Abstract
Fine scale population structure of Malays - the major population in Malaysia, has not been well studied. This may have important implications for both evolutionary and medical studies. Here, we investigated the population sub-structure of Malay involving 431 samples collected from all states from peninsular Malaysia and Singapore. We identified two major clusters of individuals corresponding to the north and south peninsular Malaysia. On an even finer scale, the genetic coordinates of the geographical Malay populations are in correlation with the latitudes (R2 = 0.3925; P = 0.029). This finding is further supported by the pairwise FST of Malay sub-populations, of which the north and south regions showed the highest differentiation (FST [North–south] = 0.0011). The collective findings therefore suggest that population sub-structure of Malays are more heterogenous than previously expected even within a small geographical region, possibly due to factors like different genetic origins, geographical isolation, could result in spurious association as demonstrated in our analysis. We suggest that cautions should be taken during the stage of study design or interpreting the association signals in disease mapping studies which are expected to be conducted in Malay population in the near future.
Electronic supplementary material
The online version of this article (doi:10.1186/s40246-015-0039-x) contains supplementary material, which is available to authorized users.
Keyword: Malay, Population sub-structure, FST, Latitude-PC correlation, GWAS simulation
Background
Malaysia, a multi-ethnic, multi-lingual, multi-cultural and multi-religious country, is located at the crossroads of Southeast Asia. It is separated by the South China Sea into two land masses namely, the Peninsular Malaysia and East Malaysia (also known as the Borneo island). Malaysia has a total population of about 30 million people, of which approximately 26 million populate the Peninsular Malaysia. Among the major populations in Peninsular Malaysia, the Malays are the largest ethnic group and make up to 63% of the total population follow by Chinese, Indians and other minority ethnic groups.
Many Malays are of Malayo-Polynesian (Austronesian) origin that are culturally and historically heterogeneous [1]. The Malays from the west coast of Peninsular Malaysia are historically linked to Sumatera across the Straits of Malacca; while those from the south are thought to have migrated from Jawa, Sulawesi and other parts of Indonesia [2]. The Malays from the north Peninsular have a closer affinity to the Malay Muslims from the Southern Thai due to geographical location. The history of Singapore has never been separated from Peninsular Malaysia since the first century until the year 1965 when Singapore became an independent republic. Therefore, it is very likely that the Malays of Singapore have a similar history of origin as those from the southern part of Peninsular Malaysia [3].
Earlier studies had indicated potential genetic sub-structure among the different groups of Malays from Peninsular Malaysia [2, 4, 5], which could be possibly attributed to the migration history of these respective sub-groups. However, fine-scale sub-structure of the Malay population remained poorly described, especially, previous studies were based on very limited sample size. Indeed, this potentially poses confounding factors to the genetic association studies, in particular genome-wide association studies (GWAS), leading to spurious association signals [6]. Being one of the major populations in the Southeast Asia, characterizing population substructure is crucial in designing, analyzing and interpreting any genetic association study in this region.
In this study, we showed that the genetic diversity and population sub-structure of the Malays from Peninsular Malaysia are correlated to the geographical latitude. Notably, we observed the main differentiations between populations corresponding to the north and south Peninsular Malaysia. In addition, simulation analyses carried out also revealed that the genetic association is greatly affected by population sub-structure, suggesting that consideration of population stratification of samples at the stage of study design and careful interpretation of the association signals are necessary when mapping complex diseases in Malay populations.
Results
Population substructure
We first compared the genetic diversity of the Peninsular Malays from a global scale with 6 populations from HapMap3 including YRI, CHB, JPT, CEU, MEX and GIH. PC plot indicated that the Malays clustered closely to the East Asian populations as expected, and showed a rather small genetic diversity. Several Malay individuals from northern Peninsular Malaysia (PMM) showed closer affinity to the South Asia populations (GIH) (Fig. 1a). We then performed PCA for the Peninsular and Singapore Malays, and revealed a seemingly homogenous cluster (Additional file 1: Figure S1). However, some level of differentiations were observed corresponding to three geographical regions (north, center and south), despite samples from center region that was scattered around (Fig. 1b). We subsequently excluded the samples form the center regions (Pahang and Selangor), and re-ran the smartPCA. Two clusters were observed representing the north and south regions, respectively (Fig. 1c).
Fig. 1.
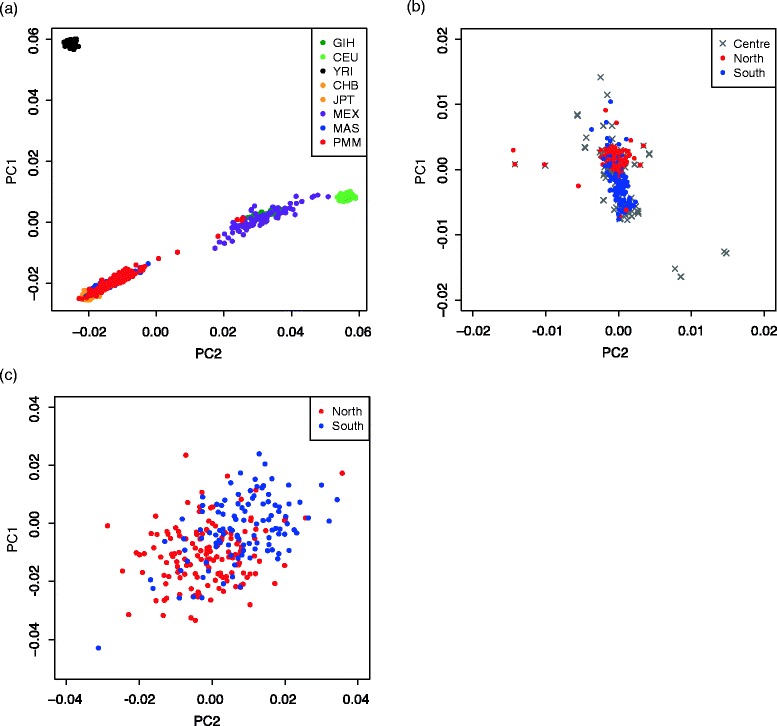
Principle Component Analysis (PCA) (a) Global PCA including populations from HapMap3. GIH, Gujarati India Houston; CEU, Northern and Western European from CEPH collection; YRI, Yoruba Ibadan from Nigeria; CHB, Chinese Beijing; JPT, Japanese Tokyo; MEX, Mexican ancestry from Los Angeles; MAS, Metropolitan Malays from Singapore; PMM, Malays from Peninsular Malay. The Malay populations are of East Asian descendant. (b) PCA plot including samples categorized into North vs Centre vs South; (c) PCA plot which included only North vs South. Symbols in red represent the northern region; symbols in blue represent southern region. Several outliers were excluded from the PCA plot
In ADMIXTURE analysis, a significant difference was observed between the Malays from the north and south in the major component, with 57% and 65% in the north and south, respectively (P < 0.0001; Fig. 2). At K=3, the newly appeared component (denoted in green) was seen slightly higher in the central Malays than in the south Malays (6.8% vs 3%; P = 0.0415).
Fig. 2.
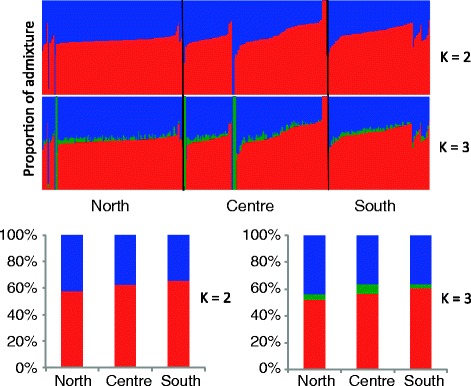
ADMXITURE analysis of the Malay populations classified according to regions. The bottom plots represented by percentages (Y-axis) indicates the average ADMIXTURE values for each region
Correlation of genetic and geographic coordinates
Given the fact that the PC1 as well as the ADMIXTURE analysis showed significant differences between northern and southern Malay samples, we then investigated if the genetic diversity between these sub-structure of Malays in Peninsular Malaysia were attributed to geographical coordinates. Average PC1 values of southern Malay samples (corresponding to Fig. 1b) were generally less than 0 (except for Johor), whilst all geographically defined northern regions with PC1 >0 (Fig. 3). When we compared the PC1 with geographical latitude of these sample locations, a significant correlation was observed (R2 = 0.3925; P = 0.029; Fig. 4). Due to the geographical nature, Peninsular Malaysia is divided into west coast and east coast by the Titiwangsa Ranges. We therefore asked if the genetic diversity could be attributed to the geographical longitude as well. Analysis between PC1 and geographical longitude, however showed no significant correlation (R2=0.0066; P = 0.7924; Addional file 1: Figure S2). We also evaluated if the genetic diversity was related to geographical distance between two populations, but found no significant correlation of FST between populations and the geographical distances between them (R2 = 0.01918; P = 0.1385; Additional file 1: Figure S3).
Fig. 3.
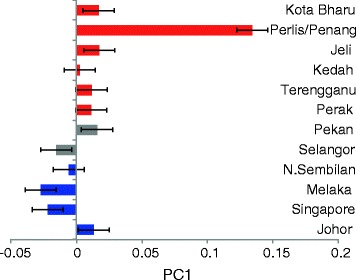
Average PC1 values of the Malay sub-populations from Peninsular Malaysia and Singapore. Standard error of each population is indicated. The PC1 values correlated well to the geographical locations of each population except for Johor
Fig. 4.
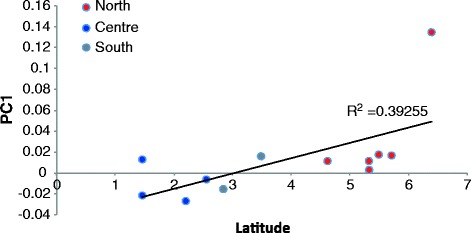
Correlation between PC1 and latitude coordinate (P = 0.029)
Genetic differentiation between northern and southern Malays
The regional FST values indicated the highest regional diversity between the north and the south after 1,000 times bootstrapping repeats (FST = 0.001; CI = 2.07E - 04) (Table 1; Additional file 2: Table S1). To further identify the genomic regions that are highly differentiated between northern and southern Peninsular Malay, we computed the FST values of the 41,400 SNPs between northern and southern samples, and identified 428 SNPs listed in the top 1% of the FST (Additional file 2: Table S2); of which 80 (0.1%) had an FST value >0.05 (Table 2). SNP with the highest FST value was rs4149264, residing in the candidate gene ABCA1 - a major gene responsible for high-density lipolipoprotein cholesterol (HDL-c) synthesis. Another highly differentiated SNP, rs4148475, is located at the candidate gene ABCC4. This gene is a member of the superfamily of ATP-binding Cassette (ABC) transporters, which may play a role in cellular detoxification [7]. A missense variant rs1056836 appears to be one of the four highly differentiated SNPs, leading to a change of valine to leucine in candidate gene CYP1B1, which had a minor allele frequency of 0.48 and 0.19 in northern and southern peninsular Malays, respectively (FST = 0.2037). This candidate gene is responsible in drug metabolism and synthesis of cholesterols, steroids and lipids. It was found to play a role in the susceptibility of glaucoma [8, 9]. We performed an enrichment analysis with DAVID (http://david.abcc.ncifcrf.gov/) by including the top 1% highly differentiated variants, but identified no significant enrichment after Benjamini correction (Additional file 2: Table S3).
Table 1.
Pairwise FST bootstrap values of the Malay between the 3 regions of Peninsular Malaysia
| North | Centre | South | |
|---|---|---|---|
| North | - | 0.00083315 (CI =2.0684E-04) | 0.00111661 (CI = 2.68108E-06) |
| Centre | - | 0.00058556 (CI = 4.0972E-06) | |
| South | - |
Pairwise FST values calculated by bootstrap resampling 1,000 replications
Table 2.
Top 0.1 % SNPs that are highly differentiated between the Malays from northern and southern region of Peninsular (total SNP = 42633)
| rsID | Chr | Position | Minor allele | FST | MAF_North | MAF_South | Gene | Category |
|---|---|---|---|---|---|---|---|---|
| rs4149264 | 9 | 107,677,211 | C | 0.2256 | 0.4856 | 0.1682 | ABCA1 | intronic |
| rs10102377 | 8 | 83,762,822 | T | 0.2251 | 0.4097 | 0.2336 | ||
| rs4148475 | 13 | 95,853,574 | A | 0.2242 | 0.4676 | 0.184 | ABCC4 | intronic |
| rs1056836 | 2 | 38,298,203 | G | 0.2037 | 0.4757 | 0.1934 | CYP1B1 | coding |
| rs1126965 | 17 | 70,642,790 | G | 0.1931 | 0.5 | 0.1822 | SLC39A11 | 3utr |
| rs17769090 | 15 | 70,630,120 | A | 0.1636 | 0.4648 | 0.2381 | ||
| rs6974363 | 7 | 47,633,187 | G | 0.1421 | 0.493 | 0.2333 | ||
| rs837395 | 1 | 47,269,338 | A | 0.1400 | 0.4897 | 0.2383 | CYP4B1 | intronic |
| rs4646430 | 2 | 38,306,415 | G | 0.1384 | 0.4621 | 0.1981 | ||
| rs215101 | 16 | 16,052,973 | G | 0.1206 | 0.4752 | 0.271 | ABCC1 | intronic |
| rs12920607 | 16 | 73,728,620 | C | 0.1183 | 0.475 | 0.2736 | ||
| rs837398 | 1 | 47,266,422 | A | 0.1124 | 0.4897 | 0.2664 | CYP4B1 | intronic |
| rs809367 | 10 | 89,741,806 | A | 0.1088 | 0.4307 | 0.2009 | ||
| rs316133 | 6 | 52,847,551 | C | 0.0957 | 0.4823 | 0.2594 | GSTA4 | intronic |
| rs6130511 | 20 | 42,681,088 | A | 0.0916 | 0.2801 | 0.1 | TOX2 | intronic |
| rs2132845 | 4 | 140,587,125 | T | 0.0910 | 0.4255 | 0.215 | MGST2 | 5utr |
| rs5761313 | 22 | 26,313,745 | T | 0.0887 | 0.4964 | 0.2804 | MYO18B | intronic |
| rs10485805 | 20 | 54,945,783 | G | 0.0853 | 0.4397 | 0.2336 | AURKA | intronic |
| rs10489142 | 1 | 7,363,310 | G | 0.0835 | 0.4507 | 0.3364 | CAMTA1 | intronic |
| rs2274928 | 13 | 24,044,546 | A | 0.0779 | 0.3601 | 0.4346 | LINC00327 | intronic |
| rs11935505 | 4 | 145,226,422 | A | 0.0758 | 0.04643 | 0.1682 | ||
| rs1566869 | 12 | 52,266,348 | A | 0.0695 | 0.3406 | 0.1682 | ||
| rs1884897 | 20 | 6,612,832 | G | 0.0695 | 0.2812 | 0.1215 | ||
| rs4530975 | 7 | 104,415,415 | T | 0.0679 | 0.1862 | 0.05607 | LHFPL3 | intronic |
| rs6024831 | 20 | 54,938,464 | G | 0.0675 | 0.4161 | 0.3915 | FAM210B | intronic |
| rs1160798 | 6 | 112,438,446 | C | 0.0661 | 0.06897 | 0.1934 | LAMA4 | intronic |
| rs2158196 | 4 | 114,416,596 | C | 0.0658 | 0.2391 | 0.09434 | CAMK2D | intronic |
| rs16961766 | 13 | 103,899,499 | A | 0.0645 | 0.3143 | 0.1524 | ||
| rs10962015 | 9 | 15,387,949 | A | 0.0638 | 0.2482 | 0.1028 | ||
| rs6884962 | 5 | 172,682,382 | A | 0.0623 | 0.4896 | 0.3271 | ||
| rs17126776 | 12 | 39,311,625 | A | 0.0603 | 0.1759 | 0.3318 | ||
| rs2755209 | 13 | 41,137,804 | C | 0.0602 | 0.25 | 0.1075 | FOXO1 | intronic |
| rs9783586 | 13 | 108,361,559 | T | 0.0601 | 0.2671 | 0.4393 | FAM155A | intronic |
| rs10968093 | 9 | 27,753,227 | A | 0.0594 | 0.06338 | 0.1776 | ||
| rs2458286 | 8 | 103,978,699 | T | 0.0591 | 0.4424 | 0.3774 | ||
| rs4875364 | 8 | 4,444,592 | C | 0.0583 | 0.3094 | 0.1557 | CSMD1 | intronic |
| rs11145506 | 9 | 80,264,584 | T | 0.0578 | 0.3821 | 0.217 | GNA14 | intronic |
| rs11604366 | 11 | 28,887,766 | C | 0.0577 | 0.2695 | 0.4387 | ||
| rs9881633 | 3 | 112,881,539 | T | 0.0574 | 0.3 | 0.472 | RP11_572M11.3 | intronic |
| rs5762448 | 22 | 28,408,444 | C | 0.0573 | 0.3514 | 0.1916 | TTC28 | intronic |
| rs6467991 | 7 | 83,954,737 | C | 0.0570 | 0.344 | 0.4811 | ||
| rs10089677 | 8 | 122,660,248 | A | 0.0569 | 0.2329 | 0.09813 | ||
| rs1923254 | 13 | 41,084,241 | G | 0.0567 | 0.3776 | 0.2143 | ||
| rs7813806 | 8 | 5,142,665 | C | 0.0567 | 0.2817 | 0.1355 | ||
| rs7625411 | 3 | 112,811,428 | A | 0.0564 | 0.3403 | 0.486 | ||
| rs2922249 | 6 | 127,954,614 | C | 0.0564 | 0.09441 | 0.2196 | ||
| rs2294088 | 8 | 124,526,607 | A | 0.0564 | 0.4514 | 0.2804 | FBXO32 | intronic |
| rs12289262 | 11 | 12,894,758 | T | 0.0560 | 0.3986 | 0.2336 | TEAD1 | intronic |
| rs4937523 | 11 | 130,347,190 | T | 0.0558 | 0.2937 | 0.4626 | ADAMTS15 | intronic |
| rs10807768 | 7 | 13,662,014 | A | 0.0554 | 0.3169 | 0.1651 | ||
| rs976272 | 14 | 61,449,328 | A | 0.0551 | 0.4897 | 0.3178 | SLC38A6 | coding |
| rs13027801 | 2 | 143,602,503 | C | 0.0551 | 0.2862 | 0.4533 | ||
| rs17701834 | 19 | 22,121,458 | G | 0.0549 | 0.2172 | 0.08879 | ||
| rs7193843 | 16 | 54,677,292 | G | 0.0548 | 0.1448 | 0.285 | ||
| rs7097885 | 10 | 16,506,501 | C | 0.0542 | 0.2832 | 0.4486 | PTER | intronic |
| rs2791398 | 1 | 245,965,551 | G | 0.0540 | 0.05944 | 0.1651 | SMYD3 | intronic |
| rs10486802 | 7 | 39,723,768 | A | 0.0531 | 0.1884 | 0.07009 | RALA | intronic |
| rs8031676 | 15 | 96,910,440 | C | 0.0529 | 0.4306 | 0.2664 | ||
| rs7186479 | 16 | 82,602,736 | C | 0.0527 | 0.2517 | 0.1168 | ||
| rs6054383 | 20 | 6,584,604 | T | 0.0526 | 0.3986 | 0.2383 | ||
| rs4460308 | 7 | 104,420,060 | C | 0.0521 | 0.1866 | 0.07009 | LHFPL3 | intronic |
| rs3775779 | 4 | 70,709,207 | A | 0.0520 | 0.476 | 0.3551 | SULT1E1 | intronic |
| rs9375877 | 6 | 132,690,239 | G | 0.0517 | 0.4862 | 0.3458 | MOXD1 | intronic |
| rs2180691 | 20 | 54,964,361 | A | 0.0517 | 0.45 | 0.2857 | AURKA | intronic |
| rs7778955 | 7 | 39,740,487 | G | 0.0517 | 0.1438 | 0.04206 | RALA | intronic |
| rs4608114 | 12 | 92,384,658 | A | 0.0517 | 0.4366 | 0.2736 | C12orf79 | intronic |
| rs6946733 | 7 | 106,670,288 | A | 0.0514 | 0.4281 | 0.2664 | ||
| rs816650 | 10 | 601,089 | T | 0.0514 | 0.114 | 0.2406 | DIP2C | intronic |
| rs6490805 | 13 | 24,084,809 | C | 0.0507 | 0.08394 | 0.1981 | ||
| rs17171480 | 7 | 35,585,669 | A | 0.0506 | 0.09286 | 0.2103 | ||
| rs17015112 | 3 | 77,319,487 | G | 0.0505 | 0.4545 | 0.3785 | ROBO2 | intronic |
| rs573186 | 3 | 124,178,276 | C | 0.0503 | 0.2483 | 0.1168 | KALRN | intronic |
| rs10752609 | 1 | 154,791,128 | A | 0.0501 | 0.3147 | 0.1698 | KCNN3 | intronic |
| rs1862737 | 16 | 75,281,964 | C | 0.0500 | 0.4155 | 0.257 | BCAR1 | 5utr |
We observed that 1,666 SNPs were presented in different minor alleles between the north and south Malays, and their allele frequencies in Malays were compared with that in South Asian (GIH) and East Asian (CHB) (Additional file 2). Although not substantial, differences in allele frequencies were observed between the South- and East- Asians, as well as the between the Malays and both South- and East- Asians. Notably, rs1126965 located at the candidate gene SLC39A11 revealed an alternative allele frequency of 0.8178 in the northern Malays and 0.4965 in the southern Malays. This gene has been reported to play a role in liver enzyme and smoking initiation [10, 11]. Whether or not this gene is under positive selection in the Malays, however, remain further investigation. We subsequently assessed if these SNPs play a role in phenotypic association, and found that 19 of these SNPs were reported in GWAS catalogue (Additional file 4).
To evaluate the potential effect of population sub-structure on a disease association study, a series of computer simulation studies were carried out with PLINK following a case–control GWAS design (Additional file 2: Table S4). The GWAS simulations revealed that the effect on false positive rate and statistical power were greater than expected [12].
Discussion
We demonstrated in this study, that the Malays from Peninsular Malaysia and Singapore are essentially sub-structured. Although genetic correlation with geographical latitude had been previously reported in the Chinese populations [12, 13], it is indeed surprising to reveal such differentiation among the Malay populations even within a small region in Peninsular Malaysia and Singapore (~800 KM from north to south). In addition to that, the FST between the north and south Malays were similar to those of the earlier report between the northern and southern Han Chinese (FST = 0.0011) [12] but lower than those within Europeans (FST = 0.0033) [14] However, we observed higher diversity within the substructures of the Malays. For instance, the FST between two northern Peninsular Malays from Kedah and Kelantan was 0.017 (Table S4), which is in line with the finding in a recent study [4]. This suggests higher heterogeneity among Malays than previously expected, possibly be due to the recent migration and gene flow from the surrounding populations in this region.
The Pahang Malays were found to have a closer affinity to the north, although they were classified as the central region in this study. This is likely due to the reason that samples were collected from the Federal Land Development Authority (FELDA) settlers in the Pahang state, of which the majority of them were originated from Kelantan. On a separate note, Selangor, being as the most advanced and most populated state of Malaysia, is where the metropolitan city Kuala Lumpur located. PCA revealed that samples from this population was scattered across both the north and south regions (Fig. 1b & c). We believe that urbanization had likely blurred the boundaries. Similar findings were observed in Xu et al. (2009), where the populations from metropolitan areas showed more complicated composition with multiple ancestral origins compared with those from the rest of the area.
Essentially, identification a panel of ancestry informative markers (AIMs) would be an ideal strategy to correct the population stratification in future genetic association studies [15]. However, the SNP coverage and the sample size in the current study are insufficient for such purpose. Those highly differentiated SNPs between the north and south Malays could be possibly due to genetic drift or, to a lesser extent, natural selection. These SNPs, however may be considered as the putative set of variants as the AIMs for the Malay populations. The candidate gene ABCA1 is a major gene that plays an important role in high-density lipoprotein cholesterol (HDL-c) synthesis and cholesterol transport [16]. However, whilst we suspect the genetic drift is likely to be the cause, the reason of this gene being highly differentiated between northern and southern Malays remains further investigated. Cautions should be taken though when positive signals of HDL-c and ABCA1 are identified in the genetic association study of Malays.
We acknowledge several limitations in this study. Sample collection from several locations were small, hence might have resulted into outliers which confounded the outcome of the correlation between genetic differentiation and geographical coordinates. In addition, self-reported ancestry might have also confounded the analysis when assigning to their respective state of origin. However, the number of samples covering all states in Peninsular Malaysia (and Singapore), and the marker utilized in our study are larger than the previous reports, thus provides further insights into the genetic structure of the Malays in Peninsular Malaysia. Notably, we revealed close relationship between genetic and geographical coordinates in the Malay populations. In addition, our results and to which extent the admixtures in Southeast Asia could impact the population stratification thus affect the genetic association studies. Therefore we call for attention to look into alternative strategies for disease mapping in genetically complex populations particularly from Southeast Asia.
Conclusion
In summary, we revealed that the population substructure of the Malays was correlated to the latitude coordinate. The genetic diversity of the Malays is more heterogeneous than previously expected, and that we proved that such population sub-structure occurred even though within a small geographical region may potentially lead to spurious signals in disease based genetic association studies. Therefore cautions should be taken when carrying out such study design.
Methods
Population and samples
A total of 431 Malay samples were included in this study. These samples were self-identified Malays from Peninsular Malaysia, 116 of which were genotyped with Affymetrix Genome-Wide Human SNP Array 6.0, whilst the remaining samples were genotyped with Illumina 660W (Sample size, N = 90) and Illumina Omni Express (N = 119). The additional 17 Malays samples from Kelantan genotyped with Affymetrix Genome-Wide Human SNP Array 6.0 [17], and 89 samples of metropolitan Malays from Singapore (SGVP) were also included in this analysis [3]. The studies were approved by the research and ethics committees of Universiti Teknologi MARA and Universiti Sains Malaysia, and the design of this study followed the Helsinki Declaration 1975, as revised in year 2000. The collected samples covered all 11 states of Peninsular Malaysia (Fig. 5), of which were divided into 3 geographical regions for the purpose of this study namely, North, South and the Centre regions, according to their respective latitude coordinate (Table 3). The number of samples and their geographical locations are listed in Table 3. Six selected populations involving 805 samples from the International HapMap Project 3 (HapMap 3) [18] were included in the analysis to characterize the genetic variation of the Malays on a global scale: YRI, GIH, CEU, CHB, JPT and MEX.
Fig. 5.
The geographical map of Peninsular Malaysia. The sampling locations are shown in red dots
Table 3.
Regional categorization of the Peninsular Malaysia states according to geographical locations and final number of sample included after QC
| Region | States | Latitude coordinate* | No. subjects |
|---|---|---|---|
| North | Perlis | 6°23′40.06″N (6.394462) | 7 |
| Kedah | 5°19′45.2″N (5.329221) | ||
| Pulau Pinang | 5°19′45.2″N (5.329221 | ||
| Perak | 4°37′8.46″N (4.619018) | 5 | |
| Kelantan (Kota Bharu) | 5°42′55.13″N (5.715314) | 56 | |
| Kelantan (Jeli) | 5°29′49.4″N (5.497056) | 74 | |
| Terengganu | 5°19′30.33″N (5.325092) | 4 | |
| Centre | Pahang (Pekan) | 3°29′32″N (3.492092) | 51 |
| Selangor | 2°50′34.7″N (2.842971) | 98 | |
| South | Negeri Sembilan | 2°33′12.75″N (2.553541) | 9 |
| Melaka | 2°12′11.81″N (2.203281) | 5 | |
| Johor | 1°27′41.98″N (1.461662) | 4 | |
| Singapore (Malay) | 1°17′13.35″N (1.287043) | 89 |
*Latitude coordinate from Yandex (http://map.yandex.com)
Data assemblage
Data QC and assemblage were carried out with PLINK 1.07. Datasets from each platform were first filtered for individuals with >10% missing rate, > 10% SNP missing rate, minor allele frequencies (MAF) < 0.05, and Hardy-Weinberg Equilibrium (HWE) P < 0.002. Then the filtered datasets were subsequently merged, consisting 42,633 SNPs shared among all the 402 Malay samples. The dataset was further pruned down by removing any SNP with r2>0.8, leaving a total SNP of 41,400 for further analyses.
Analysis of population structure
Principal Component Analysis (PCA) was first carried out using the smartPCA in EIGENSOFT (ver 4.0) package. The genetic component of the Malay populations was inferred with ADMIXTURE ver 1.22 (Alexander et al., 2009) [19], with the 41,400 SNPs overlapped across all samples.
Latitude-PC correlation
Pearson’s correlation coefficient was calculated to evaluate the relationship between the genetic coordinates (PC values) and the geographic latitudes.
Pairwise FST
Unbiased estimation of FST was calculated according to Weir and Hill (2002) [20], with confidence intervals estimated by bootstrapping with 1,000 replications.
GWAS simulation
Simulations on genome-wide association study (GWAS) were performed using PLINK 1.07, following the procedure of Xu et al. (2009) [12].
Acknowledgent
The authors would like to acknowledge the medical staffs from Hospital Kota Bharu, Hospital Sungai Buloh, and Hospital Selayang, as well as the staffs and scientists from the PURE-REDISCOVER Study whom assisted in the sample collection. The authors also thank all subjects who had participated this study. B.P. Hoh is supported by the Chinese Academy of Sciences Fellowship for Visiting Scientists from the Developing Countries (2914FFSA0009). The computational analyses of this study were supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB13040100), and by the National Science Foundation of China (NSFC) grants (91331204 and 31171218). The experimental funding of this study was supported by the Long-term Research Grant Scheme (LRGS) for Infectious Disease 2011 [600-RMI/LRGS 5/3(3/2011)] funded by Ministry of Education Malaysia, and the Long-term Research Grant Scheme (LRGS) for the REsponDing to IncreaSing CardiOVascular disease pRevalence (REDISCOVER) [600-RMI/LRGS 5/3(2/2011)] funded by Ministry of Education Malaysia. We also would like to acknowledge the contributions the staffs of Human Genome Center, Universiti Sains Malaysia (USM). This study was also supported by USM Apex grant: 1002 / PPSP / 910343 and USM short term grant: 304 / PPSP / 61311034. S.X. is Max-Planck Independent Research Group Leader and member of CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the National Program for Top-notch Young Innovative Talents of the “Wanren Jihua” Project.
Abbreviations
- GWAS
Genome wide association
- FELDA
Federal land development authority
- SNP
Single nucleotide polymorphism
- AIMs
Ancestry informative markers
- HDL-c
High density lipoprotein cholesterol
- HDL-c
High density lipoprotein cholesterol
- HapMap 3
Hapmap project 3
- Punadj
Unadjusted P value
- GC
Genomic control
- Bonf
Bonfferoni correction
- FDR
False discovery rate
Additional files
PCA plot including samples from the 11 states of Peninsular Malaysia. Figure S2. Correlation between PC1 and longitude. Figure S3. Correlation between geographical distance and FST.
Regional pair-wise FST of the populations collected from different states in Malaysia. Table S2. Top 1% SNPs that are highly differentiated between the Malays from northern and southern region of Peninsular. Table S3. Gene ontology and enrichment analysis of the candidate genes underlying the top 0.1% highly differentiated SNPs between the Malays from the north and south Peninsular Malaysia. Table S4. Simulation analysis for GWAS before and after removing 1,666 SNPs with different minor allele present between the north and south Malays.
SNPs with different minor allele between northern and southern Malays.
GWAS annotation.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author’s contribution
BPH and SX conceived and designed the study; BPH and LD performed the data analysis; MJJA, ZZ, MNH, NSAR, WIH, IE, ABZ, YK involved in sample collection, BPH, SX and YK funded the experimental and computational costs. BPH, SX and LD prepared and wrote the manuscript. All authors have read and approved the manuscript.
Contributor Information
Boon-Peng Hoh, Email: hoh.boonpeng@gmail.com.
Shuhua Xu, Email: xushua@picb.ac.cn.
References
- 1.Bellwood P, Gamble C, Le Blanc SA, Pluciennik M, Richards M, Terrell JE. First Farmers: the Origins of Agricultural Societies, by Peter Bellwood. Malden (MA): Blackwell, 2005; ISBN 0-631-20565-9 hardback £60; ISBN 0-631-20566-7 paperback £17.99, xix+360 pp., 59 figs., 3 tables. Cambridge Archaeological Journal 2007:87
- 2.Hatin WI, Nur-Shafawati AR, Zahri M-K, Xu S, Jin L, Tan S-G, Rizman-Idid M, Zilfalil BA. Population genetic structure of peninsular Malaysia Malay sub-ethnic groups. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo Y-Y, Sim X, Ong RTH, Tan AKS, Chen J, Tantoso E, Small KS, Ku C-S, Lee EJD, Seielstad M, Chia K-S. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 2009;19:2154–62. doi: 10.1101/gr.095000.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatin W, Nur-Shafawati A, Etemad A, Jin W, Qin P, Xu S, Jin L, Tan S-G, Limprasert P, Feisal M, Rizman-Idid M, Zilfalil B. A genome wide pattern of population structure and admixture in peninsular Malaysia Malays. Hugo J. 2014;8:5. doi: 10.1186/s11568-014-0005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium HP-AS. Mapping human genetic diversity in Asia. Science. 2009;326:1541–5. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 6.Helgason A, Yngvadóttir B, Hrafnkelsson B, Gulcher J, Stefánsson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37:90–5. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- 7.Nandi S, Ma L, Denis M, Karwatsky J, Li Z, Jiang X-C, Zha X. ABCA1-mediated cholesterol efflux generates microparticles in addition to HDL through processes governed by membrane rigidity. J Lipid Res. 2009;50:456–466. doi: 10.1194/jlr.M800345-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Micheal S, Ms HA, Frcs SNZ, Bs BB, Fcps MA, Fcps FA, Fcps FI, Khan MI, Qamar R, Den Hollander AI. Original Article Identification of novel CYP1B1 gene mutations in patients with primary congenital and primary open-angle glaucoma. Clin Experiment Ophthalmol. 2015;43(1):31–39. doi: 10.1111/ceo.12369. [DOI] [PubMed] [Google Scholar]
- 9.Bouyacoub Y, Ben Yahia S, Abroug N, Kahloun R, Kefi R, Khairallah M, Abdelhak S. CYP1B1 gene mutations causing primary congenital glaucoma in Tunisia. Ann Hum Genet. 2014;78:255–263. doi: 10.1111/ahg.12069. [DOI] [PubMed] [Google Scholar]
- 10.Park T-J, Hwang J-Y, Go MJ, Lee H-J, Jang HB, Choi Y, Kang JH, Park KH, Choi M-G, Song J, Kim B-J, Lee J-Y. Genome-wide association study of liver enzymes in korean children. Genomics Inform. 2013;11:149–54. doi: 10.5808/GI.2013.11.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argos M, Tong L, Pierce BL, Rakibuz-zaman M, Islam T, Rahman M, Paul-brutus R, Rahaman R, Roy S, Jasmine F, Kibriya MG, Ahsan H. Genome-wide association study of smoking behaviours among Bangladeshi adults. J Med Genet. 2014;51:327–333. doi: 10.1136/jmedgenet-2013-102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Yin X, Li S, Jin W, Lou H, Yang L, Gong X, Wang H, Shen Y, Pan X, He Y, Yang Y, Wang Y, Fu W, An Y, Wang J, Tan J, Qian J, Chen X, Zhang X, Sun Y, Zhang X, Wu B, Jin L. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 2009;85:762–74. doi: 10.1016/j.ajhg.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suo C, Xu H, Khor C-C, Ong RT, Sim X, Chen J, Tay W-T, Sim K-S, Zeng Y-X, Zhang X, Liu J, Tai E-S, Wong T-Y, Chia K-S, Teo Y-Y. Natural positive selection and north–south genetic diversity in East Asia. Eur J Hum Genet. 2012;20:102–10. doi: 10.1038/ejhg.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, Balascakova M, Bertranpetit J, Bindoff LA, Comas D, Holmlund G, Kouvatsi A, Macek M, Mollet I, Parson W, Palo J, Ploski R, Sajantila A, Tagliabracci A, Gether U, Werge T, Rivadeneira F, Hofman A, Uitterlinden AG, Gieger C, Wichmann HE, Rüther A, Schreiber S, Becker C, Nürnberg P, et al. Correlation between Genetic and Geographic Structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 15.Qin P, Li Z, Jin W, Lu D, Lou H, Shen J, Jin L, Shi Y, Xu S. A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet. 2014;22(2013):248–53. doi: 10.1038/ejhg.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Waite LL, Jackson AU, Sheu WH-H, Buyske S, Absher D, Arnett DK, Boerwinkle E, Bonnycastle LL, Carty CL, Cheng I, Cochran B, Croteau-Chonka DC, Dumitrescu L, Eaton CB, Franceschini N, Guo X, Henderson BE, Hindorff LA, Kim E, Kinnunen L, Komulainen P, Lee W-J, Le Marchand L, Lin Y, Lindström J, Lingaas-Holmen O, Mitchell SL, Narisu N, Robinson JG, et al. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 2013;9:e1003379. doi: 10.1371/journal.pgen.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, Hoh BP, Lu D, Fu R, Phipps ME, Li S, Nur-Shafawati AR, Hatin WI, Ismail E, Mokhtar SS, Jin L, Zilfalil BA, Marshall CR, Scherer SW, Al-Mulla F, Xu S. The population genomic landscape of human genetic structure, admixture history and local adaptation in Peninsular Malaysia. Hum Genet. 2014;133:1169–85. doi: 10.1007/s00439-014-1459-8. [DOI] [PubMed] [Google Scholar]
- 18.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PIW, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]


