Abstract
By monitoring the fragmentation of a GST-BHMT (a protein fusion of glutathionine S-transferase N-terminal to betaine-homocysteine S-methyltransferase) reporter in lysosomes, the GST-BHMT assay has previously been established as an endpoint, cargo-based assay for starvation-induced autophagy that is largely nonselective. Here, we demonstrate that under nutrient-rich conditions, proteasome inhibition by either pharmaceutical or genetic manipulations induces similar autophagy-dependent GST-BHMT processing. However, mechanistically this proteasome inhibition-induced autophagy is different from that induced by starvation as it does not rely on regulation by MTOR (mechanistic target of rapamycin [serine/threonine kinase]) and PRKAA/AMPK (protein kinase, AMP-activated, α catalytic subunit), the upstream central sensors of cellular nutrition and energy status, but requires the presence of the cargo receptors SQSTM1/p62 (sequestosome 1) and NBR1 (neighbor of BRCA1 gene 1) that are normally involved in the selective autophagy pathway. Further, it depends on ER (endoplasmic reticulum) stress signaling, in particular ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1) and its main downstream effector MAPK8/JNK1 (mitogen-activated protein kinase 8), but not XBP1 (X-box binding protein 1), by regulating the phosphorylation-dependent disassociation of BCL2 (B-cell CLL/lymphoma 2) from BECN1 (Beclin 1, autophagy related). Moreover, the multimerization domain of GST-BHMT is required for its processing in response to proteasome inhibition, in contrast to its dispensable role in starvation-induced processing. Together, these findings support a model in which under nutrient-rich conditions, proteasome inactivation induces autophagy-dependent processing of the GST-BHMT reporter through a distinct mechanism that bears notable similarity with the yeast Cvt (cytoplasm-to-vacuole targeting) pathway, and suggest the GST-BHMT reporter might be employed as a convenient assay to study selective macroautophagy in mammalian cells.
Keywords: BHMT, cargo receptors SQSTM1/p62 and NBR1, MTOR, PRKAA/AMPK, proteasome inhibition, selective macroautophagy
Abbreviations: ACACA/B, acetyl-CoA carboxylase α/β; ACTB, actin, β; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; ATG7, autophagy-related 7; Baf A1, bafilomycin A1; BCL2, B-cell CLL/lymphoma 2; BECN1, Beclin 1, autophagy-related; BHMT, betaine-homocysteine S-methyltransferase; CTNNB1, catenin (cadherin-associated protein), β 1, 88kDa; Cvt, cytoplasm-to-vacuole-targeting; DDIT3, DNA-damage-inducible transcript 3; EBSS, Earle's Balanced Salt Solution; EIF2AK3, eukaryotic translation initiation factor 2-α, kinase 3; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; ER, endoplasmic reticulum; ERN1, endoplasmic reticulum to nucleus signaling 1; GST, glutathionine S-transferase; GST-BHMT, a fusion protein of glutathionine S-transferase N-terminal to betaine-homocysteine S-methyltransferase; GST-BHMT(FRAG), an autophagy-mediated cleavage product of the GST-BHMT reporter; HA, hemagglutinin; HSPA5, heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa); LSCS, linker-specific cleavage site; MAP1LC3, microtubule-associated protein 1 light chain 3; MAP2K7, mitogen-activated protein kinase kinase 7; MAPK8, mitogen-activated protein kinase 8; MTOR, mechanistic target of rapamycin (serine/threonine kinase); MTORC1, MTOR complex 1; NBR1, neighbor of BRCA1 gene 1; P4HB, prolyl 4-hydroxylase, β polypeptide; PRKAA, protein kinase, AMP-activated, α catalytic subunit; RHEB, Ras homolog enriched in brain; RM, rich medium; RPS6KB1, ribosomal protein S6 kinase, 70kDa, polypeptide 1; SQSTM1, sequestosome 1; TSC1/2, tuberous sclerosis 1/2; ULK1, unc-51 like autophagy activating kinase 1; UPR, unfolded protein response; UPS, ubiquitin proteasome system; XBP1, X-box binding protein 1
Introduction
Autophagy is a catabolic process characterized by the delivery of cytosolic constituents into the lysosome for degradation and recycling.1 Depending on how the substrates are delivered, autophagy in general can be categorized into 3 classes: microautophagy, chaperone-mediated autophagy, and macroautophagy.1 Macroautophagy, the most well-studied autophagic process, is characterized by the engulfment of cytoplasmic materials within a double-membrane vesicle known as an autophagosome for eventual fusion and delivery of its targets to the lysosome. Based on the nature of its targets, macroautophagy (hereafter referred to as autophagy) can be further subdivided into nonselective and selective pathways.2,3 Typically, nutrition deprivation induces a strong adaptive autophagic response that is considered to be nonselective, as it involves bulk engulfment and degradation of nonessential cellular components including proteins and organelles to provide energy and critical building blocks such as amino acids to sustain basic cell metabolism and survival. Conversely, selective autophagy targets specific substrates including protein aggregates (also called aggrephagy) and damaged organelles such as mitochondria (mitophagy) and peroxisomes (pexophagy).2,3
Extensive studies in different model systems have provided critical insights into the mechanisms of autophagic activation and have uncovered many components of the highly conserved nonselective autophagy pathway. For example, in both yeast and mammals, the conserved Atg1/ULK1 (unc-51 like autophagy activating kinase 1) complex plays a critical role in the initiation of the autophagy cascade and the subsequent bulk degradation process, where its kinase activity is tightly regulated to accommodate different cellular demands.4 The serine/threonine kinase MTORC1 (mechanistic target of rapamycin [serine/threonine kinase] complex 1), one well-studied upstream regulator, is the master sensor of cellular nutrient status and controls the activation of the starvation-induced nonselective autophagic response.5 Under favorable conditions, active MTORC1 suppresses ULK1 by phosphorylating it at Ser757.6 Upon nutrient starvation, MTORC1 activity becomes subdued, which in turn activates ULK1 leading to subsequent autophagic initiation. Similarly, energy depletion leads to the activation of the master energy sensor PRKAA/AMPK (protein kinase, AMP-activated, α catalytic subunit), which in turn promotes autophagy through stimulatory phosphorylation of ULK1 at Ser555.6
Although the selective autophagy process shares many components with the nonselective autophagic pathway, it is often regulated differently, showing less conservation among different model organisms. Selective autophagy is still not well-defined, especially in mammalian systems. One of the best characterized selective autophagy pathways is the cytoplasm-to-vacuole targeting (Cvt) pathway in yeast.7 This process requires the assembly of the precursor Ape1 (prApe1) into a dodecamer in the cytoplasm before its delivery to the vacuole, the yeast equivalent of the lysosome, for its final maturation. Yeast genetic screens have revealed that in addition to a common set of genes involved in starvation-induced autophagy, the Cvt pathway also requires several Cvt pathway-specific components such as Atg19 and Atg11. Atg19 functions as a receptor for the prApe1 dodecamer cargos, while Atg11 acts as a scaffold protein to facilitate the interaction between Atg19 and the autophagosome protein Atg8.7 Similarly, a genome-wide RNAi screen in C. elegans led to the identification of another group of novel components required for the autophagy-dependent degradation of P-granules.8 Notably, in both the Cvt and P-granule pathways, sequestration of cargos into autophagosomes is likely ubiquitin-independent,7,9 whereas in the mammalian system, cargos that are sequestered by the selective pathway often contain specific modifications such as ubiquitination.10 In particular, selective autophagy often requires the presence of receptor proteins such as SQSTM1/p62 (sequestosome 1) and NBR1 (neighbor of BRCA1 gene 1), the mammalian ortholog of yeast Atg19, which contains both a ubiquitin binding domain and a MAP1LC3 (microtubule-associated protein 1 light chain 3)-interacting motif to bridge the sequestration of ubiquitin-modified cargos into the autophagosome.11
Another important role of the autophagic response is to maintain intracellular quality control and counteract cellular stress.12 The autophagy-lysosome pathway works together with the ubiquitin-proteasome system (UPS), another cellular clearance mechanism, to degrade misfolded or unwanted proteins. In agreement with the important roles of these pathways in preserving protein homeostasis (proteostasis) in the cell, dysfunction in both pathways has been linked to abnormal accumulation of ubiquitinated protein aggregates in the cell. For example, inactivating basal levels of cellular autophagy by depleting ATG5 (autophagy-related 5) or ATG7 in mouse brain leads to protein aggregation and neurodegeneration.13,14 Similarly, disruption of proteasomal function also results in the accumulation of abnormal protein aggregates.15
Available evidence supports the existence of intercommunication between these 2 important cellular protective mechanisms.16 For example, application of the chemical compound MG132, a specific and reversible proteasome inhibitor, can induce autophagy.17,18 It is assumed that this MG132-induced autophagic activation is an indirect cellular compensatory response, possibly mediated by ER (endoplasmic reticulum) stress or MAPK11/12/13/14 (mitogen-activated protein kinase 11/12/13/14) signaling pathways, to offset compromised proteasomal activity and maintain proper proteostasis.17,19 However, the detailed mechanism of this MG132-induced autophagic activation is still unclear.
The GST-BHMT (a fusion protein of GST [glutathionine S-transferase] tagged to the N terminus of BHMT [betaine-homocysteine S-methyltransferase]) reporter has recently been developed as an endpoint, cargo-based assay for the study of autophagy.20,21 The endogenous BHMT enzyme is highly expressed in liver and kidney cells. BHMT as a cargo is delivered through the autophagy pathway into the lysosome where it is cleaved at its N-terminal loop site by asparaginyl endopeptidase LGMN (legumain) to produce a specific proteolytic fragment (BHMT(FRAG)).22 Further, this specific cleavage event responds to amino acid or serum starvation in an autophagy-dependent manner. Accordingly, the accumulation of a GST-tagged version of the cleaved BHMT product (GST-BHMT(FRAG)) has been successfully used as a cargo-based, endpoint reporter to monitor starvation-induced autophagy activity in different cell lines.21 However, whether the GST-BHMT assay is applicable to study nonstarvation-induced autophagy has not been fully examined.
Here, we found that under nutrient-rich conditions, inactivation of proteasome function induced a similar BHMT cleavage that was autophagy dependent, but through a mechanism different from that induced by starvation. In particular, although the BHMT processing relied on the critical autophagy components ATG7 and ULK1, it did not involve the upstream autophagy regulators MTORC1 or PRKAA. Instead, proteasome inhibition induced elevated ER stress while knockdown of ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1), one of the signaling mediators of the unfolded protein response (UPR), as well as its main downstream effector MAPK8/JNK1 (mitogen-activated protein kinase 8) but not XBP1 (X-box binding protein 1), blocked this autophagy-mediated BHMT processing. Further, upregulation of ERN1 induces strong MAPK8 activation and subsequent multisite phosphorylation of BCL2 (B-cell CLL/lymphoma 2), resulting in its dissociation from its inhibitory binding with the essential autophagy activator BECN1 (Beclin 1, autophagy related). Intriguingly, the proteasome inhibition-induced BHMT processing, but not the starvation-induced one, requires the cargo receptor proteins SQSTM1 and NBR1 and also depends on the functional multimerization domain of the BHMT protein itself. However, BHMT and SQSTM1 failed to interact, BHMT was not stabilized, and we did not observe increased ubiquitination of BHMT upon proteasome inhibition. Together, these data support a scenario in which, under nutrient-rich environments, proteasome inhibition can induce autophagic activation through a distinct mechanism that bears resemblance to the selective Cvt pathway in the yeast. Our data also support the use of the GST-BHMT assay to study mammalian selective autophagy.
Results
Proteasome inhibitor MG132 induces BHMT processing
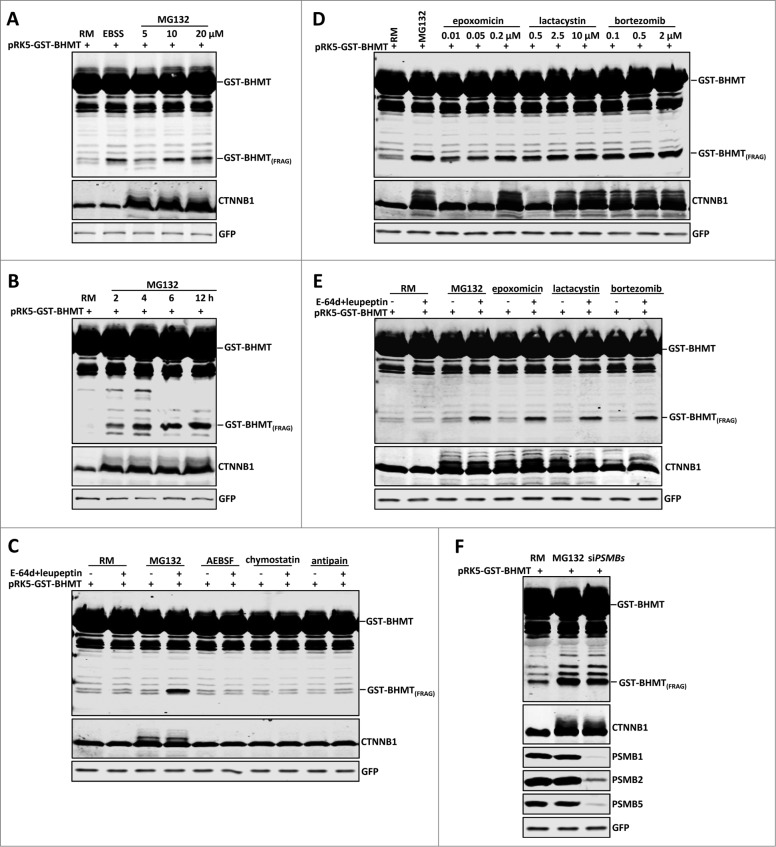
It has been established that upon cellular starvation, for example after prolonged incubation in Earle's Balanced Salt Solution (EBSS), the GST-BHMT reporter releases a cleaved product with a predicted molecular mass of 35 kDa (GST-BHMT(FRAG)) in an autophagy-dependent manner.21 Considering potential crosstalk between the UPS and autophagy pathways, we tested whether treating GST-BHMT-expressing cells with MG132, a widely used proteasome inhibitor, would activate autophagy and induce similar GST-BHMT processing. Indeed, resembling EBSS treatment, MG132 induced a dose-dependent accumulation of GST-BHMT(FRAG) (Fig. 1A). In addition, the accumulation of GST-BHMT(FRAG) was also time dependent, with levels of the processed product increasing with time and plateauing at around 12 h of continued treatment (Fig. 1B). Accordingly, in subsequent experiments, we treated cells using 10 µM MG132 for 6 h unless otherwise indicated.
Figure 1 (See previous page).
Proteasome inhibition induces GST-BHMT fragmentation. HEK293T cells were transfected with 2 µg pRK5-GST-BHMT plasmids and incubated in the indicated medium followed by coimmunoprecipitation with anti-GST antibody and western blot analyses to detect the proteolytic processing of the GST-BHMT reporter. The prominent accumulation of CTNNB1, an endogenous proteasome substrate, confirmed the effective inactivation of the proteasome by MG132. GFP-MYC, whose expression is driven by the internal ribosome binding sites in the pRK5-GST-BHMT plasmid, was revealed by anti-MYC antibody and served as normalization control, as reported.20 (A) MG132 treatment induces GST-BHMT fragmentation. HEK293T cells were incubated in EBSS or in nutrient-rich medium (RM) containing MG132 at the indicated concentrations from 5 µM to 20 µM for 6 h, before being processed for the GST-BHMT assay. (B) Time-dependent analysis of GST-BHMT processing. After incubation in nutrient-rich medium with 10 µM MG132, transfected HEK293T cells were harvested at indicated time points from 2 h to 12 h. Notice the gradual accumulation of GST-BHMT(FRAG) product overtime, as revealed by western analysis with anti-GST antibody. (C) Inhibition of lysosomal proteases does not induce GST-BHMT fragmentation. HEK293T cells transfected with GST-BHMT reporter were treated with MG132 (10 µM) or with lysosome cysteine protease inhibitors AEBSF (2 mM), chymostatin (100 µM) or antipain (50 µg/ml), as indicated. Note the apparent accumulation of GST-BHMT(FRAG) only in the sample treated with both MG132 and lysosome protease inhibitors E-64d and leupeptin (lane 4). (D) Multiple proteasome inhibitors, including epoxomicin, lactacystin, and bortezomib, induce similar GST-BHMT processing as that by MG132. (E) The proteasome inhibition-induced accumulation of GST-BHMT(FRAG) requires the presence of lysosomal protease inhibitors E-64d and leupeptin. The transfected cells were treated in the presence or absence of E-64d and leupeptin together with different proteasome inhibitors, as indicated: MG132 (10 µM), epoxomicin (0.1 µM), lactacystin (2.5 µM), and bortezomib (0.5 µM). (F) Genetic interference of proteasomal function leads to similar BHMT fragmentation as that induced by MG132 (compare lanes 3 with 2). For sample in lane 3, cells were cotransfected with 5 nM of siRNAs against proteasome catalytic subunits PSMB1, PSMB2 and PSMB5, and their knockdown efficiency was verified by western blotting analysis, as indicated.
Proteasome inhibition, not lysosomal inactivation, is responsible for GST-BHMT processing
In the established GST-BHMT assay, 2 lysosomal protease inhibitors, E-64d and leupeptin, are included to prevent further degradation of GST-BHMT(FRAG), the cleaved reporter product,21 so as to preserve it as the readout for cellular autophagic activity. It has been reported that in addition to inactivating the proteasome, MG132 can also directly block lysosome function by inactivating lysosomal serine and cysteine proteases.15 This raises the possibility that MG132-induced GST-BHMT fragmentation is not due to proteasome inhibition, but instead is due to an off-target effect of MG132, because of its lysosomal inhibitory activity, similar to E-64d and leupeptin. To address this question, we tested whether MG132 alone could replace E-64d and leupeptin in the GST-BHMT assay. Upon MG132 treatment but in the absence of E-64d and leupeptin, we did not observe an obvious accumulation of GST-BHMT(FRAG), which was in sharp contrast to its prominent production with the presence of these 2 lysosomal protease inhibitors (Fig. 1C, comparing lane 3 with lane 4). Therefore any lysosomal inhibitory activity of MG312 is not sufficient to induce the observed accumulation of GST-BHMT(FRAG).
To further confirm that lysosomal inactivation alone cannot account for the increased levels of GST-BHMT(FRAG), we treated the cells with other known lysosomal serine and cysteine protease inhibitors, including AEBSF, chymostatin, and antipain. Even at the maximum reported working concentrations, none of these inhibitors led to a buildup of GST-BHMT(FRAG) (Fig. 1C, from lane 5 to lane 10). Together, these results indicate that MG132-induced GST-BHMT fragmentation is not due to MG132 inactivating lysosomal serine and cysteine proteases.
To further establish that MG132-induced GST-BHMT processing is due to proteasome inhibition, we next tested several other known proteasome inhibitors that act through different mechanisms. MG132 is a tripeptide aldehyde that effectively inactivates the proteolytic activity of the 26S proteasome by covalently binding to and blocking the active site of its β subunits.23 In comparison, epoxomicin and lactacystin are naturally occurring nonpeptide proteasome inhibitors.24 Bortezomib is a clinically proven dipeptide boronic acid analog that can potently and reversibly block the 26S proteasome.25 All these proteasome inhibitors had a similar effect on GST-BHMT(FRAG) stability as MG132, dramatically inducing the accumulation of GST-BHMT(FRAG) even at the lowest dosage we tested (Fig. 1D). For example, consistent with reports that epoxomicin is about 100-fold more potent than lactacystin in inactivating the proteasome,24 the lowest concentration of epoxomicin (10 nM) we tested was capable of eliciting robust GST-BHMT(FRAG) accumulation, similar to the accumulation seen with 500 nM lactacystin (Fig. 1D, compare lanes 3 with 6). Moreover, resembling responses to MG132, none of these proteasome inhibitors alone led to GST-BHMT fragmentation (Fig. 1E, lanes 5, 7, and 9) unless E-64d and leupeptin were included in the assay (Fig. 1E, lanes 6, 8, and 10).
To further circumvent any possible unknown off-target effect from these chemical-based proteasome inhibitors, we genetically inactivated the proteasome by knocking down the 3 essential catalytic proteasome subunits PSMB1 (proteasome [prosome, macropain] subunit, β type, 1), PSMB2, and PSMB5 using small interfering RNA (siRNA), which effectively depleted target gene expression (Fig. 1F). Again, siRNA-mediated inactivation of the proteasome induced significant accumulation of GST-BHMT(FRAG) (Fig. 1F, lane 3). Together, these results establish that proteasome inhibition is sufficient to induce specific GST-BHMT processing.
Proteasome inhibition-induced GST-BHMT fragmentation is autophagy dependent
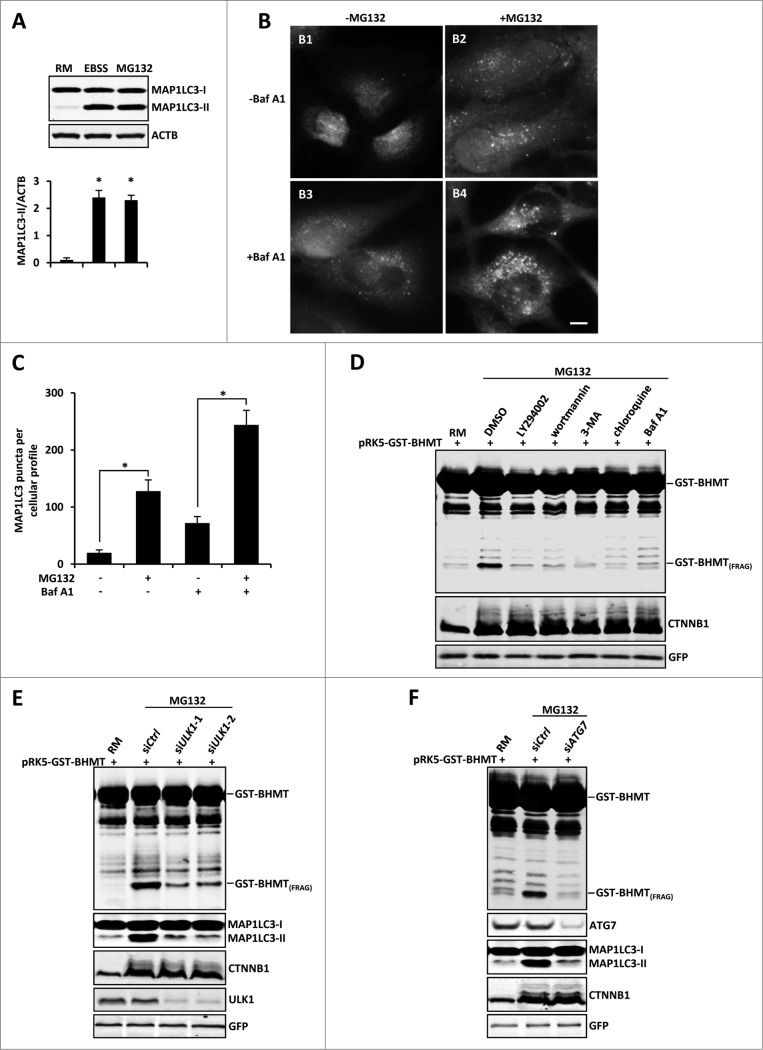
BHMT is normally delivered through the autophagy pathway to the lysosome for its maturation, which is the basis of the GST-BHMT assay used to examine cellular autophagic activity upon starvation.20 Given that proteasome inhibition generated a similar pattern of GST-BHMT processing as that induced by EBSS starvation, we hypothesized that proteasome inhibition activates an autophagic response, which in turn leads to an increased delivery of GST-BHMT to the lysosome and its subsequent processing. To test this possibility, we first used a standard MAP1LC3 (microtubule-associated protein 1 light chain 3) lipidation assay to examine whether there is activation of autophagic response upon proteasome inhibition. Indeed, upon MG132 treatment in nutrient-rich medium, the amount of lipidated MAP1LC3 (MAP1LC3-II) was significantly increased, comparable to starvation-induced MAP1LC3 activation (Fig. 2A). Consistent with this observation, immunofluorescent staining also revealed a significant increase in MAP1LC3-positive puncta in the presence of MG132 (Fig. 2B, comparing panel 2B1 with 2B2).
Figure 2 (See previous page).
Proteasome inhibition-induced GST-BHMT processing is autophagy-dependent. (A-C) Proteasome inhibition induces increased levels of MAP1LC3 lipidation and MAP1LC3-positive puncta formation. (A) For MAP1LC3 lipidation assay, HEK293T cells were treated with 10 µM MG132 in the presence of the lysosome inhibitor Baf A1 (100 nM). Both MAP1LC3-I and MAP1LC3-II were enriched by immunoprecipitation using antibody against MAP1LC3. Note that MG132 induced a similar level of MAP1LC3-II as by EBSS treatment. (B-C) For MAP1LC3-positive puncta formation assay, HeLa cells were treated with 10 µM MG132 in the absence or presence of lysosome inhibitor Baf A1 (100 nM), as indicated. Scale bar: 5 µm. (C) Quantitative analysis of cellular MAP1LC3-positive puncta profile. For each sample, MAP1LC3-positive puncta in about 200 cells were counted. The data was presented as the average number of MAP1LC3-positive puncta per cell. (D-F) Proteasome inhibition-induced BHMT processing is autophagy-dependent. (D) HEK293T cells were treated with MG132 (10 µM) together with DMSO control or the following pharmacological inhibitors of the autophagy pathway: LY294002 (100 µM), 3-methyladenine (3-MA; 10 mM), wortmannin (1 µM), chloroquine (100 µM), and Baf A1 (100 nM). Compared to the mock-treated sample using solvent DMSO, all other samples that were simultaneously treated with the indicated autophagy inhibitors showed reduced accumulations of GST-BHMT(FRAG). (E and F) GST-BHMT fragmentation was inhibited following the depletion of essential autophagy components. HEK293T cells were cotransfected with 10 nM siRNA against ULK1 (E) or ATG7 (F). Note that knockdown of either ULK1 (E) or ATG7 (F), but not treatment with control siRNA (siCtrl), significantly reduced the production of GST-BHMT(FRAG) and MAP1LC3 lipidation.
MAP1LC3 itself is also a direct substrate of autophagy and is degraded by the lysosome. Therefore there is a constant turnover of cellular MAP1LC3, termed autophagic flux.26 Accordingly, the observed elevated level of MAP1LC3-II could be due to either an increased autophagic biogenesis or compromised lysosomal function, such as defective autophagosome-lysosome fusion. To distinguish between these 2 possibilities, we compared the effect of lysosome inhibitor baflomycin A1 (Baf A1) on MAP1LC3-puncta formation under different assay conditions. Inclusion of Baf A1 led to an additional increase of MAP1LC3-positive puncta both in the presence and absence of MG132, indicating that autophagic flux was normal in MG132-treated cells (Fig. 2B and C, comparing panels B3 with B1, and B4 with B2). Thus, in nutrient-rich conditions, proteasome inhibition activates cellular autophagic responses without affecting cellular autophagic flux.
To establish that autophagic activation is responsible for the induced GST-BHMT processing and accumulation of GST-BHMT(FRAG), we next carried out MG132 treatment in the presence of known pharmacologic inhibitors targeting different steps of the autophagy cascade. LY294002, 3-methyladenine (3-MA) and wortmannin act by blocking macroautophagic cargo sequestration, while chloroquine and Baf A1 function in later autophagic steps by preventing lysosomal acidification and its ability to degrade engulfed cargos. Importantly, all of these autophagy inhibitors almost completely abolished MG132-induced GST-BHMT fragmentation, validating that autophagic activation is responsible for MG132-induced GST-BHMT processing (Fig. 2D).
To provide further evidence for the conclusion that GST-BHMT is fragmented by autophagic activation, we next examined the effect of genetically blocking the autophagy pathway. ULK1 is a kinase critical for autophagosome initiation, whereas ATG7 is essential for autophagosome formation.4,6 siRNAs against ULK1 or ATG7 effectively knocked down the expression of respective protein products, and significantly reduced MG132-induced GST-BHMT cleavage (Fig. 2E and F). Collectively, these results indicate that GST-BHMT fragmentation upon proteasome inhibition is autophagy dependent.
Proteasome inhibition-induced autophagy is mechanistically different from starvation-induced autophagy
We next investigated whether similar mechanisms mediate autophagic activation by proteasome inhibition as those that activate autophagy in response to starvation. MTOR kinase complex 1 (MTORC1) is the central sensor of cellular nutrition status, while kinase PRKAA is essential for monitoring cellular energy status. Both can activate autophagy mainly through differential phosphorylation of ULK1.
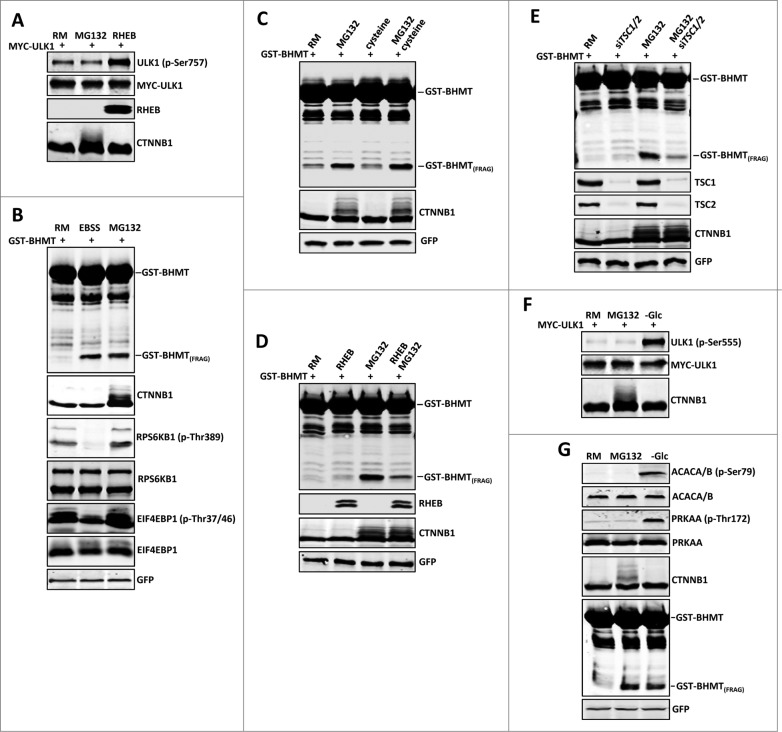
Under nutrient-rich conditions, MTORC1 is active and inhibits ULK1 by phosphorylating Ser757 (p-Ser757), thus inactivating autophagy.27 Inclusion of MG132 did not significantly change the phosphorylation status of ULK1 at Ser757 (Fig. 3A, comparing lane 2 with lane 1), whereas cotransfection with the positive control RHEB (Ras homolog enriched in brain), a MTORC1 activator, significantly increased Ser757 phosphorylation (Fig. 3A, comparing lane 3 with lane 1), indicating that proteasome inhibition does not interfere with MTOR kinase activity. Consistent with this finding, the phosphorylation status of RPS6KB1 (ribosomal protein S6 kinase, 70kDa, polypeptide 1) and EIF4EBP1 (eukaryotic translation initiation factor 4E binding protein 1), 2 authentic substrates of MTORC1,28 was not affected (Fig. 3B, comparing lane 3 to lane 1). This is in sharp contrast to EBSS-induced starvation, which effectively suppresses MTORC1 activity and the subsequent phosphorylation of RPS6KB1 at Thr389 (p-Thr389) and EIF4EBP1 at Thr37 and Thr46 (p-Thr37/Thr46); the second lane in Fig. 3B). Amino acid shortage such as cysteine depletion has been reported in some MG132-treated cells,29 which could lead to autophagic activation through MTORC1. However, supplementing MG132-treated cells with cysteine did not affect GST-BHMT fragmentation (Fig. 3C). Altogether, these data support the conclusion that proteasome inhibition-induced autophagy is not mediated through the nutrient sensor MTORC1.
Figure 3.
Proteasome inhibition-induced GST-BHMT fragmentation is MTOR- and PRKAA-independent. (A to C) Proteasome inhibition does not affect MTORC1 activity. Western blot analysis of the phosphorylation status of MTORC1 substrates (A) ULK1 as well as (B) RPS6KB1 and EIF4EBP1. (A) HEK293T cells were treated with MG132 (10 µM) for 6 h. The phosphorylation status of ULK1 at Ser757 was detected by an antibody specifically against ULK1 (p-Ser757). (B) Compared to the control of untreated cells in nutrient-rich medium (lane 1), MG132 treatment in rich medium did not affect the phosphorylation levels of RPS6KB (p-Thr389) and EIF4EBP1 (p-Thr37/46) (lane 3), whereas EBSS starvation led to significantly reduced levels of phosphorylation at these target sites (lane 2). (C) Cysteine levels do not affect the cellular autophagy activity. In the nutrition-rich medium, addition of cysteine (1 mM) had no effect on the basal level of GST-BHMT fragmentation (compare lane 3 with lane 1) or on MG132-induced GST-BHMT fragmentation (compare lane 4 with lane 2). (D and E) MTOR activation suppresses the proteasomal inhibition-induced autophagy GST-BHMT assays. GST-BHMT (2 µg) was cotransfected with (D) FLAG-RHEB (2 µg) or (E) a mixture of 10 nM siRNAs against TSC1 and TSC2 either in the absence or presence of 10 µM MG132, as indicated. Activation of MTOR, either by (D) overexpression of its activator RHEB or by (E) depletion of its inhibitory TSC1/2 complex, both significantly diminished the MG132-induced fragmentation of the GST-BHMT reporter. The ectopic expression of RHEB and the knockdown efficiency of TSC1 and TSC2 were verified by western blotting against RHEB and TSC1 and TSC2 proteins. (F and G) Proteasome inhibition does not affect PRKAA activity. HEK293T cells treated with MG132 (10 µM) for 6 h were analyzed by western blotting to detect the endogenous phosphorylation levels of (F) ULK1 (p-Ser555), (G) ACACA/B (p-Ser79) and PRKAA (p-Thr172), the known targets of PRKAA. Compared to the controls of untreated samples (lane 1), glucose starvation led to significantly increased phosphorylations of these endogenous PRKAA substrates (lanes 3) whereas MG132 treatment (lane 2) showed little effect, although both treatments similarly resulted in the increased accumulation of GST-BHMT(FRAG).
If proteasome inhibition and starvation indeed induce autophagy through different pathways, it would be expected that simultaneous activation of both pathways should lead to a stronger autophagic response than by either pathway alone. To test this assertion, we incubated GST-BHMT-expressing cells undergoing EBSS starvation with MG132. This led to a more pronounced accumulation of GST-BHMT(FRAG) (Fig. S1, compare lane 4 with lanes 2 and 3). Similarly, coexpression of a kinase-dead MTOR mutant (MTOR KD), which acts as a dominant negative to suppress MTOR kinase activity (Fig. S1, lane 5),30 combined with MG132 treatment also resulted in a stronger autophagic response (Fig. S1, compare lane 6 with lanes 5 and 3). Collectively, these results support the conclusion that different mechanisms mediate proteasome inhibition-induced and starvation-induced autophagy.
Given the above observations, we further tested the effect of MTOR activation on proteasome inhibition-induced GST-BHMT processing. Quite interestingly, MTOR activation, either by overexpression of the MTOR activator RHEB (Fig. 3D) or by siRNA-mediated knockdown of the tuberous sclerosis 1/2 (TSC1/2) inhibitory complex (Fig. 3E), both significantly suppressed the MG132-induced GST-BHMT fragmentation. Thus, under the assay condition, although MTOR inhibition is not necessary for MG132-induced autophagy, its activation is sufficient to suppress the amplitude of the induction. Such a result suggests that the level of GST-BHMT processing may reflect the overall net autophagic activity of the cell. It also raises an intriguing possibility of a potential overlapping or converging regulatory mechanisms downstream of MTOR pathway and the proteasomal inhibition-induced autophagy.
Next, we tested whether proteasome inhibition affects the activity of the cellular energy sensor kinase PRKAA, which positively regulates autophagy by phosphorylating ULK1 at Ser555 (p-Ser555).31 In the presence of MG132, the phosphorylation level of ULK1 (p-Ser555) was not altered (the second lane in Fig. 3F). MG132 treatment also had no effect on the phosphorylation status of 2 other known PRKAA targets, Thr172 in the PRKAA catalytic subunit (p-Thr172) and Ser79 in ACACA/B (acetyl-CoA carboxylase α/β; p-Ser79) (the second lane in Fig. 3G).32 In contrast, upon energy depletion by glucose deprivation, there was a dramatic induction of ULK1 (p-Ser555) together with PRKAA (p-Thr172) and ACACA/B (p-Ser79), and interestingly also a significant accumulation of GST-BHMT(FRAG) (the third lane in Fig. 3F and G). Together, these data suggest that proteasome inhibition induces autophagy through a mechanism different from that by starvation, independent of MTORC1 and PRKAA pathways.
The ERN1-mediated ER stress response is important for proteasome inhibition-induced autophagy
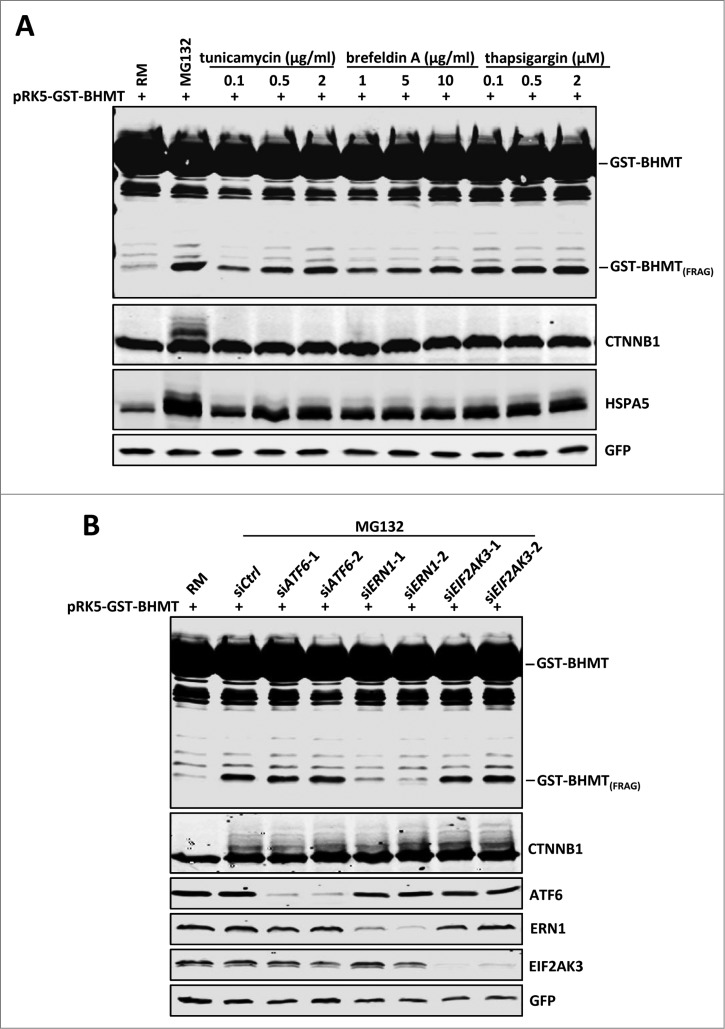
As proteasome inhibition-induced autophagic activation does not rely on regulation of MTORC1 and PRKAA, we explored alternative pathways that linked proteasome inhibition with autophagic activation. It has been established that proteasome inhibition leads to accumulation of misfolded proteins, which in turn causes ER stress.33 Further, ER stress induces autophagy in both yeast and mammalian cells.33,34 These findings led us to hypothesize that an ER stress signaling pathway might be the connection between proteasome inhibition and autophagic activation. To test this, we treated cells in parallel with several known ER stress inducers including tunicamycin, brefeldin A, and thapsigargin, which induce expression of several ER stress markers such as HSPA5/GRP78 (heat shock 70kDa protein 5 [glucose-regulated protein, 78kDa]), DDIT3/CHOP (DNA-damage-inducible transcript 3), P4HB/PDI (prolyl 4-hydroxylase, β polypeptide) and ATF4 (activating transcription factor 4). MG132 treatment also caused a dramatic induction of HSPA5, DDIT3, P4HB and ATF4, indicating heightened ER stress upon proteasome inhibition (Fig. 4A, lane 2 and Fig. S2). Notably, the induction of HSPA5 by MG132 is even stronger than when treated by other known ER stress inducers such as tunicamycin, brefeldin A, and thapsigargin, possibly due to reduced turnover of HSPA5 caused by proteasome inhibition. Importantly, each ER stress inducer caused significant accumulation of GST-BHMT(FRAG) in a dosage-dependent manner (Fig. 4A), consistent with ER stress inducing autophagic activation.
Figure 4.
Proteasome inhibition-induced GST-BHMT fragmentation involves the ERN1 branch of the ER stress signaling. (A) ER stress induces GST-BHMT fragmentation. HEK293T cells were treated with MG132 (10 µM) or different ER stress inducers at indicated concentrations, followed by western blotting analyses to examine the proteolytic processing of GST-BHMT reporter and the expression levels of HSPA5, a marker of ER stress. Compared to the untreated control (lane 1), all ER stressors induced robust accumulation of GST-BHMT(FRAG) similar as that by MG132 treatment. (B) ERN1 is critical for the proteasome inhibition-induced BHMT processing. siRNAs against ATF6, ERN1 and EIF2AK3 were transfected into HEK293T cells together with pRK5-GST-BHMT, followed by the GST-BHMT assay. The effectiveness of siRNA-mediated target knockdown was verified by western blotting using the corresponding specific antibodies, as indicated.
ER stress is sensed and transduced by 3 ER-localized transmembrane proteins, which in turn induce discrete aspects of the unfolded protein response.35 Briefly, activation of EIF2AK3/PERK (eukaryotic translation initiation factor 2-α kinase 3) leads to translational attenuation in the cell, while ATF6 (activating transcription factor 6) and ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1) upregulate the cellular machineries involved in protein folding and clearance.35 To investigate the involvement of these 3 UPR pathways in proteasome inhibition-induced autophagy, we applied siRNA to knock down each of the ER stress sensors and examined their effect on MG132-induced GST-BHMT(FRAG) accumulation. Interestingly, although depletion of ATF6 or EIF2AK3 showed no effect, siRNA against ERN1 significantly blocked GST-BHMT fragmentation (Fig. 4B, lanes 5 and 6), suggesting that ERN1-mediated UPR pathway induction is essential for proteasome inhibition-induced autophagy.
The MAPK8-BCL-BECN1 pathway is the main mediator downstream of ERN1 in proteasome inhibition-induced autophagy
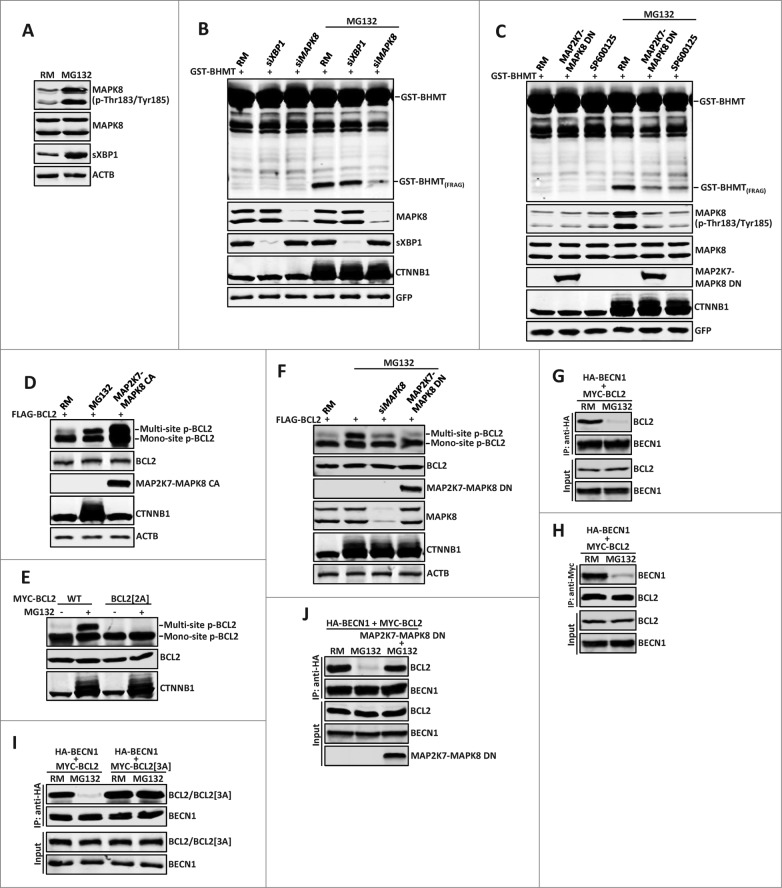
We next investigated how ERN1 is involved in proteasome inhibition-induced autophagy. ERN1 mediates the UPR mainly through its 2 enzymatic domains: the N-terminal endoribonuclease domain controls the ER stress-induced splicing of XBP1 (X-box binding protein 1), whereas its C-terminal serine/threonine kinase domain activates MAPK8/JNK1 (mitogen-activated protein kinase 8).35 Consistent with the elevated ER stress and the involvement of ERN1 signaling in proteasome inhibition-induced autophagy, MG132 treatment led to robust XBP1 splicing and MAPK8 activation, as revealed by the significantly increased levels of spliced XBP1 (sXBP1) and active MAPK8 isoforms with Thr183/Tyr185 dual phosphorylation (p-Thr183/Tyr185) recognized by a phospho-specific antibody, respectively (Fig. 5A). To examine whether both XBP1 and MAPK8 signaling are required for MG132-induced autophagy, we tested the effect of siRNA-mediated knockdown of XBP1 or MAPK8 on GST-BHMT fragmentation in MG132-treated samples. Strikingly, while knockdown of XBP1 showed no obvious effect, depletion of MAPK8 significantly blocked MG132-induced fragmentation of GST-BHMT (Fig. 5B, lanes 5 and 6). Consistent with this finding, inclusion of MAP2K7-MAPK8 DN, a dominant-negative (DN) MAPK8 construct,36 or SP600125, a potent MAPK8 inhibitor, both led to a dramatically reduced accumulation of GST-BHMT(FRAG) (Fig. 5C, lanes 5 and 6). Together, they support MAPK8 activation as a critical signaling event downstream of ERN1 in mediating proteasome inhibition-induced autophagy.
Figure 5 (See previous page).
The MAPK8-BCL2-BECN1 pathway is the main mediator downstream of ERN1 in proteasome inhibition-induced autophagy. (A) MG132 activates main downstream effectors of ERN1 signaling: MAPK8 and XBP1. 10 µM MG132 treatment for 2 h induced strong increase of dual-phosphorylated MAPK8 and spliced XBP1 (sXBP1). (B and C) MAPK8 but not XBP1 is required for MG132-induced autophagy. GST-BHMT assays. HEK293T cells were cotransfected with GST-BHMT reporter (2 µg) and (B) siRNAs against XBP1 (30 nM) or MAPK8 (20 nM), or (C) a dominant-negative MAPK8 (MAP2K7-MAPK8 DN, 2 µg) or were treated with SP600125 for 1 h before MG132 treatment. (D and E) MG132 induces multisite phosphorylation of BCL2. (D) Similar to that catalyzed by the constitutively active MAPK8 (MAP2K7-MAPK8 CA, 2 µg), MG132 significantly induced multisite phosphorylation of wild-type BCL2, (E) but not a mutant BCL2 with Thr69Ala and Ser87Ala substitutions. (F) MPAK8 is responsible for the MG132-induced multisite phosphorylation of BCL2. HEK293T cells were treated with 10 µM MG132 for 2 h. Both siRNA-mediated depletion of MAPK8 and expression of MAP2K7-MAPK8 DN significantly reduced the level of multisite phosphorylation in BCL2. (G and H) MG132 treatment induced dissociation between BCL2 and BECN1. HEK293T cells were cotransfected with 2 µg each of MYC-BCL2 and HA-BECN1. Reciprocal coimmunoprecipitation (coIP) assays were performed with (G) anti-HA or (H) anti-MYC antibodies in the absence or the presence of 10 µM MG132 (2 h incubation), as indicated. Note the dramatically reduced interaction between BCL2 and BECN1 in MG132-treated samples. (I and J) MAPK8-mediated multisite phosphorylation of BCL2 is required for the MG132-induced dissociation of BCL2 and BECN1. HA-BECN1 was cotransfected with wild-type MYC-BCL2 or in parallel with (I) mutant MYC-BCL2[3A] (T69A, S70A, S87A), or (J) a dominant-negative MAPK8 (MAP2K7-MAPK8 DN) into HEK293T cells. (I) Contrary to wild-type BCL2, mutant BCL2[3A] remained associated with BECN1 after MG132 treatment. (J) MAP2K7-MAPK8 DN blocked MG132-induced disassociation of BCL2 from BECN1. The knockdown efficiency against target proteins by different siRNAs were verified by western blotting assays. Proteasome inhibition by MG132 was confirmed by the increased accumulation of CTNNB1.
The observed link between the ERN1-MAPK8 pathway and proteasome inhibition-induced autophagy is rather intriguing. As in mammalian cells, MAPK8 is a known regulator of starvation-induced autophagy by controlling the phosphorylation-dependent interaction between BCL2 and an essential autophagy activator BECN1.37 To investigate whether proteasomal inhibition employs the similar mechanism downstream of MAPK8 to activate autophagy, we first tested whether MG132 induced the MAPK8-dependent multisite phosphorylation of BCL2 at residues Thr69, Ser70 and Ser87.37 Indeed, resembling that induced by a constitutive active (CA) MAPK8 (Fig. 5D),36 MG132 treatment led to robust multisite phosphorylation of wild-type BCL2 (Fig. 5D) but not a mutant BCL2 in which 2 target phosphorylation sites are mutated to nonphosphorylatable alanine (T69A and S87A; BCL2[2A]) (Fig. 5E). Further, downregulation of MAPK8 activity, either by siRNA-mediated knockdown of MAPK8 or by overexpressing the dominant negative MAP2K7-MAPK8 DN, all significantly inhibited the MG132-induced multisite phosphorylation of BCL2 (Fig. 5F). Collectively, they support that proteasomal inhibition induces ERN1 activation and MAPK8-dependent phosphorylation of BCL2. Such a result would predict that MG132 treatment also leads to the similar phosphorylation-dependent disassociation of BCL2 from BECN1. Indeed, in reciprocal coIP assays, MG132 incubation largely abolished the strong physical interaction of BECN1 with wild-type BCL2 (Fig. 5G and H), but not with a mutant BCL2 containing alanine substitutions for all 3 of its MAPK8 phosphorylation sites (BCL2[3A]; Fig. 5I). Finally, inclusion of the dominant negative MAP2K7-MAPK8 DN almost completely blocked the MG132-induced disassociation of BCL2 from BECN1 (Fig. 5J). Combined, these data support that proteasome inhibition-induced autophagy is mediated through the ER stress-induced ERN1 signaling and its downstream MAPK8-BCL2-BECN1 pathway branch. Specifically, proteasome inhibition induces ER stress and upregulation of ERN1 signaling, which leads to MAPK8 activation and subsequent multisite phosphorylation of BCL2 and its dissociation from BECN1, thus releasing BECN1 from the inhibitory BCL2-BECN1 complex to stimulate autophagy.
Proteasome inhibition but not starvation-induced autophagy requires SQSTM1 and NBR1
It is well established that starvation often induces nonselective autophagy through the nutrient and energy sensors MTORC1 or PRKAA. Our results establish that in a nutrient-rich environment, proteasome inhibition also leads to autophagic activation, but likely through a distinct mechanism that is independent of MTORC1 and PRKAA but requires the ERN1 branch of the UPR and its downstream MAPK8-BCL2-BECN1 signaling pathway. As the proteasome normally functions to degrade ubiquitinated proteins, its dysfunction can lead to an accumulation of ubiquitinated substrates that are known to be recognized and cleared by autophagy. The selective autophagy pathway, such as aggrephagy, acts as an alternative or compensatory mechanism to maintain proper cellular proteostasis.16,17 Considering this, we examined whether known components of the selective autophagy pathway might play a role in proteasome inhibition-induced autophagy.
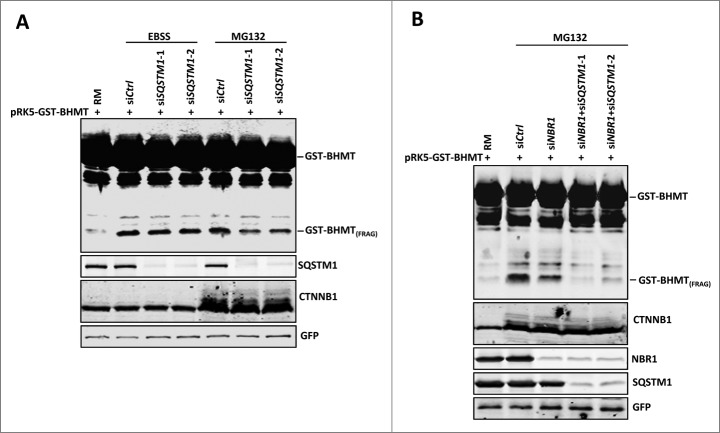
Clearance of ubiquitinated targets through selective autophagy is dependent on cargo receptors such as SQSTM1 and NBR1,3 both of which contain C-terminal UBA (ubiquitin-associated domain) for binding ubiquitinated substrates and N-terminal PB1 (Phox and Bem 1) domains for homo-polymerization, in addition to a MAP1LC3 binding motif that localizes cargos to the autophagosome.11,38,39 To test whether the autophagic response induced by proteasome inhibition relies on cargo receptors, we used siRNAs to specifically knock down the expression of these 2 cargo receptors, SQSTM1 and NBR1, and examined the resulting effect on GST-BHMT fragmentation. Western blotting analyses confirmed the effectiveness of siRNA-mediated knockdown of both SQSTM1 and NBR1 proteins (Fig. 6A and B). Notably, in nutrient-rich medium, knockdown of SQSTM1 by 2 independent siRNAs, but not by control siRNA, significantly attenuated MG132-induced accumulation of GST-BHMT(FRAG) (Fig. 6A, compare lane 5 with lanes 6 and 7). By contrast, SQSTM1 reduction had a negligible effect on starvation-induced accumulation of GST-BHMT(FRAG) (Fig. 6A, compare lanes 3 and 4 with lanes 6 and 7), which is consistent with a role of SQSTM1 in selective cargo recognition but not in starvation-induced autophagy. Similarly, knockdown of NBR1 expression also reduced MG132-induced GST-BHMT(FRAG) accumulation, as compared to control siRNA-treated samples (Fig. 6B, compare lanes 2 with 3). Further, simultaneous knockdown of both SQSTM1 and NBR1 almost completely abolished MG132-induced GST-BHMT fragmentation, indicating that SQSTM1 and NBR1 function redundantly in this autophagy response (Fig. 6B, compare lanes 4 and 5 with lanes 2 and 3). Together, these results suggest that autophagic activation induced by proteasome inhibition is dependent on cargo receptors SQSTM1 and NBR1.
Figure 6.
Proteasome inhibition-induced GST-BHMT fragmentation requires cargo receptor SQSTM1 and NBR1. (A) SQSTM1 knockdown specifically reduces MG132-induced but not starvation-induced GST-BHMT fragmentation. HEK293T cells were incubated either in EBSS or in nutrient-rich medium (RM) with 10 µM MG132 to induce an autophagic response. (B) NBR1 knockdown reduced MG132-induced GST-BHMT fragmentation (lane 3). Simultaneous knockdown of both NBR1 and SQSTM1 led to a more prominent reduction of GST-BHMT(FRAG) accumulation (lanes 4 and 5). In (A) and (B), 10 nM siRNA against SQSTM1 or NBR1 were cotransfected with the GST-BHMT reporter into the HEK 293T cells and incubated in medium as indicated for 6 h, followed by GST-BHMT assay as described in Figure 1.
BHMT is not recruited into phagophores through ubiquitination or direct association with SQSTM1
We further examined how SQSTM1 and NBR1 are involved in proteasomal inhibition-induced processing of GST-BHMT. Considering the important role of the ERN1-MAPK8-BCL2-BECN1 pathway in this process, we tested whether these cargo receptors affect MAPK8 activation, a critical step in proteasomal inhibition-induced autophagy. However, depletion of SQSTM1 by 2 independent siRNAs had no effect on MG132-induced dual-phosphorylation of MAPK8 (Fig. S3), suggesting that these cargo receptors are involved in a later step in the autophagic activation process and/or cargo recruitment into the phagophore.
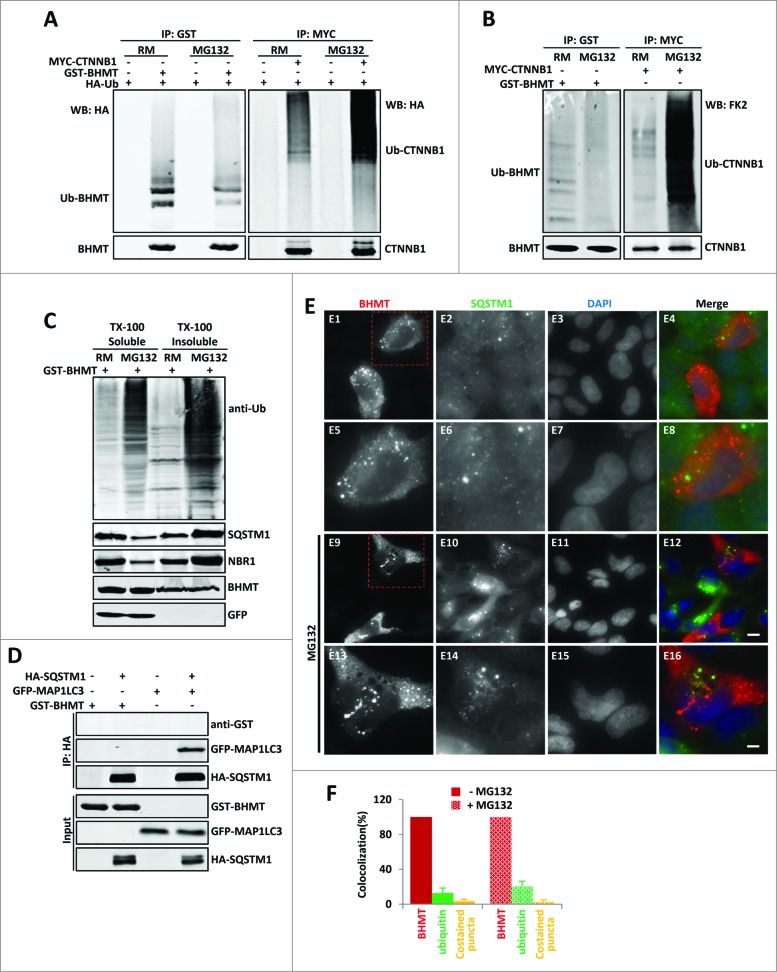
Next, we examined the hypothesis that GST-BHMT accumulates and becomes ubiquitinated upon proteasome inhibition, which is then recognized by cargo receptors and subsequently delivered to the autophagosome. At basal levels, GST-BHMT is only lightly ubiquitinated, as detected by cotransfected HA-tagged ubiquitin, in contrast to the smeared bands of high-molecular weight ubiquitinated CTNNB1/β-catenin (catenin [cadherin-associated protein], β 1, 88 kDa), a canonical substrate of the proteasome (Fig. 7A, comparing lane 2 with lane 6). To our surprise, in the presence of MG132, the level of ubiquitinated GST-BHMT did not increase but instead was clearly reduced, in contrast to the prominent stabilization of ubiquitinated CTNNB1 (Fig. 7A, comparing lane 4 with lane 2, and lane 8 with lane 6). To further confirm this observation, we evaluated the levels of GST-BHMT ubiquitination by endogenous ubiquitin and obtained similar results: while the level of ubiquitinated CTNNB1 (Ub-CTNNB1) was significantly increased in the presence of MG132, the level of ubiquitinated GST-BHMT (Ub-BHMT) was instead decreased (Fig. 7B).
Figure 7 (See previous page).
GST-BHMT does not show increased ubiquitination and physical interaction with SQSTM1 in response to proteasome inhibition. (A- C) The ubiquitination patterns of GST-BHMT reporter in response to MG132 treatment. (A and B) HEK293T cells were transfected with pRK5-GST-BHMT reporter (left panels) or the CMV-MYC-CTNNB1 control (right panels), followed by immunoprecipitation with anti-GST or anti-MYC antibodies, as indicated. In (A), cells were cotransfected with CMV-HA-Ub, and the levels of the ubiquitinated GST-BHMT (Ub-BHMT; left panel) or MYC-CTNNB1 (Ub-CTNNB1; right panel) proteins from the cell were detected with anti-HA antibody. In (B), the levels of the ubiquitinated GST-BHMT (Ub-BHMT; left panel) or MYC-CTNNB1 (Ub-CTNNB1; right panel) by endogenous ubiquitin were revealed by FK2 antibody. (C) MG132 does not affect the distribution of GST-BHMT protein in Triton X-100-soluble fractions. The control or MG132-treated HEK293T cells transfected with pRK5-GST-BHMT were harvested into 2 fractions: Triton X-100-soluble (TX-100 soluble) and Triton X-100-insoluble (TX-100 insoluble). The distribution of GST-BHMT, total ubiquitinated proteins, SQSTM1 and NBR1 in both fractions was examined by western blotting using anti-GST, anti-ubiquitin (FK2), anti-SQSTM1 and anti-NBR1 antibodies, respectively, as indicated. The soluble GFP protein, which was hardly detectable in Triton X-100 insoluble fraction, was included as the control for the fractionation procedure. (D) GST-BHMT does not interact with SQSTM1. HEK293T cells were transfected with HA-SQSTM1 together with GST-BHMT or with the positive control GFP-MAP1LC3, followed by immunoprecipitation using anti-HA antibody and western blot detection, as indicated. (E and F) GST-BHMT does not colocalize with SQSTM1. (E) HeLa cells transfected with pRK5-GST-BHMT were immunostained with antibodies against both GST (red) and endogenous SQSTM1 (green) in the absence or presence of MG132, as indicated. Scale bar: 5 µm for E1-E4 and E9-E12; 2 µm for E5-E8 and E13-E16. (F) The quantification of GST-BHMT- and SQSTM1-positive puncta as well as their colocalization profile as described in Materials and Methods.
As ubiquitinated proteins tend to form aggregates that are resistant to mild detergents such as Triton X-100, we also examined the distribution of GST-BHMT in both Triton X-100 soluble and Triton X-100 insoluble fractions. While MG132 treatment led to a significant increase in total levels of ubiquitinated proteins in both fractions as well as a redistribution of both SQSTM1 and NBR1 from Triton X-100-soluble to -insoluble fractions, presumably due to an increased accumulation of protein aggregates, the distribution of GST-BHMT in the 2 fractions was not affected (Fig. 7C). Further, immunofluorescence staining also failed to reveal clear colocalization of GST-BHMT with ubiquitin regardless of MG132 treatment (Fig. S4). Together, these results show that GST-BHMT is not normally turned over by the proteasome. They also do not support the hypothesis that upon proteasome inactivation, GST-BHMT accumulates and become ubiquitinated, subsequently being recognized by the cargo receptors SQSTM1 and NBR1 and delivered to the autophagosome.
It was reported that SQSTM1 could deliver cargos, such as mutated SOD1 (superoxide dismutase 1, soluble) protein, to the phagophore in a ubiquitin-independent manner.40 To examine whether SQSTM1 physically associates with GST-BHMT to deliver it to the phagophore, we performed coimmunoprecipitation (coIP) analysis. We found no detectable interaction between GST-BHMT and SQSTM1, in contrast to the strong interaction observed between the positive control MAP1LC3 and SQSTM1 (Fig. 7D, compare lanes 4 and 2). In addition, we did not find observable colocalization between GST-BHMT and endogenous SQSTM1 either in the presence or absence of MG132, as revealed by immunofluorescence staining against GST-BHMT and SQSTM1 protein (Fig. 7E and F). Noticeably, in transfected cells, in addition to its diffuse cytoplasmic distribution, GST-BHMT protein also showed localization to distinct puncta that clearly did not overlap with SQSTM1-positive structures. Although MG132 treatment led to an increased number of SQSTM1-positive structures, it did not have a visible effect on the number of GST-BHMT puncta and had no impact on its colocalization with SQSTM1 (Fig. 7E and F).
Proteasome inhibition-induced GST-BHMT fragmentation requires the multimerization domain
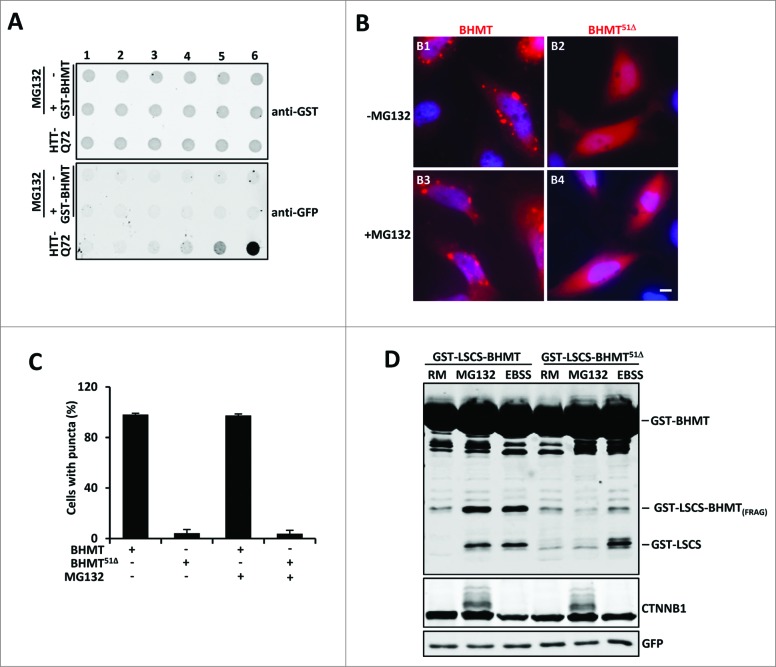
To examine the nature of subcellular GST-BHMT-positive puncta, we performed filter trap assays to see if GST-BHMT forms protein aggregates similar as those formed by mutant HTT (huntingtin) with expanded polyglutamine (polyQ) tracts.41 While polyQ-containing mutant HTT formed tight protein aggregates resistant to harsh chemicals such as 6 M urea (Fig. 8A), we did not detect such aggregates in cells expressing GST-BHMT with or without MG132 treatment. Thus, GST-BHMT puncta are not composed of compact protein aggregates.
Figure 8.
The multimerization domain of GST-BHMT is required for its autolysosomal loading in response to proteasome inhibition. (A) GST-BHMT does not form tight aggregates. Extracts of HEK293T cells transfected with either pRK5-GST-BHMT or CMV-EGFP-Q72-HTT-exon1 were analyzed by filter trap assay using antibodies against GST or GFP, respectively, as indicated. The amount of total proteins loaded in each well was 0, 0.1, 0.4, 2, 20, and 100 µg from lane 1 to lane 6, respectively. While mutant HTT protein formed tight aggregates that were resistant to 6M urea, hardly any such aggregates were detected in extracts from GST-BHMT expressing cells regardless of MG132 treatment. (B and C) Formation of GST-BHMT-positive puncta is dependent on its multimerization domain. (B) HeLa cells transfected with either GST-BHMT or mutant GST-BHMT51Δ were immunostained with anti-GST antibody. Whereas wild-type GST-BHMT protein showed prominent puncta-like subcellular localization, GST-BHMT51Δ was diffusely distributed throughout the cell. Scale bar: 5 µm. (C) Quantification of GST-BHMT-positive puncta, which revealed little change in puncta formation by either wild-type GST-BHMT or mutant GST-BHMT51Δ in response to MG132 treatment. (D) The multimerization domain of GST-BHMT is essential for its autolysosomal loading in response to proteasome inhibition. HEK293T cells were transfected with either pRK5-GST-LSCS-BHMT or mutant pRK5-GST-LSCS-BHMT-Δ51 constructs, followed by EBSS or MG132 treatment and GST-BHMT assay, as indicated.
It has been reported that BHMT is a homotetrameric protein, with a multimerization domain at its C terminus, as removal of the last 51 amino acids (GST-BHMT51Δ) completely abolishes its multimerization ability.20 We thus tested whether puncta formation by GST-BHMT is linked to its multimerization capacity. Indeed, in the absence of the C-terminal multimerization domain (GST-BHMT51Δ), there were no visible GST-BHMT-positive puncta. Instead, GST-BHMT51Δ protein was diffusely localized throughout the cytoplasm (Fig. 8B and C), suggesting that the GST-BHMT-positive puncta are composed of the highly polymerized form of GST-BHMT. A similar diffusive subcellular pattern was also observed for a mutated version of GST-BHMTW352A, which harbors a point mutation, W352A, that disrupts the multimerization capacity of BHMT (data not shown).
The close association between the C-terminal multimerization domain of BHMT and its distinct subcellular puncta formation provoked an intriguing possibility that the multimerization of BHMT plays a role in its fragmentation through proteasome inhibition-induced autophagy. In the yeast Cvt pathway, one of the best studied selective autophagy process, prApe1 needs to form a dodecamer in order to be recognized by its receptor Atg19 and subsequent sequestration by the autophagosome. Interestingly, it has been shown that under starvation conditions, multimerization of BHMT is dispensable for its sequestration by the autophagosome, but is necessary for its proteolytic processing in the lysosome.20 In order to monitor the successful autolysosomal loading of the GST-BHMT reporter regardless of its multimerization status, a GST-LSCS-BHMT construct was elegantly designed that included an additional cleavage site termed LSCS (linker-specific cleavage site) between the GST and BHMT sequences. Inclusion of the LSCS cleavage site causes this construct to be responsive to autolysosomal loading and release an extra proteolytic fragment GST-LSCS.20 Thus, upon autophagic activation in response to starvation, the GST-LSCS-BHMT reporter releases 2 fragments, the smaller GST-LSCS and the larger GST-LSCS-BHMT(FRAG), while the GST-LSCS-BHMT51Δ mutant produces only the smaller GST-LSCS fragment.20
Using the GST-LSCS-BHMT reporter, we investigated whether multimerization of BHMT is important for its proteasome inhibition-induced autophagic processing. As expected, MG132 treatment led to the production of 2 reporter fragments for the wild-type GST-LSCS-BHMT, the smaller GST-LSCS and the larger GST-LSCS-BHMT(FRAG), similar to the fragments produced under EBSS starvation (Fig. 8D, compare lanes 2 with 3). However, for the GST-LSCS-BHMT51Δ mutant, which is deleted for the multimerization domain, although EBSS starvation indeed induced the expected production of the smaller GST-LSCS fragment (Fig. 8D, compare lanes 6 with 3), reflecting its successful autolysosomal loading, only background levels of the GST-LSCS fragment were present in MG132-treated samples (Fig. 8D, compare lane 5 with lanes 2, 3, and 6). Thus, in contrast to its dispensable role for autophagosome sequestration of BHMT in response to starvation, multimerization of BHMT is necessary for its autolysosomal loading in a nutrient-rich environment under proteasome inhibition. Together, these data reveal that starvation and proteasome inactivation elicit distinct mechanisms of autophagic activation as well as autolysosomal loading of proteins slated for autophagic processing.
Discussion
Different mechanisms mediate starvation- and proteasome inhibition-induced autophagy
Previous findings suggest crosstalk between the UPS and autophagosome-lysosome pathways,42 2 major cellular protective mechanisms for maintaining cellular homeostasis. In support of this, genetic or pharmacological interference of proteasome function both lead to a strong activation of an autophagy response (Figs. 1 and 2A to C). Interestingly, our data show that the GST-BHMT assay, which has been developed as an endpoint, cargo-based assay to study starvation-induced autophagy, also responds to proteasome inhibition-induced autophagy, as its processing is blocked by the depletion of core autophagy components such as ULK1 or ATG7 (Fig. 2D to F). However, further studies suggest distinct mechanisms underlying proteasome inhibition-induced autophagy activation. First, unlike in starvation conditions, proteasome inhibition does not affect the activities of MTORC1 and PRKAA, 2 essential upstream regulators of nutrient and energy-starvation induced autophagy, as the phosphorylated status of their substrates, ULK1 (p-Ser757) and EIF4EBP1 as well as ULK1 (p-Ser555) and ACACA/B, are not affected (Fig. 3). Instead, proteasome inhibition induces robust ER stress (Fig. 4A), as reported.43 Further, ERN1, one of the mediators of ER stress-associated UPRs, but not ATF6 or EIF2AK3, is critical for the proteasome inhibition-induced BHMT processing (Fig. 4B). Interestingly, contrary to starvation-promoted BHMT fragmentation, the proteasome inhibition-induced response also relies on the cargo receptors SQSTM1 and NBR1, as their depletion largely abolishes the accumulation of GST-BHMT(FRAG) (Fig. 6). Finally, there is a differential requirement for the multimerization domain in BHMT between the starvation- vs. the proteasome inhibition-induced processing of the BHMT reporter (Fig. 8). Together, these findings suggest that in a nutrient-rich environment, proteasome inactivation can activate the autophagy-dependent processing of GST-BHMT reporter through a mechanism distinct from that induced by starvation.
Intriguingly, our data further show that of the 2 main signaling events downstream of ERN1, MAPK8 activation is critical for this proteasome inhibition-induced autophagy while XBP1 splicing only plays a dispensable role. In addition, proteasome inhibition also induces MAPK8-dependent multisite phosphorylation of BCL2 and its disassociation from BECN1, an event that is known to activate autophagy (Fig. 5).44 Together they suggest a signaling cascade where proteasome inhibition leads to elevated UPR and ERN1 signaling, which in turn induces MAPK8 activation and subsequent phosphorylation of BCL2, thus freeing the essential autophagy regulator BECN1 from the inactive BCL2-BECN1 complex to stimulate autophagy. Notably, a similar MAPK8-BCL2-BECN1 pathway has also been linked to starvation-induced autophagy.37 Under normal growth conditions, ER-localized BCL2 binds to BECN1 and inhibits BECN1-stimulated autophagy. Starvation induces strong MAPK8 activation and eventual dissociation of BECN1 from the phosphorylated BCL2 for autophagic induction. Thus, although unlike nutritional stress, proteasome inhibition does not engage MTOR pathway, both utilize a common stimulatory mechanism involving MAPK8-BCL2-BECN1 pathway to activate autophagic response.
Consistent with this finding, simultaneous stress with both nutritional starvation and proteasome inhibition results in greater autophagic response (Fig. S1), while MTOR activation by either RHEB overexpression or depletion of the TSC1-TSC2 complex both blunt the proteasome inhibition-induced autophagy (Fig. 3D and E), implying an overlapping regulatory mechanism(s) governing both the proteasomal inhibition- and starvation-induced autophagy. In such a case, it also raises another interesting question as to the exact point of intersection between the 2 pathways. Do they all converge on the phosphorylation-dependent MAPK8 activation or through additional layers of regulation? Noticeably, an earlier study showed that in both the TP53/p53 (tumor protein p53)-null H1299 cell line derived from human nonsmall cell lung carcinoma and the PTEN (phosphatase and tensin homolog)-deficient U87-MG cell line of human glioblastoma origin, active proteasomal function is specifically required for proper MTOR activity, as MG132 treatment inhibits both the initiation and maintenance of MTORC1 signaling.45 As similar MG132 treatment in our study showed little effect on the MTOR activity in the HEK293 cells (Fig. 3A and B), which contain both wild-type TP53 and PTEN,46,47 it raises a possibility that signal inputs from TP53 and PTEN, 2 essential mediators of cellular stress and growth conditions with demonstrated functional links to MAPK8 and MTOR pathways,48–50 play such regulatory roles in mediating the crosstalk between different autophagic pathways. It is likely that faced with a plethora of extra- and intra-cellular stresses such as mitogen stimulation, starvation, and protein misfolding, mammalian cells command multiple avenues of regulation to sense and incorporate these information, so as to fine-tune the overall cellular autophagic response and survival.
Another puzzling question is the exact role of the cargo receptors SQSTM1 and NBR1 in the proteasome inhibition-induced autophagy. Although SQSTM1 and NBR1 are necessary for the proteasome inhibition-induced processing of the GST-BHMT reporter, their depletion does not affect MAPK8 activation (Fig. S3), suggesting their involvement either in a later step during autophagic activation or in the recruitment of GST-BHMT into the autophagosome. Surprisingly, we could not detect a meaningful level of ubiquitinated GST-BHMT or its physical interaction with SQSTM1 (Fig. 7), raising an intriguing possibility that these cargo receptors play a regulatory and/or indirect role in the autophagic induction in response to proteasomal dysfunction. Existing evidence indeed support such a scenario. For example, it has been shown that BHMT is highly enriched on autophagosomal membranes,51,52 suggesting a potential direct or indirect physical interaction with autophagosome proteins including MAP1LC3. Further, cargo receptors SQSTM1 and NBR1 not only recognize and bind to ubiquitinated proteins, but are also required for their assembly into aggregate-like structures (aggresomes) that are important for their subsequent recognition and sequestration by the MAP1LC3-containing autophagosome structures, as supported by the findings that loss of SQSTM1 significantly suppresses the accumulation of ubiquitinated proteins as well as the formation of inclusion bodies both in autophagy-deficient neurons and hepatocytes, and also in hepatocytes with impaired proteasome activity that otherwise show elevated levels of ubiquitinated aggregates.53,54 Given these observations, one putative scenario is that upon proteasomal impairment, cargo receptors SQSTM1 and NBR1 recruit ubiquitinated proteins into aggresomes, a prerequisite step for their recognition by the MAP1LC3-containing membranes that subsequently assemble into mature autophagosomes. During this process, as BHMT are normally highly enriched on the membranes of autophagosomes or their precursors,51,52 they are passively packaged into autophagosomes and subsequently processed. In the absence of the cargo receptors, aggresomes cannot form,52,53 thereby abolishing the MAP1LC3-mediated autophagosome formation and the parallel sequestration of BHMT. Many other possibilities exist and many questions remain, including whether and how the multimeric BHMT are preferentially associated with autophagosome membranes, that need to be further addressed in the future.
The above data also raise another intriguing question of how crosstalk signaling is integrated from the dysfunctional proteasome to the autophagy machinery, especially how ERN1, MAPK8, and BCL2, coordinate with cargo receptors SQSTM1 and NBR1, and with ULK1 to activate autophagy. Notably, ER localization stands out as a converging theme among these different players: ERN1 is a type I transmembrane kinase on ER while activated MAPK8 targets only the ER-localized pool of BCL2 for BECN1-dependent autophagy;37 Further, both ULK1 and SQSTM1 have been reported to localize to the ER;55 Coincidently, ER has been proposed to be one of sources for the autophagosome membrane.56 Do they act in a single linear pathway or work in parallel in a concerted manner? Among many possibilities, one putative scenario is that proteasome inhibition causes abnormal buildup of ubiquitinated proteins in the cell which are gradually sequestrated into aggresomes through the cargo receptors SQSTM1 and NBR1 (Figs. 7C, E and 8D). The accumulating aggresomes in turn might provoke an independent stress signal, potentially sensed and transduced by the cargo receptors SQSTM1 and NBR1 who have been suggested to play a signaling role,57,58 and act in concert with the ER stress-activated ERN1-MAPK8-BCL2-BECN1 pathway to stimulate ULK1 and initiate the autophagy cascade. More studies are needed to clarify the detailed mechanism to answer this interesting question.
Proteasome inhibition induces a Cvt-like autophagy response in mammalian cells
Intriguingly, in nutrient-rich conditions, BHMT processing induced by proteasome inhibition bears remarkable similarity to the Cvt pathway in yeast. In particular, they both require specific cargo receptors (SQSTM1 and NBR1 in mammalian and their counterpart Atg19 in yeast), which, by contrast, are not essential in starvation-induced nonselective autophagy.7 Moreover, reminiscent of the yeast Cvt pathway, in which the formation of dodecamer prApe1 is a prerequisite step for cargo sequestration by the autophagosome,7 the C-terminal multimerization domain of BHMT is necessary for its autophagic processing in response to proteasome inhibition, although this domain is dispensable for its autolysosomal loading during starvation (Fig. 8D). Together, these findings raise the possibility that in mammalian cells, proteasome dysfunction in nutrient-rich conditions induces a Cvt-like selective autophagy response.
Compared with starvation-induced nonselective autophagy, which is an adaptive cellular response that sacrifices nonessential cellular components to sustain survival, selective macroautophagy mainly targets particular substrates such as unwanted and/or damaged proteins and organelles through specific receptors. As one of the main cellular clearance mechanisms, interference with proteasome function leads to the accumulation of ubiquitinated and often misfolded proteins that can serve as cognate targets for selective autophagy.3 Thus, it is reasonable to assume that confronted by a dysfunctional proteasome, the stress-challenged cell employs the selective autophagy pathway as a preferred and effective compensatory mechanism to minimize cellular stress and maintain proper proteostasis. In support of this, proteasome inhibition does not affect the activities of MTORC1 and PRKAA (Fig. 3), the essential upstream regulators of nonselective autophagy.
It is important to compare the apparent difference between the established selective autophagy pathways in yeast and mammalian cells with the cascade mediating the proteasome inhibition-induced BHMT processing. In particular, the cargo receptors SQSTM1 and NBR1 are known to bind to ubiquitinated substrates and facilitate their aggregation and sequestration into the autophagosome. However, we could not detect an obvious physical association between the GST-BHMT reporter and Ubiquitin or the cargo receptor SQSTM1 by either coimmunoprecipitation or coimmunostaining analyses (Fig. 7 and Fig. S4). This could be due to the insufficient sensitivity of our experiments, or it might suggest that SQSTM1 does not serve as the cargo receptor for delivery of GST-BHMT into the autophagosome. In agreement with the latter possibility, we found that BHMT itself is weakly ubiquitinated and is not stabilized by proteasome inhibition (Fig. 7A-C). This raises a possibility that BHMT might utilize a SQSTM1 and NBR1- and ubiquitin-independent mechanism for its delivery to the autophagosome. Similar precedents exist in both yeast and mammalian cells. In the Cvt pathway, the maturation of the cargo protein prApe1 does not involve its ubiquitination.7 In mammalian cells, mutant SOD protein is also degraded through the autophagy pathway in a ubiquitin-independent manner.40 As GST-BHMT is a cytosolic protein enriched within the autophagosome and autolysosome membrane,51 it might be directly sequestered into the autophagosome without the requirement for an adaptor or through alternative receptor proteins such as OPTN (optineurin),59 which is known to recognize cargos in a ubiquitin-independent manner. Thus, although many questions remain to be answered, considering the notable similarity between the Cvt pathway and the autophagy-dependent BHMT processing that occurs in response to proteasome inhibition, it is plausible that proteasome dysfunction induces a compensatory autophagic response that is selective.
Interestingly, cargo polymerization emerges as a shared feature among several proposed selective autophagy mechanisms, including the dodecamer prApe1 in the yeast Cvt pathway, aggregates formation in aggrephagy and now the multimerization of BHMT in proteasome inhibition-induced BHMT processing. An intriguing possibility is that there might exist an “aggregate sensor,” potentially the cargo receptors SQSTM1 and NBR1 who have been postulated to play additional signaling roles,57,58 in the selective autophagy pathway that facilitates the interaction between polymerized cargos and the autophagosome. It is equally likely that the cargo in its polymerized form can directly facilitate autophagosomal assembly and sequestration, as has been proposed for the dodecamer prApe1 in the yeast Cvt pathway.7
In mammalian cells, selective autophagy has been gaining more attention due to its increasing importance in human conditions such as liver and brain degenerative diseases.12 However, few established assays are available that allow detailed examination of this important protective cellular pathway. Our data suggest that the GST-BHMT assay, which has been effective in monitoring starvation-induced nonselective autophagy, might also be applied to study selective autophagy in mammalian systems. Technically, as the processing of BHMT only occurs in the lysosome, this endpoint-based autophagy assay does not require fractionation or autophagosome-lysosome enrichment. As versatile proteasome inhibitors with different properties are available and easily applicable, such as the reversible inhibitor MG132 and irreversible inhibitor lactacystin, this proteasome inhibition-based assay might provide a convenient platform for future dissection of the selective autophagy pathway. Further, it is also possible that by developing modified versions of GST-BHMT reporter, for example a reporter with organelle-specific localization signals, this convenient biochemical assay might also be useful for analyzing other types of selective autophagy such as pexophagy and mitophagy.
Materials and Methods
GST-BHMT assay
The GST-BHMT assay was performed as described previously.20,21 Briefly, HEK293T cells transfected with pRK5-GST-BHMT and various siRNAs were treated with 10 µM MG132 or EBSS medium together with 11 µM leupeptin and 6 µM E-64d for 6 h prior to immunoprecipitation (IP) lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100 (Sigma-Aldrich, T8787), 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM sodium orthovanadate, 1 µg/ml leupeptin (Sigma-Aldrich, L2884), 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich, P7626). After brief sonication, whole cell lysate was centrifuged for 18506 g at 4°C for 30 min and the supernatant fraction was incubated with precleared glutathione agrose for 3 h. The immuoprecipitated GST fusion proteins were washed 3 times with IP lysis buffer before denaturation in 2X SDS sample buffer for western blotting. Anti-GST antibody (Santa Cruz Biotechnolgy, sc-138) was used for detecting both GST-BHMT and GST-BHMT(FRAG). GFP expression detected by anti-MYC antibody (Santa Cruz Biotechnolgy, sc-40) was used as a loading control.
siRNAs
All the following siRNAs with published sequence are synthesized from Sigma-Aldrich: siATG7 (ccaacacacucgagucuuu);60 siULK1-1 (ggauacgucuuguaaucuu); siULK1-2 (gaggcaguucuuuguucaa);20 siSQSTM1-1 (gcauugaaguugauaucgau);11 siSQSTM1-2 (ccgaaucuacauuaaagagaa);61 siPSMB1 (gacuaaagauguauaagca), siPSMB2 (caccgacuaucucacguga), siPSMB5 (ugauagagaucaacccaua). Additional siRNAs were: siNBR1 (Santa Cruz Biotechnology, sc-94187),62 and siMAPK8/JNK1 siRNA (Cell Signaling Technology, 6232). The following siRNAs were from Life Technologies: siATF6 (s223544), siEIF2AK3 (s18102), siERN1/IRE1 (s200432), siXBP1 (s14915), siTSC1 (s14433) and siTSC2 (s14438).
Plasmids and reagents
pRK5-GST-BHMT and pRK5-GST-BHMTΔ, pRK5-GST-LSCS-BHMT and pRK5-GST-LSCS-BHMT51Δ were provided by Dr. P.B. Dennis (Wright State University).20 HA-SQSTM1 (28027), GFP-MAP1LC3 (24920), HA-Ub (18712) HA-RHEB (19310), FLAG-MAP2K7-MAPK8 DN (19730), FLAG-MAP2K7-MAPK8/JNK CA (19726), FLAG-BCL2 (18003), HA-BECN1 /beclin 1(24399), and MTOR KD (8482) were purchased from Addgene; MYC-BCL2, MYC-BCL2T69A,S87A (MYC-BCL2[2A]), and MYC-BCL2T69A,S70A,S87A (MYC-BCL2[3A]) were gifts from Dr. Yongjie Wei (The University of Texas Southwestern Medical Center at Dallas. LY294002 (LC laboratories, L-7962), wortamannin (LC laboratories, W-2990), bortezomib (LC laboratories, B-1408), AEBSF (Enzo Life Sciences, 89147-060), MG132 (Enzo Life Sciences, BML-PI102-0005), chymostatin (Fisher Scientific, 50-751-7222), antipain (Fisher Scientific, 50-147-298), 3-methyladenine (Cayman Chemical, 13242), epoxomicin (Cayman Chemical, 101962-668), lactacystin (Cayman Chemical, 101955-536), leupeptin (Sigma-Aldrich, L2884), E-64d (Sigma-Aldrich, E8640), sodium pyrophosphate (Sigma-Aldrich, P8010), β-glycerolphosphate (Sigma-Aldrich, 50020), sodium orthovanadate (Sigma-Aldrich, S6508), phenylmethylsulfonyl fluoride (Sigma-Aldrich, P7626), chloroquine (Sigma-Aldrich, C6628), Baf A1 (Sigma-Aldrich, B1793), SP600125 (Sigma-Aldrich, S5567), tunicamycin (Sigma-Aldrich, T7765), thapsigargin (Sigma-Aldrich, T9033), brefeldin A (Sigma-Aldrich, B7651).
Antibodies
The following primary antibodies were used: GST (for immunoprecipitation: sc-57753), MYC (sc-40), SQSTM1/p62 (sc-28359), ATF6 (sc-22799), NBR1 (sc-130380), and ULK1 (sc-33182) were from Santa Cruz Biotechnology; HA (Roche Life Science, 12CA5); MAP1LC3 (MBL International, PM036); Ub (EMD Millipore, ST1200 [FK2]); FLAG (Sigma-Aldrich, F3165); PSMB1 (BML-PW8140), PSMB2 (BML-PW8145), and PSMB5 (BML-PW8895) were from Enzo Life Sciences. ATG7 (8558), XBP1 (12782), p-MAPK8 (Thr183/Tyr185; 4668), MAPK8 (3708), p-BCL2 (Ser70; 2827), CTNNB1/β-Catenin (9582), RPS6KB1 (9202), p-RPS6KB1 (Thr389; 9234), EIF4EBP1 (9452), p-EIF4EBP1 (Thr37/46; 9459), p-ULK1 (Ser555; 5869), p-ULK1 (Ser757; 6888), ACACA/B (3676), p-ACACA/B (Ser79; 3661), PRKAA (2603), p-PRKAA (Thr172; 2535), HSPA5 (4592), GST (2625); GFP (2956), ERN1 (3294), EIF2AK3/PERK (3192), TSC1 (6935), TSC2 (3990), DDIT3 (5554), P4HB (2446), and ATF4 (11815) were from Cell Signaling Technology. Secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594 for immunofluorescence imaging were from Life Technologies (A-11029; A-11037). Secondary antibodies including IRdye 680RD and IRdye 800CW (926-68072; 925-68073; 926-32212; 926-32213) for the Odyssey western system were from LI-COR Biosciences (Lincoln, NE).
Cell culture and transfection
HEK293T cells were maintained in DMEM medium (Corning, 10-013-CV) containing 10% fetal bovine serum (Invitrogen, 10082147), 100 IU penicillin, and 100 µg/ml streptomycin. Transfections of small interfering RNAs and plasmid DNA were performed using lipofectamine 2000 (Life Technologies, 11668027) according to the manufacturer's instructions. For inducing starvation-mediated autophagy, DMEM as nutrition-rich medium was replaced by EBSS (Sigma-Aldrich, E2888) medium for further incubation from 2 h to 6 h as indicated. For studying compensated autophagy induced by blocking proteasome function, MG132 was used in DMEM at a final concentration from 5 µM to 20 µM for indicated time of incubation. Cell lysates were prepared using IP lysis buffer as described previously.
Coimmunoprecipitation and western blotting
Transiently transfected 293T cells in 60-mm dishes were lysed in an IP-lysis buffer and sonicated briefly before centrifuged at 18506 g for 30 min at 4°C. For detection of protein ubiquitination, 10 mM N-ethylmaleimide (Fisher Scientific, O2829-25) was added for inhibiting de-ubiquitination enzymes. GST-BHMT proteins or MYC-CTNNB1 proteins were immunoprecipitated from the cell lysate with anti-GST, anti-MYC and protein A/G plus agarose beads (Santa Cruz Biotechnology, sc-2003). Immunoprecipitates or total cell lysates were added by 2X SDS sample buffer and then subjected to discontinuous SDS-PAGE analysis. Proteins were transferred to nitro-cellulose membranes using a Bio-Rad (Hercules, CA) mini transfer apparatus followed by blocking with 5% nonfat milk. Primary antibodies and secondary antibodies were used usually at 1:1000 and 1:10000 dilutions respectively before using an Odyssey system to detect the fluorescence signal.
Immunostaining
HeLa cells were grown on glass coverslips in the DMEM medium for 16 h. Expression plasmids of GST-BHMT was transfected into HeLa cells. Approximately 24 h after transfection, cells were left untreated or treated with 10 µM MG132 for another 6 h, and fixed with 4% formaldehyde-phosphate-buffered saline (Electronic Microscopy Sciences, 15710; Fisher Scientific, BP399-500) for 10 min. Cells were incubated with primary antibodies against GST, ubiquitin or SQSTM1 overnight at 4°C after 1 h blocking by 5% goat serum. Secondary antibodies conjugated with Alexa Fluor 488 or 594 were used to visualize the localization. DAPI was used to indicate the nuclear. Image was taken with a Zeiss Compound Microscope (Thornwood, NY) and processed by Amira software for statistical analysis. Colocalization quantification: about 30 to 80 cells with BHMT puncta were chosen to calculate the numbers of 3 colors, red for BHMT, green for ubiquitin or SQSTM1, and yellow for colocalized signal. As the number of BHMT puncta is normally much higher than that of ubiquitin or SQSTM1, we choose the former as the denominator to calculate the ratio of colocalization.
Filter trap assay
Filter trap assay was performed as previously described.41 Briefly, cells transiently expressed with GST-BHMT or GFP-HTT-exon1-Q72 were lysed in phosphate-buffered saline by sonication. Bradford method was used to calculate protein concentration. The lysate was applied with different concentrations to the dot-blotting apparatus (Bio-Rad, Hercules, CA) and western blotting was used to detect the trapped proteins on the nitrocellular membranes.
Statistics analysis
Statistical analysis was performed with Microsoft Excel and P values were calculated by the Student t test. Data are represented as mean+s.e.m. P < 0.05 is considered significant.
Supplementary Material
Acknowledgment
We are thankful to Dr. Patrick B. Dennis (Wright State University) for the BHMT plasmids and Dr. Yongjie Wei (The University of Texas Southwestern Medical Center) for the BCL2 plasmids. We also appreciate the critical reading of this manuscript by Dr. Nicholas J. Justice (The University of Texas Health Science Center at Houston).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work is partially supported by NIH grant R01-NS069880.
References
- 1.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124–31; PMID:20034776; http://dx.doi.org/ 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X, et al.. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297–305; PMID:19258318; http://dx.doi.org/ 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol 2012; 2012:736905; PMID:22518139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67–93; PMID:19653858; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett 2010; 584:1287–95; PMID:20083114; http://dx.doi.org/ 10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunlop EA, Tee AR. The kinase triad, AMPK, mTORC1 and ULK1, maintains energy and nutrient homoeostasis. Biochem Soc Trans 2013; 41:939–43. [DOI] [PubMed] [Google Scholar]
- 7.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett 2010; 584:1359–66; PMID:20146925; http://dx.doi.org/ 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, Miao L, et al.. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 2009; 136:308–21; PMID:19167332; http://dx.doi.org/ 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 9.Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J Androl 2010; 31:53–60; PMID:19875490; http://dx.doi.org/ 10.2164/jandrol.109.008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 2010; 12:836–41; PMID:20811356; http://dx.doi.org/ 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- 11.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131–45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 12.Jing K, Lim K. Why is autophagy important in human diseases? Exp Mol Med 2012; 44:69–72; PMID:22257881; http://dx.doi.org/ 10.3858/emm.2012.44.2.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al.. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885–9; PMID:16625204; http://dx.doi.org/ 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- 14.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al.. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880–4; PMID:16625205; http://dx.doi.org/ 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- 15.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 1998; 8:397–403; PMID:9789328; http://dx.doi.org/ 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- 16.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al.. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007; 447:859–63; PMID:17568747 [DOI] [PubMed] [Google Scholar]
- 17.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513–24; PMID:17620365; http://dx.doi.org/ 10.2353/ajpath.2007.070188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Q, Su H, Tian Z, Wang X. Proteasome malfunction activates macroautophagy in the heart. Am J Cardiovasc Dis 2011; 1:214–26; PMID:22081794 [PMC free article] [PubMed] [Google Scholar]
- 19.Choi CH, Lee BH, Ahn SG, Oh SH. Proteasome inhibition-induced p38 MAPK/ERK signaling regulates autophagy and apoptosis through the dual phosphorylation of glycogen synthase kinase 3beta. Biochem Biophys Res Commun 2012; 418:759–64; PMID:22310719; http://dx.doi.org/ 10.1016/j.bbrc.2012.01.095 [DOI] [PubMed] [Google Scholar]
- 20.Mercer CA, Kaliappan A, Dennis PB. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy 2008; 4:185–94; PMID:18059170; http://dx.doi.org/ 10.4161/auto.5275 [DOI] [PubMed] [Google Scholar]
- 21.Dennis PB, Mercer CA. The GST-BHMT assay and related assays for autophagy. Methods Enzymol 2009; 452:97–118; PMID:19200878; http://dx.doi.org/ 10.1016/S0076-6879(08)03607-0 [DOI] [PubMed] [Google Scholar]
- 22.Furuya N, Kanazawa T, Fujimura S, Ueno T, Kominami E, Kadowaki M. Leupeptin-induced appearance of partial fragment of betaine homocysteine methyltransferase during autophagic maturation in rat hepatocytes. J Biochem 2001; 129:313–20; PMID:11173534; http://dx.doi.org/ 10.1093/oxfordjournals.jbchem.a002859 [DOI] [PubMed] [Google Scholar]
- 23.Groll M, Huber R. Inhibitors of the eukaryotic 20S proteasome core particle: a structural approach. Biochim Biophys Acta 2004; 1695:33–44; PMID:15571807 [DOI] [PubMed] [Google Scholar]
- 24.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A 1999; 96:10403–8; PMID:10468620; http://dx.doi.org/ 10.1073/pnas.96.18.10403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curran MP, McKeage K. Bortezomib: a review of its use in patients with multiple myeloma. Drugs 2009; 69:859–88; PMID:19441872; http://dx.doi.org/ 10.2165/00003495-200969070-00006 [DOI] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445–544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132–41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009; 122:3589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 2012; 48:242–53; PMID:22959274; http://dx.doi.org/ 10.1016/j.molcel.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilella-Bach M, Nuzzi P, Fang Y, Chen J. The FKBP12-rapamycin-binding domain is required for FKBP12-rapamycin-associated protein kinase activity and G1 progression. J Biol Chem 1999; 274:4266–72; PMID:9933627; http://dx.doi.org/ 10.1074/jbc.274.7.4266 [DOI] [PubMed] [Google Scholar]
- 31.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al.. Phosphorylation of ULK1 (hATG1; by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456–61; PMID:21205641; http://dx.doi.org/ 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251–62; PMID:22436748; http://dx.doi.org/ 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol 2012; 34:286–97; PMID:23070014 [PubMed] [Google Scholar]
- 34.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 2006; 281:30299–304; PMID:16901900; http://dx.doi.org/ 10.1074/jbc.M607007200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012; 13:89–102; PMID:22251901 [DOI] [PubMed] [Google Scholar]
- 36.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, Davis RJ. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol Cell Biol 2002; 22:4929–42; PMID:12052897; http://dx.doi.org/ 10.1128/MCB.22.13.4929-4942.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678–88; PMID:18570871; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009; 5:732–3; PMID:19398892; http://dx.doi.org/ 10.4161/auto.5.5.8566 [DOI] [PubMed] [Google Scholar]
- 39.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al.. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505–16; PMID:19250911; http://dx.doi.org/ 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 40.Gal J, Ström AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem 2009; 111:1062–73; PMID:19765191; http://dx.doi.org/ 10.1111/j.1471-4159.2009.06388.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol 1999; 309:375–86; PMID:10507036; http://dx.doi.org/ 10.1016/S0076-6879(99)09026-6 [DOI] [PubMed] [Google Scholar]
- 42.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584:1393–8; PMID:20040365; http://dx.doi.org/ 10.1016/j.febslet.2009.12.047 [DOI] [PubMed] [Google Scholar]
- 43.Park HS, Jun do Y, Han CR, Woo HJ, Kim YH. Proteasome inhibitor MG132-induced apoptosis via ER stress-mediated apoptotic pathway and its potentiation by protein tyrosine kinase p56lck in human Jurkat T cells. Biochem Pharmacol 2011; 82:1110–25; PMID:21819973; http://dx.doi.org/ 10.1016/j.bcp.2011.07.085] [DOI] [PubMed] [Google Scholar]
- 44.Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res 2012; 2:214–21; PMID:22485198 [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh P, Wu M, Zhang H, Sun H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle 2008; 7:373–81; PMID:18235224; http://dx.doi.org/ 10.4161/cc.7.3.5267 [DOI] [PubMed] [Google Scholar]
- 46.Sun L, Shen X, Liu Y, Zhang G, Wei J, Zhang H, Zhang E, Ma F. The location of endogenous wild-type p53 protein in 293T and HEK293 cells expressing low-risk HPV-6E6 fusion protein with GFP. Acta Biochim Biophys Sin (Shanghai) 2010; 42:230–5; PMID:20213049; http://dx.doi.org/ 10.1093/abbs/gmq009 [DOI] [PubMed] [Google Scholar]
- 47.Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB. PCAF modulates PTEN activity. J Biol Chem 2006; 281:26562–8; PMID:16829519; http://dx.doi.org/ 10.1074/jbc.M605391200 [DOI] [PubMed] [Google Scholar]
- 48.Lorin S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy 2010; 6:153–4; PMID:19949306; http://dx.doi.org/ 10.4161/auto.6.1.10537 [DOI] [PubMed] [Google Scholar]
- 49.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A 2005; 102:8204–9; PMID:15928081; http://dx.doi.org/ 10.1073/pnas.0502857102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cogwheels of cancer. Autophagy 2006; 2:67–73; PMID:16874041; http://dx.doi.org/ 10.4161/auto.2.2.2458 [DOI] [PubMed] [Google Scholar]
- 51.Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E, et al.. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem 1999; 274:15222–9; PMID:10329731; http://dx.doi.org/ 10.1074/jbc.274.21.15222 [DOI] [PubMed] [Google Scholar]
- 52.Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 2007; 3:300–22; PMID:17377489; http://dx.doi.org/ 10.4161/auto.3910 [DOI] [PubMed] [Google Scholar]
- 53.Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, Yamamoto M, et al.. Proteasome dysfunction activates autophagy and the keap1-nrf2 pathway. J Biol Chem 2014; 289:24944–55; PMID:25049227; http://dx.doi.org/ 10.1074/jbc.M114.580357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al.. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131:1149–63; PMID:18083104; http://dx.doi.org/ 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- 55.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 2011; 192:17–27; PMID:21220506; http://dx.doi.org/ 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12:831–5; PMID:20811355; http://dx.doi.org/ 10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- 57.Liu XD, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, Ishii T, Komatsu M, Eissa NT. Transient aggregation of ubiquitinated proteins is a cytosolic unfolded protein response to inflammation and endoplasmic reticulum stress. J Biol Chem 2012; 287:19687–98; PMID:22518844; http://dx.doi.org/ 10.1074/jbc.M112.350934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci 2007; 32:95–100; PMID:17174552; http://dx.doi.org/ 10.1016/j.tibs.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 59.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci 2013; 126:580–92; PMID:23178947; http://dx.doi.org/ 10.1242/jcs.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 2006; 281:8518–27; PMID:16415355; http://dx.doi.org/ 10.1074/jbc.M506182200 [DOI] [PubMed] [Google Scholar]
- 61.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem 2010; 285:27879–90; PMID:20573959; http://dx.doi.org/ 10.1074/jbc.M110.119537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mardakheh FK, Yekezare M, Machesky LM, Heath JK. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J Cell Biol 2009; 187:265–77; PMID:19822672; http://dx.doi.org/ 10.1083/jcb.200905118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.