via multiple mechanisms. First, in adipocytes, Cav1 deficiency suppresses insulin and lipolytic responses, which will lead to autophagy induction.19 Second, loss of CAV1 promotes autophagy via enhanced oxidative stress and NFKB1/NF-κB activation.20,42 Third, a recent study provides direct evidence to show a competitive interaction of CAV1 and the ATG12–ATG5 system, which suppresses the formation and function of the ATG12–ATG5 complex in lung epithelial cells.43 In contrast, some components of lipid rafts are reported to promote autophagy. For instance, a paradigmatic component of lipid rafts, GD3 ganglioside, plays a positive role in autophagosome formation.25 In addition, another component of lipid rafts, ceramides, has also been implicated in autophagy induction via blockage of AKT activation, upregulation of BECN1 or suppression of nutrient transporter expression.24,26,44 Notably, these studies do not provide the direct evidence to show whether the function of those lipids in autophagy regulation is lipid rafts dependent and whether CAV1 is involved in the regulation of these lipids. It remains to be further determined whether those individual components of lipid rafts act through lipid rafts per se in the regulation of autophagy and whether the CAV1- dependent lipid rafts are affected by the changes of these lipids.
In our attempt to understand the molecular mechanisms underlining the regulatory role of CAV1 in autophagy, we focused our attention on lysosomes. The roles of lipid rafts in different cellular organelles have been reported previously. For instance, the lipid raft adaptor CDKN2C/p18 controls endosome dynamics by anchoring the MAP2K1/7-MAPK1/3 (formerly also known as MEK–ERK) pathway to late endosomes.45 Lipid rafts also play an important role in the trans-Golgi network for membrane trafficking.46 Furthermore, several lysosome membrane proteins associate with lipid rafts, such as V-ATPase, chloride channels CLCN6-CLCN7, and LAMP2A, suggesting a possible role of lipid rafts in regulation of lysosomal functions.35,47 However, the involvement of CAV1 in lysosome regulation is unknown. Here, we provided clear evidence to show the involvement of CAV1 and lipid rafts in the regulation of lysosomal function. Disruption of lipid rafts by CAV1 deficiency and MBCD treatment enhances lysosome function via promotion of V-ATPase assembly (Figs. 2 and 3). It has been reported that CAV1 and lipid rafts localize to late endosome and lysosome membranes.9 CAV1 moves to and from the lysosome membrane during intracellular cholesterol trafficking based on the evidence of increased lysosomal CAV1 accumulation after perturbation of lysosome pH and blockage of cholesterol trafficking.9 Therefore, lysosomes might be critical for the regulation of CAV1 and cholesterol trafficking. However, the functions of CAV1 on lysosomes are not well understood. In some lysosomal storage diseases, a higher level of lipid rafts is found on late endosomes or lysosomes.48,49 A recent study also demonstrates that the abnormal accumulation of 'cholesterol-enriched regions' leads to lysosomal disease.37 Such observations thus suggest the possibility that lipid rafts may act as a negative regulator of lysosomal function. Data from our study are indeed supportive of such a hypothesis. It is thus believed that modulation of CAV1 and lipid rafts regulates lysosome function. Interestingly, the degradation of CAV1 requires acidic lysosomal or late endosomes.49 Here, we showed that the CAV1 deficiency will promote lysosomal function, indicating a positive feedback loop between CAV1 and lysosomal function.
In order to understand the molecular mechanisms underlying the regulatory roles of CAV1 and lipid rafts on lysosomal function, we examined the changes of V-ATPase assembly. V-ATPase is the main determinant in lysosome acidification and functions.34 The assembly of V1 and V0 domains is critical for its enzymatic activity34 and the V-ATPase complex localizes to lipid rafts.35 We found that deficiency of CAV1 and disruption of lipid rafts by MBCD increased the rate of V-ATPase assembly (Fig. 3), suggesting that CAV1 and lipid rafts may hinder the assembly process of the V-ATPase complex. In addition, we have provided evidence showing that both lipid rafts and amino acid starvation lead to the decrease between CAV1 and a V-ATPase V0 subunit (Fig. S7), which provides a possible explanation for the promoted V-ATPase assembly. The membrane bonded V0 subunit localized in lipid rafts and might interact with CAV1 directly. The CAV1 and lipid rafts might prevent the V0 and V1 subunit-assembly process. Thus, disruption of these lipid rafts will release the V0 subunits to be assembled with the V1 subunits. However, this hypothesis still needs to be further confirmed and assessed as to exactly how the lipid rafts regulate the V-ATPase assembly.
Autophagy has been well established as a cell survival mechanism under stress conditions such as starvation.50 Consistently, in this study we found that autophagy mediated by CAV1 deficiency via lipid raft disruption protects against cell death induced by starvation (Figs. 5, 6I, and J). The prosurvival function of autophagy is implicated in cancer development. Autophagy functions as a tumor suppressor during the initiation stage of cancer progression; on the contrary, once a tumor is formed, autophagy in the tumor cells will provide a survival advantage to resist cell death induced by cancer therapeutic agents.51 Based on this understanding, suppression of autophagy using chemical inhibitors becomes a novel approach in cancer therapy, including combinational therapy with autophagy inhibitors and established therapeutic agents.38 CAV1 possesses a tumor suppression function in several types of cancer, especially in breast cancer,10 based on observations that CAV1 is downregulated or exhibits loss of function in mutants, in breast cancer cells.10,14,52 Moreover, in breast cancer, lipid rafts and CAV1 are crucial for some key processes, including apoptosis, cell fate regulation, growth factor receptor signaling, and several important signaling pathways.53–55 For instance, the oncogenic proteins EGFR, ERBB2/HER2, and IGFL are associated with lipid rafts.55-57 Targeting lipid rafts may have an anticancer effect by promoting the internalization and degradation of those oncogenic proteins.58 In this study, we also found downregulated CAV1 levels in both breast cancer cell lines and tissue samples (Figs. 6A, 7A, and D). Our data also indicate that a higher autophagy level correlated with lower CAV1 expression in the breast cancer tissues. Therefore, breast cancer cells use autophagy as a prosurvival mechanism to resist cell death, especially those cancer cells with lower levels of CAV1 and impaired lipid raft function. The results presented in this study also suggest a possible explanation for the poor clinical outcomes of those breast cancer patients with CAV1 deficiency, as reported previously.10,52,59 At present, there is no reported biomarker in the selection of cancer patients for autophagy-targeted chemotherapies. Thus, our study indicates that those breast cancer patients with CAV1 deficiency might be more suitable for autophagy-inhibitor therapies.
In summary, our data reveal a novel function of CAV1 in cell stress responses and breast cancer development via regulation of lysosomal functions and autophagy, a process dependent on lipid rafts but independent of caveolae. Understanding such a novel function of CAV1 provides a clue for suppressing autophagy as a potential therapeutic approach, especially for breast cancer patients with deficient CAV1 expression.
Materials and Methods
Reagents and antibodies
The chemicals used in our experiments were: methyl-β-cyclodextrin (MBCD; Sigma, C4555), cholesterol-water soluble (Sigma, C4951), chloroquine diphosphate (CQ, Sigma, C6628), bafilomycin A1 (Baf, Sigma, B1793), rapamycin (Sigma, R8781), cholera toxin subunit B conjugated with Alexa Fluor 594 (CTxB, Invitrogen, C34777), LysoTracker® Green DND-26 (Invitrogen, L7526), LysoTracker® Red DND-99 (Invitrogen, L7528), Magic RedTM CTSL reagent (Immunochemistry Technologies, LLC, 942), DQTM Red BSA (dye quenched; Molecular Probes, D12051). The antibodies used in our experiments were: anti-MAP1LC3B/LC3B (Sigma, L7543), anti-SQSTM1/p62 (Abnova, H00008878–2C11), anti-ACTB/β-actin (Sigma, A5441), p-RPS6 (S235/236) (Cell Signaling Technology, 2211), anti-TFRC/transferrin receptor (Invitrogen, 136800), anti-CAV1/caveolin-1 (BD Pharmingen, 610059), anti-TSC2 (Cell Signaling Technology, 3990s), mouse anti-ATP6V1B2/V-ATPase B2 (Santa Cruz Biotechnology, sc-166122), goat anti-ATP6V0D1/V-ATPase D1 (Santa Cruz Biotechnology, sc-69105), anti-PTRF/Cavin-1 (Merck, ABT131).
Cell culture
MEFs, MCF7, and L929 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma, D7777) with 10% fetal bovine serum (Hyclone, SH30071.03) and 1% penicillin-streptomycin (Pan-Biotech, P06–07100) (defined as normal medium in this study) in a 5% CO2 incubator at 37°C. The Cav1+/+ and cav1−/− MEFs were described previously.27 MCF-7 cells (1 × 106 cells) were transfected with 5 μg pEGFP DNA and 5 μg pEGFP-CAV1 DNA using Lipofectamine 2000 (Life Technologies, 11668) according to the manufacturer's instructions. G418 (Sigma Aldrich, A1720) was used as a selection drug at 500 μg/ ml for 3 wk to establish stable lines. PTRF WT and KO MEFs have been previously described.60 The Atg5+/+ and atg5−/− and Atg5 Tet-off inducible MEFs (m5–7) with stably expressing GFP-LC3B were all kindly provided by Dr. N. Mizushima (Tokyo Medical and Dental University). The Tsc2+/+ and tsc2−/− MEFs were a generous gift from Dr. D. Kwiatkowski (Harvard Medical School). The tfLC3B stably transfected L929 cells have been reported previously.61
Plasmids
The mRFP-GFP tandem fluorescence-tagged LC3B construct (tfLC3B) was a kind gift provided by Dr. T. Yoshimori (Osaka University).36
Western blotting
After the designated treatments, Laemmli SDS buffer were used to lyse the collected samples (62.5 mM Tris, pH 6.8, 25% glycerol, 2% SDS [Vivantis, PB0640], phosphatase inhibitor [Thermo Scientific, 78428] and proteinase inhibitor cocktail [Roche, 11697498001]). An equal amount of protein of each sample was resolved by SDS-PAGE, and then the SDS-PAGE gels were transferred onto PVDF membrane (Bio-Rad, 162–0177). Membranes were blocked for 30 min with blocking buffer (Thermo, 37538) and followed by incubations with the indicated primary and secondary antibodies. Then the membranes were visualized with the enhanced chemiluminescence method (Thermo Scientific, 34076) by the ImageQuant LAS 500 (GE).
Quantitative real-time RT-PCR
The CAV1 primer set (CAV1: forward, CGTAGACTCGGAGGGACATC and reverse, ACTTGCTTCTCGCTCAGCTC) were used for quantitative RT-PCR. SsoFast EvaGreen (Bio-Rad, 1725201AP) was used in a thermal cycler (model C1000TM, Bio-Rad). All samples were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression levels.
Measurement of intralysosomal function using LysoTracker
The intralysosomal function was estimated by LysoTracker.62,63 Cells were cultured on coverglass slide chamber, and then followed with the designated treatments. After 30 min incubation with 50 nM LysoTracker® Green DND-26 or LysoTracker® Red DND-99, the fluorescence intensity was observed under a confocal microscope (Olympus Fluoview FV1000, Shinjuku-ku, Tokyo, Japan) and representative cells from 3 fields of view were selected and photographed. All the images were single focal planes.
Cathepsin activity assay
The CTSL enzymatic activity was measured by Magic RedTM reagent (ImmunoChemistry technologies, 942) with the method described earlier with slight modifications.64 Cells were cultured in 24-well plates and treated with indicated treatments. Then cells were further loaded with Magic RedTM CTSL reagent for 15 min. The fluorescence intensities of 10,000 cells per sample were quantified by a BD FACS cytometer (BD Biosciences).
Cell fractionation
Isolation of detergent-resistant lipid raft fraction from the detergent-soluble fraction was performed based on a previously conducted study with minor modifications.31 Briefly, after indicated treatments, cells were washed with ice-cold phosphate-buffered saline (PBS [Vivantis, PB0344]; 137mM NaCl, 2.7mM KCl and 10mM Phosphate Buffer, pH 7.4) and collected by centrifugation at 400 g at 4°C for 3 min. Then cells were homogenized with TNE buffer (25 mM Tris HCl, pH 7.4, 150 mM NaCl, 3 mM EDTA, and protease inhibitor cocktail) with 1% Triton X-100 (Sigma, X100) by passage through a 27-gauge needle for 20 times on ice. Cells were collected, lysed and kept on ice for 30 min. Afterwards, samples were spun down at 16,000 g at 4°C for 20 min, and the supernatant fractions were collected as the DSF. The insoluble pellet fractions were resuspended and lysed in Laemmli SDS buffer, and were regarded as the DRF. The isolation of lysosomes was performed according to the manufacturer's protocol of the Lysosome Enrichment Kit for Tissue and Cultured Cells (Thermo Scientific, 89839). Three × 150-mm2 cell culture dishes were used for each indicated treatment. The isolated lysosome fractions were then subjected to western blot analysis.
Visualization of lipid rafts by CTxB staining
The lipid rafts were stained with CTxB conjugated with Alexa Fluor 594 as reported previously with minor modifications.28 Briefly, MEFs were seeded on coverglass slide chambers. After the incubation and indicated treatments, cells were loaded with 1 μg/mL CTxB for 15 min on ice and then washed with PBS and followed by a 30 min incubation at 37°C. The fluorescence intensity was observed under a confocal microscope (Olympus Fluoview FV1000) and representative cells from 3 fields of view were photographed. All the images were single focal planes.
Visualization of lipid rafts by filipin staining
For cholesterol staining, the cells were labeled by Filipin III (Sigma, F4767) as reported previously.65 Briefly, cells were seeded and treated on coverglass slide chambers. After treatments, cells were fixed with 4% paraformaldehyde for 30 min and quenched in 50 mM NH4Cl3 for 10 min. Then cells were blocked, permeabilized and stained in the solution which containing 0.2% BSA (Sigma, A7906), 0.2% fish skin gelatin (Sigma, G7041) with 50 μm of filipin III for 20 min. After 3 washes in PBS, the fluorescence intensity was detected under a confocal microscope (Olympus Fluoview FV1000). Representative cells from 3 fields of view were selected and photographed. All the images were single focal planes.
Proteolysis activity assay
Proteolysis activity was performed by DQTM Red BSA staining, conducted with the methods reported previously.66 MEFs were cultured on coverglass slide chambers and preloaded with DQ-BSA (10 μg/ml) for 1 h. After 3 washes with PBS, the medium was changed to the designated treatments. Confocal microscopy (Olympus Fluoview FV1000) was used to detect the fluorescence intensity. The representative cells from 3 fields of view were selected and photographed. All the images were single focal planes.
Proximity ligation assay (PLA)
Cells were cultured in coverglass slide chambers and treated as indicated. After fixation with 4% paraformaldehyde and permeabilization by 0.1% saponin (Sigma, 47036), the cells were subjected to PLA by Duolink detection kit (Olink Bioscience, [PLA Probe Anti-Goat MINUS, 92006; PLA Probe Anti-Mouse PLUS, 92001; Detection Reagent Red, 92008]).67 Briefly, permeabilized cells were blocked and incubated with mouse anti-ATP6V1B2 and goat anti-ATP6V0D1 antibodies overnight at 4°C. Then after incubation with secondary antibodies conjugated to unique DNA probes (anti-mouse and anti-goat for 2 primary antibodies, provided by the kit), a rolling circle amplification step was used to subject to the proximity ligation (<40 nm) and circularization of the DNA. After the amplification process, replications of the DNA circle were labeled by complementary oligonucleotide probes and the signals were observed under a confocal microscope (Olympus Fluoview FV1000). Representative cells from 3 fields of view were selected and photographed. All the images were single focal planes.
CAV1 immunohistochemistry
The paraffin-embedded tissue sections of 7 breast cancer samples and their adjacent normal tissue samples were collected from a Tissue Repository (NUH, Singapore) for determination of CAV1 expression. Sections are dewaxed and rehydrated using standard dewaxing protocol. Then the samples were subjected to heat antigen retrieval by exposure to 10 mM citric acid buffer (pH 6.0 for 15 min at 105°C). Endogenous peroxidase activity was blocked by a 10 min incubation with 3% H2O2. Then the samples were blocked by 3% BSA and incubated with the 1:100 dilution of the anti-CAV1 primary antibody overnight at room temperature (RT) in a humid chamber. Envision HRP anti-rabbit polymer (Dako, K401011) was added for 30 min at RT. Color is developed by incubation with DAB (Dako, K401011) for 5 min, followed a counterstain with Mayer haematoxylin (Sigma, MHS1). Then the sections were dehydrated through ascending graded alcohols, cleared in xylene, mounted and visualized. Representative regions were selected and photographed. To quantify the immunohistochemistry result of positive staining intensity, 3 observations of each sample were analyzed by an experienced pathologist. Motic Images Advanced software (version 3.2, Motic China Group) was used to analyze the protein level. Mean values ± SD were presented. Student t tests were used to identify the significance of differences.
Detection of cell death
Several methods were used to detect cell death quantitatively and qualitatively, which are (i) morphological changes under phase-contrast microscopy, (ii) PtdIns exclusion assay coupled with flow cytometry (5 μg/ml). (iii) PtdIns staining observed under fluorescence microscopy. For the PtdIns exclusion assay, trypsinized cells with the medium in each well were collected. The pellets were resuspended into PBS containing 5 μg/ml PtdIns. Then 10,000 cells from each sample were analyzed by using a BD FACS cytometer (BD Biosciences). For PtdIns staining assay, cells stained with PtdIns (5 μg/ml). Then, the cells with PtdIns staining were observed through fluorescent microscope. The percentage of PtdIns-positive cells were counted and analyzed by ImageJ.
Measurement of lysosome pH
Several methods were used to evaluate lysosome pH, which are (1) LysoSensor Green DND189 (1 μM; Life Technologies, L7535) staining observed under a confocal microscope; (2) LysoSensor Green DND189 (1 μM) staining coupled with flow cytometry; (3) lysosome pH quantification. For lysosome pH quantification, briefly, cells were seeded in 96-well plates and labeled with 2 μM LysoSensor Yellow/Blue DND160 (Life Technologies, L7545) for 45 min in regular medium, then washed in HEPES solution (10 mM HEPES, 133.5 mM NaCl, 2.0 mM CaCl2, 4.0 mM KCl, 1.2 mM MgSO4, 1.2 mM NaH2PO4, 11 mM glucose, pH 7.4). The fluorescence emitted at 440 and 535 nm in response to excitation at 340 and 380 nm was measured immediately by a microplate reader at 37°C. The ratio of light emitted with 340 and 380 nm excitation was calculated for each well. The pH calibration curve for the fluorescence probe was generated by using the 2-(N-morpholino) ethanesulfonic acid (MES) calibration buffer (10 μM monensin [Sigma, M5273], 10 M nigericin [Sigma, N7143], 25 mM MES [Sigma, M2933], 5 mM NaCl, 115 mM KCl and 1.2 mM MgSO4, pH 3.5 to 6.0).
Statistical analysis
All image and western blot data presented above are representatives from 3 independent experiments. The numeric data are presented as means ± SD from at least 3 independent experiments and analyzed by the Student t test.
Acknowledgments
We thank NUH-NUS Tissue Repository (TR) for providing the tissue samples, and the personnel of the NUHS Research Core Facility for technical assistance. We thank Dr. David J. Kwiatkowski for providing Tsc2+/+ and tsc2−/− MEFs. We thank Dr. N Mizushima for providing the atg5−/− MEFs and Atg5 Tet-off inducible MEFs with stable expression of GFP-LC3B.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by research grants from Singapore National Medical Research Council (NMRC-1260/2010 and CIRG/1346/2012). The work in YSK's lab was supported in part by the National Research Foundation of Korea (NRF, No. 2011–0030043).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Nassar ZD, Hill MM, Parton RG, Parat MO. Caveola-forming proteins caveolin-1 and PTRF in prostate cancer. Nat Rev Urol 2013; 10:529-36; PMID:23938946; http://dx.doi.org/ 10.1038/nrurol.2013.168 [DOI] [PubMed] [Google Scholar]
- 2.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010; 327:46-50; PMID:20044567; http://dx.doi.org/ 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- 3.Allen JA, Halverson-Tamboli RA, Rasenick MM. Lipid raft microdomains and neurotransmitter signalling. Nat Rev Neurosci 2007; 8:128-40; PMID:17195035; http://dx.doi.org/ 10.1038/nrn2059 [DOI] [PubMed] [Google Scholar]
- 4.Bosch M, Mari M, Herms A, Fernandez A, Fajardo A, Kassan A, Giralt A, Colell A, Balgoma D, Barbero E, et al.. Caveolin-1 deficiency causes cholesterol-dependent mitochondrial dysfunction and apoptotic susceptibility. Curr Biol 2011; 21:681-6; PMID:21497090; http://dx.doi.org/ 10.1016/j.cub.2011.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence JC, Saslowsky DE, Edwardson JM, Henderson RM. Real-time analysis of the effects of cholesterol on lipid raft behavior using atomic force microscopy. Biophys J 2003; 84:1827-32; PMID:12609884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 2010; 49:6305-16; PMID:20583817; http://dx.doi.org/ 10.1021/bi100882y [DOI] [PubMed] [Google Scholar]
- 7.Schlegel A, Arvan P, Lisanti MP. Caveolin-1 binding to endoplasmic reticulum membranes and entry into the regulated secretory pathway are regulated by serine phosphorylation. Protein sorting at the level of the endoplasmic reticulum. J Biol Chem 2001; 276:4398-408; PMID:11078729; http://dx.doi.org/ 10.1074/jbc.M005448200. [DOI] [PubMed] [Google Scholar]
- 8.Robenek MJ, Severs NJ, Schlattmann K, Plenz G, Zimmer KP, Troyer D, Robenek H. Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J 2004; 18:866-8; PMID:15001554; http://dx.doi.org/ 10.1096/fj.03-0782fje [DOI] [PubMed] [Google Scholar]
- 9.Mundy DI, Li WP, Luby-Phelps K, Anderson RG. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Mol Biol Cell 2012; 23:864-80; PMID:22238363; http://dx.doi.org/ 10.1091/mbc.E11-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan EK, Ciocca DR, Pouliot N, Natoli A, Restall C, Henderson MA, Fanelli MA, Cuello-Carrion FD, Gago FE, Anderson RL. Stromal cell expression of caveolin-1 predicts outcome in breast cancer. A J Pathol 2009; 174:2035-43; PMID:19411449; http://dx.doi.org/ 10.2353/ajpath.2009.080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman JA, Wykoff CC, Yasuhara S, Song KS, Okamoto T, Lisanti MP. Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem 1997; 272:16374-81; PMID:9195944 [DOI] [PubMed] [Google Scholar]
- 12.Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG, Lisanti MP. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J 1998; 17:6633-48; PMID:9822607; http://dx.doi.org/ 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sotgia F, Williams TM, Schubert W, Medina F, Minetti C, Pestell RG, Lisanti MP. Caveolin-1 deficiency (−/−) conveys premalignant alterations in mammary epithelia, with abnormal lumen formation, growth factor independence, and cell invasiveness. A J Pathol 2006; 168:292-309; PMID:16400031; http://dx.doi.org/ 10.2353/ajpath.2006.050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai L, Deng X, Li Q, Wang M, An W, Deli A, Gao Z, Xie Y, Dai Y, Cong YS. Down-regulation of the cavin family proteins in breast cancer. J Cell Biochem 2012; 113:322-8; PMID:21913217; http://dx.doi.org/ 10.1002/jcb.23358. [DOI] [PubMed] [Google Scholar]
- 15.Li T, Sotgia F, Vuolo MA, Li M, Yang WC, Pestell RG, Sparano JA, Lisanti MP. Caveolin-1 Mutations in Human Breast Cancer. A J Pathol 2006; 168:1998-2013; http://dx.doi.org/ 10.2353/ajpath.2006.051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H, Park DS, Razani B, Russell RG, Pestell RG, Lisanti MP. Caveolin-1 Mutations (P132L and Null) and the Pathogenesis of Breast Cancer. A J Pathol 2002; 161:1357-69; http://dx.doi.org/ 10.1016/s0002-9440(10)64412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, et al.. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell 2011; 146:148-63; PMID:21729786; http://dx.doi.org/ 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709-30; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Lay S, Briand N, Blouin CM, Chateau D, Prado C, Lasnier F, Le Liepvre X, Hajduch E, Dugail I. The lipoatrophic caveolin-1 deficient mouse model reveals autophagy in mature adipocytes. Autophagy 2010; 6:754-63; PMID:20574167 [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Outschoorn UE, Whitaker-Menezes D, Lin Z, Flomenberg N, Howell A, Pestell RG, Lisanti MP, Sotgia F. Cytokine production and inflammation drive autophagy in the tumor microenvironment: Role of stromal caveolin-1 as a key regulator. Cell Cycle 2011; 10:1784-93; http://dx.doi.org/ 10.4161/cc.10.11.15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calay D, Vind-Kezunovic D, Frankart A, Lambert S, Poumay Y, Gniadecki R. Inhibition of Akt signaling by exclusion from lipid rafts in normal and transformed epidermal keratinocytes. J Invest Dermatol 2010; 130:1136-45; PMID:20054340; http://dx.doi.org/ 10.1038/jid.2009.415. [DOI] [PubMed] [Google Scholar]
- 22.Grider MH, Park D, Spencer DM, Shine HD. Lipid raft-targeted Akt promotes axonal branching and growth cone expansion via mTOR and Rac1, respectively. J Neurosci Res 2009; 87:3033-42; PMID:19530170; http://dx.doi.org/ 10.1002/jnr.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res 2013; 23:508-23; PMID:23337583; http://dx.doi.org/ 10.1038/cr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peralta ER, Edinger AL. Ceramide-induced starvation triggers homeostatic autophagy. Autophagy 2009; 5:407-9; PMID:19202357 [DOI] [PubMed] [Google Scholar]
- 25.Matarrese P, Garofalo T, Manganelli V, Gambardella L, Marconi M, Grasso M, Tinari A, Misasi R, Malorni W, Sorice M. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 2014; 10(5):750-65; PMID:24589479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem 2004; 279:18384-91; PMID:14970205; http://dx.doi.org/ 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 27.Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, et al.. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem 2001; 276:38121-38; PMID:11457855; http://dx.doi.org/ 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi H, Takeo Y, Yoshida S, Kouchi Z, Nakamura Y, Fukami K. Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res 2009; 69:8594-602; PMID:19887621; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2305. [DOI] [PubMed] [Google Scholar]
- 29.Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, et al.. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 2008; 132:113-24; PMID:18191225; http://dx.doi.org/ 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilsdonk EP, Yancey PG, Stoudt GW, Bangerter FW, Johnson WJ, Phillips MC, Rothblat GH. Cellular cholesterol efflux mediated by cyclodextrins. J Biol Chem 1995; 270:17250-6; PMID:7615524 [DOI] [PubMed] [Google Scholar]
- 31.Lingwood D, Simons K. Detergent resistance as a tool in membrane research. Nat Protoc 2007; 2:2159-65; PMID:17853872; http://dx.doi.org/ 10.1038/nprot.2007.294. [DOI] [PubMed] [Google Scholar]
- 32.Ng S, Wu YT, Chen B, Zhou J, Shen HM. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy 2011; 7:1173-86; PMID:21808151; http://dx.doi.org/ 10.4161/auto.7.10.16681. [DOI] [PubMed] [Google Scholar]
- 33.Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci 2014; 39:61-71; PMID:24369758; http://dx.doi.org/ 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 34.Mindell JA. Lysosomal acidification mechanisms. Ann Rev Physiol 2012; 74:69-86; PMID:22335796; http://dx.doi.org/ 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- 35.Yoshinaka K, Kumanogoh H, Nakamura S, Maekawa S. Identification of V-ATPase as a major component in the raft fraction prepared from the synaptic plasma membrane and the synaptic vesicle of rat brain. Neurosci Lett 2004; 363:168-72; PMID:15172108; http://dx.doi.org/ 10.1016/j.neulet.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; PMID:17534139 [DOI] [PubMed] [Google Scholar]
- 37.Fraldi A, Annunziata F, Lombardi A, Kaiser HJ, Medina DL, Spampanato C, Fedele AO, Polishchuk R, Sorrentino NC, Simons K, et al.. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J 2010; 29:3607-20; PMID:20871593; http://dx.doi.org/ 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 2013; 73:3-7; PMID:23288916; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 39.Shen HM, Codogno P. Autophagy is a survival force via suppression of necrotic cell death. Exp Cell Res 2012; 318:1304-8; PMID:22366289; http://dx.doi.org/ 10.1016/j.yexcr.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy 2013; 9:1270-85; PMID:23846383; http://dx.doi.org/ 10.4161/auto.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen H-M, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy 2011; 7:457-65; http://dx.doi.org/ 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- 42.Shiroto T, Romero N, Sugiyama T, Sartoretto JL, Kalwa H, Yan Z, Shimokawa H, Michel T. Caveolin-1 is a critical determinant of autophagy, metabolic switching, and oxidative stress in vascular endothelium. PloS one 2014; 9:e87871; PMID:24498385; http://dx.doi.org/ 10.1371/journal.pone.0087871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, Li W, Choi AM, Shen HH. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol 2014; 306(11):L1016-25; PMID:24727585; http://dx.doi.org/ 10.1152/ajplung.00268.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert KM, Scheid MP, Duronio V. Ceramide inhibits protein kinase B/Akt by promoting dephosphorylation of serine 473. J Biol Chem 2000; 275:13330-5; PMID:10788440 [DOI] [PubMed] [Google Scholar]
- 45.Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J 2009; 28:477-89; PMID:19177150; http://dx.doi.org/ 10.1038/emboj.2008.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, et al.. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol 2009; 185:601-12; PMID:19433450; http://dx.doi.org/ 10.1083/jcb.200901145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A 2003; 100:5813-8; PMID:12724530; http://dx.doi.org/ 10.1073/pnas.0631608100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons K, Gruenberg J. Jamming the endosomal system: lipid rafts and lysosomal storage diseases. Trends Cell Biol 2000; 10:459-62; PMID:11050411 [DOI] [PubMed] [Google Scholar]
- 49.Hayer A, Stoeber M, Ritz D, Engel S, Meyer HH, Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J Cell Biol 2010; 191:615-29; PMID:21041450; http://dx.doi.org/ 10.1083/jcb.201003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meijer AJ, Codogno P. Autophagy: regulation and role in disease. Crit Rev Clin Lab Sci 2009; 46:210-40; PMID:19552522; http://dx.doi.org/ 10.1080/10408360903044068 [DOI] [PubMed] [Google Scholar]
- 51.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12:401-10; PMID:22534666; http://dx.doi.org/ 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witkiewicz AK, Dasgupta A, Sammons S, Er O, Potoczek M, Guiles F, Sotgia F, Brody JR, Mitchell EP, Lisanti MP. Loss of stromal caveolin-1 expression predicts poor clinical outcome in triple negative and basal-like breast cancers. Cancer Biology & Therapy 2010; 10:135-43; http://dx.doi.org/ 10.4161/cbt.10.2.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gajate C, Mollinedo F. Cytoskeleton-mediated death receptor and ligand concentration in lipid rafts forms apoptosis-promoting clusters in cancer chemotherapy. J Biol Chem 2005; 280:11641-7; PMID:15659383; http://dx.doi.org/ 10.1074/jbc.M411781200. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Resh MD. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J Biol Chem 2002; 277:49631-7; PMID:12397069; http://dx.doi.org/ 10.1074/jbc.M208327200 [DOI] [PubMed] [Google Scholar]
- 55.Freeman MR, Cinar B, Kim J, Mukhopadhyay NK, Di Vizio D, Adam RM, Solomon KR. Transit of hormonal and EGF receptor-dependent signals through cholesterol-rich membranes. Steroids 2007; 72:210-7; PMID:17173942; http://dx.doi.org/ 10.1016/j.steroids.2006.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chinni SR, Yamamoto H, Dong Z, Sabbota A, Bonfil RD, Cher ML. CXCL12/CXCR4 transactivates HER2 in lipid rafts of prostate cancer cells and promotes growth of metastatic deposits in bone. Mol Cancer Res 2008; 6:446-57; PMID:18337451; http://dx.doi.org/ 10.1158/1541-7786.MCR-07-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manes S, Mira E, Gomez-Mouton C, Lacalle RA, Keller P, Labrador JP, Martinez AC. Membrane raft microdomains mediate front-rear polarity in migrating cells. The EMBO Journal 1999; 18:6211-20; PMID:10562533; http://dx.doi.org/ 10.1093/emboj/18.22.6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee EJ, Yun UJ, Koo KH, Sung JY, Shim J, Ye SK, Hong KM, Kim YN. Down-regulation of lipid raft-associated onco-proteins via cholesterol-dependent lipid raft internalization in docosahexaenoic acid-induced apoptosis. Biochim Biophys Acta 2014; 1841:190-203; PMID:24120917; http://dx.doi.org/ 10.1016/j.bbalip.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 59.Ma X, Liu L, Nie W, Li Y, Zhang B, Zhang J, Zhou R. Prognostic role of caveolin in breast cancer: A meta-analysis. Breast 2013; 22(4):462-9; PMID:23639584; http://dx.doi.org/ 10.1016/j.breast.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 60.Liu L, Brown D, McKee M, Lebrasseur NK, Yang D, Albrecht KH, Ravid K, Pilch PF. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab 2008; 8:310-7; PMID:18840361; http://dx.doi.org/ 10.1016/j.cmet.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu YT, Tan HL, Huang Q, Ong CN, Shen HM. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy 2009; 5:824-34; PMID:19556857 [DOI] [PubMed] [Google Scholar]
- 62.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7:167-78; PMID:15296714 [DOI] [PubMed] [Google Scholar]
- 64.Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 2011; 7:188-204; PMID:21107021; http://dx.doi.org/ 10.4161/auto.7.2.14181 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inder KL, Zheng YZ, Davis MJ, Moon H, Loo D, Nguyen H, Clements JA, Parton RG, Foster LJ, Hill MM. Expression of PTRF in PC-3 Cells modulates cholesterol dynamics and the actin cytoskeleton impacting secretion pathways. Mol Cell Proteomics 2012; 11:M111 012245; PMID:22030351; http://dx.doi.org/ 10.1074/mcp.M111.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ha SD, Ham B, Mogridge J, Saftig P, Lin S, Kim SO. Cathepsin B-mediated autophagy flux facilitates the anthrax toxin receptor 2-mediated delivery of anthrax lethal factor into the cytoplasm. J Biol Chem 2010; 285:2120-9; PMID:19858192; http://dx.doi.org/ 10.1074/jbc.M109.065813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JY, Park JH, Lee S. GLTSCR2 contributes to the death resistance and invasiveness of hypoxia-selected cancer cells. FEBS Lett 2012; 586(19):3435-40; PMID:22850112; http://dx.doi.org/ 10.1016/j.febslet.2012.07.064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.