Abstract
Multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins are a heterogeneous group of toxins found in a number of Vibrio species and other Gram-negative bacteria. The toxins are composed of conserved repeat regions and an autoprocessing protease domain that together function as a delivery platform for transfer of cytotoxic and cytopathic domains into target eukaryotic cell cytosol. Within the cells, the effectors can alter biological processes such as signaling or cytoskeletal structure, presumably to the benefit of the bacterium. Ten effector domains are found in the various Vibrio MARTX toxins, although any one toxin carries only two to five effector domains. The specific toxin variant expressed by a species can be modified by homologous recombination to acquire or lose effector domains, such that different strains within the same species can express distinct variants of the toxins. This review examines the conserved structural elements of the MARTX toxins and details the different toxin arrangements carried by Vibrio species and strains. The catalytic function of domains and how the toxins are linked to pathogenesis of human and animals is described.
INTRODUCTION
Multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins are large single polypeptide toxins produced by various Gram-negative bacteria that interact with a wide range of insect, mammalian, and aquatic animal hosts. The toxins range in size from 3,500 to 5,300 amino acids and are frequently encoded by the largest open reading frames of a bacterial genome (1). A search of the National Center for Biotechnology Information database revealed that, within the Vibrio genus, an rtxA gene that could encode a full-length MARTX toxin are present in the genomes of six different species: Vibrio cholerae, V. vulnificus, V. anguillarum, V. ordalli, V. splendidus, and V. caribbeanicus. Toxin genes are also present on large plas-mids isolated from V. vulnificus and V. nigripulchritudo.
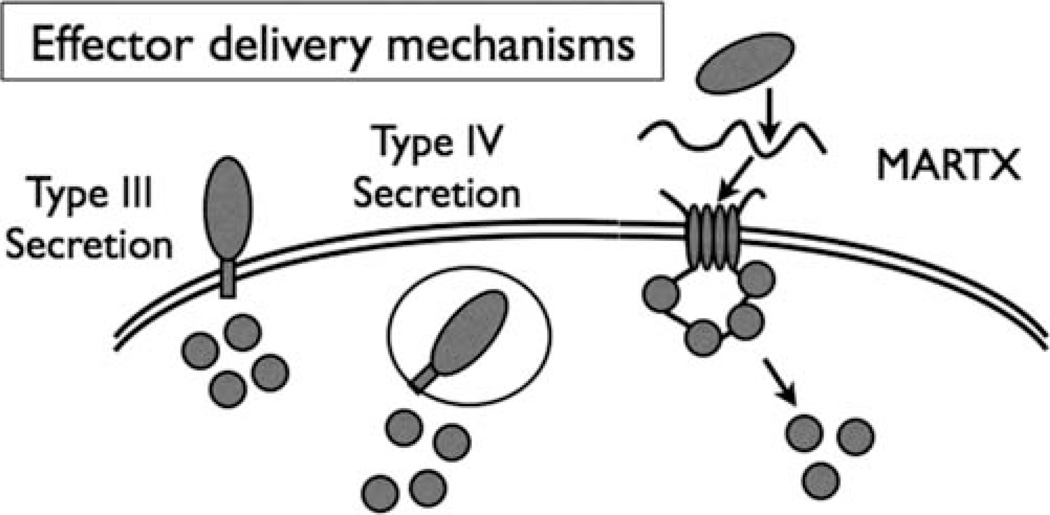
Similar to contact-dependent bacterial secretion mechanisms, such as type III secretion (T3S) and type IV secretion (T4S), the ultimate result of delivery of a MARTX toxin to target eukaryotic cells is transfer of cytotoxic and cytopathic effectors that disrupt normal eukaryotic cell biology (2, 3), presumably to the benefit of the infectious or commensal organism (Fig. 1). Consistent with the concept that MARTX toxins are an alternate effector delivery platform utilized in bacteria lacking T3S and T4S systems, the common seafood-borne pathogens V. parahaemolyticus (4) and V. alginolyticus (5) do not have rtxA genes, and presumably, instead use their T3S contact-dependent system to deliver effectors to target cells (6). V. anguillarum strain 96F interestingly has both a T3S system and encodes a MARTX toxin while other V. anguillarum strains have lost many of their T3S genes (7). Thus, it seems probable that trans-location of effectors by vibrios is often important and that either T3S and/or MARTX toxins are used for this function.
FIGURE 1.
The multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins are a form of effector delivery similar in concept to contact-dependent type III secretion and type IV secretion. The major difference is that the toxin is secreted from the bacterium by type I secretion and then the large single polypeptide toxin delivers effectors directly across the plasma membrane with delivery occurring by polypeptide autoprocessing. Similar to type III and type IV secretion, the effector themselves confer cytopathic and cytotoxic activities that then alter host-cell biology to the benefit of the bacterium. doi:10.1128/microbiolspec.VE-0002-2014.f1
MARTX TOXIN STRUCTURE, SECRETION, AND EFFECTOR TRANSLOCATION
MARTX Toxin Repeat Structure Is Conserved across the Vibrios
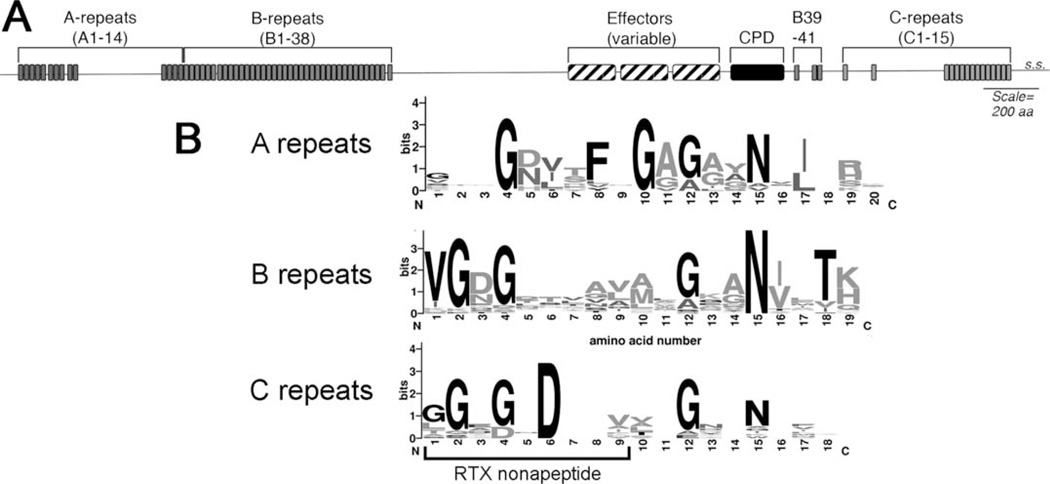
All MARTX toxins have regions at the amino- and carboxyl-termini of strong amino acid sequence similarity that are characterized by extensive glycine rich repeats (1, 8). The amino-terminal conserved region is usually 1,950 to 1,970 amino acids in length and can be separated into distinct regions. (i) The N-terminal extension of ~73 amino acids; (ii) the 19 amino acid A-repeats comprised of 14 arrayed repeats interrupted by a 290 amino acid insertion between repeats A10 and A11; (iii) 38 copies of the 20 amino acid B-repeats; and (iv) the B-repeat/effector junction zone that is a region of high sequence variability between toxins both between species and within the same species (Fig. 2). The carboxyl-terminus conserved region includes two to three additional copies of the B-repeat and 15 copies of a C-repeat (1). The C-repeats are similar to the classical nonapeptide calcium-binding repeat of all repeats-in-toxin (RTX) family proteins (9, 10), except the repeat is 18 amino acids with only half the repeat representing the GD-rich RTX repeat (8). The structural implications of the longer repeat are as yet uncharacterized.
FIGURE 2.
(A) The general structure of a multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin showing the number and position of various repeat sequences, auto-processing cysteine protease domain (CPD), and variable region containing the effector domains. The secretion signal (s.s.) is shown at the extreme C-terminus. (B) Graphical representation of the different repeat sequences generated by Weblogo 2.8.2 [(113); weblogo.berkeley.edu]. The sequences used were repeat sequences from Vibrio vulnificus CMCP6 identified based on the alignment of sequence to the repeat annotation of V. cholerae (1). The portion of the C-repeat that aligns to the calcium-binding beta roll nonapeptide repeat of other RTX family proteins is indicated. Note conservation of a G-7x–G-4x–N repeat in all of the repeats. doi:10.1128/microbiolspec.VE-0002-2014.f2
MARTX Toxin Secretion from the Bacterium
The extreme carboxyl-terminus of a MARTX toxin, like all RTX proteins, presumably is the site of the type I secretion (T1S) signal for export, such that even small deletions that affect the carboxy-terminus can disrupt function of the entire toxin (11). Secretion from the bacterium is mediated by an atypical T1S secretion system comprised of ATPases RtxB and RtxE, a trans-membrane linker RtxD, and the outer membrane porin TolC for direct transfer of the toxin from the cytosol to the extracellular environment (12, 13, 14, 15). The extracellular intermediate of toxin delivery has been verified by detection of toxin in bacterial-free culture supernatant fluids by activity, western blotting, and mass spectrometry (12, 13, 16, 17, 18, 19). The MARTX toxin from V. cholerae has also been suggested to be associated with outer membrane vesicles, but this is apparently not the case with V. vulnificus MARTX (12, 20).
Toxin Translocation into Eukaryotic Target Cells
MARTX toxins have been shown to be functional against a wide range of eukaryotic cells types, including erythrocytes, epithelial cells, and phagocytic cells and cells of human, mouse, fish, and eel origins (16, 19, 21, 22, 23, 24, 25, 26, 27). Once secreted from the bacterium, the repeat regions of the toxins are postulated to bind to eukaryotic receptors on target cells where they have been shown to form a pore for trans-location of the central portion of the toxin across the plasma membrane to the eukaryotic cell cytoplasm (26, 28, 29). The portion of the toxin translocated is comprised of domains with low thermostability, suggesting they are easily unfolded for translocation across the plasma membrane (26, 30).
One domain delivered across the plasma membrane is the cysteine protease domain (CPD) (31), which is activated after translocation to the eukaryotic cell cytoplasm by binding of inositol hexakisphosphate (InsP6) (32). Upon binding InsP6, folding of the CPD is completed and the protease is able to autoprocess the MARTX toxin after leucine residues present in extended unfolded regions found between “effector domains” that confer the cytotoxic functions of the toxin (3, 17, 33). In total, ten different effector domains have been recognized among the various MARTX toxins, although any one toxin carries only two to five effectors (Fig. 3). The domains are assorted across different strains and species and they can be exchanged by homologous recombination. This swapping of rtxA effector domain gene sequences results in heterogeneity of the toxins even between different isolates within the same species (11, 34, 35).
FIGURE 3.
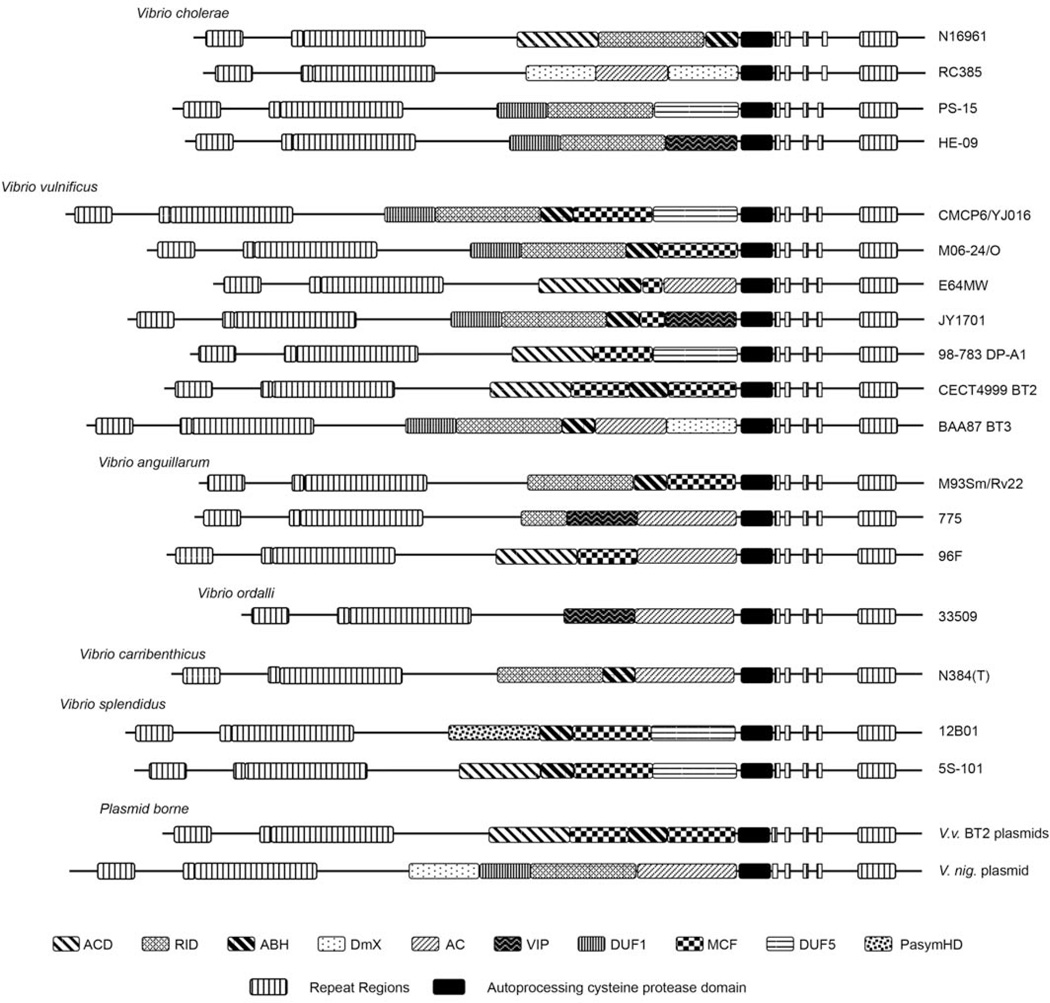
Schematic representation of all multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins described in text. Toxins are identified by species at left with representative strain isolate designation at the right. BT2 and BT3 refer to Vibrio vulnificus biotype 2 and 3, respectively, and Vnig_pA refers to the large pA1066 plasmid of V. nigripulchritudo. Legend for different domains is at the bottom. All amino acid sequences are from published papers referenced in the text and the sequences were downloaded from the NCBI website (www.ncbi.nlm.nih.gov). All effector domain arrangements are as annotated previously (1, 11, 29, 34, 35, 63) except for new sequences done specifically for this article, including V. vulnificus JY1701, V. anguillarum 775 and 96F, V. ordalli 33509, and V. splendidus 5S-101. doi:10.1128/microbiolspec.VE-0002-2014.f3
MARTX TOXINS OF V. CHOLERAE
Structure and Function of Most Common MARTX of V. cholerae
V. cholerae is the pathogen that causes the human diarrheal disease cholera (36). The MARTX toxin of V. cholerae El Tor strain N16961 was the first to be discovered and is the representative member of this toxin family (8). This representative toxin, which is produced by most strains of V. cholerae, is 4,545 amino acids in length and shows remarkable conservation in sequence (11). An analysis of nearly 70 genome sequences shows that among the El Tor and El Tor-like strains that have caused cholera during the most recent century, the gene has been highly stable with individual isolates showing only one to six SNPs over the course of 100 years resulting in only one to four amino acid changes. This lack of genetic diversity does not reflect specialized conservation of the rtxA gene, but rather the stability of the El Tor genomes as a whole (11). The rtxA gene in environmental strains also shows strong conservation (only 1% to 2% amino acid differences), with the overall length and organization of the toxin highly conserved (11, 37).
This common variant of V. cholerae MARTX toxin carries three effector domains (17, 33). In the first position immediately downstream of the MARTX conserved region is the actin crosslinking domain (ACD), an enzyme related to glutamine sythetases that covalently crosslinks G-actin protomers via introduction of an isopeptide bond between lysine 50 and glutamic acid 270 (16, 38, 39, 40). Similar to the catalytic action of glutamine sythetases, the reaction involves the initial formation of an activated acyl phosphate intermediate formed by the transfer of phosphate from the hydrolysis of ATP to glutamic acid 270 followed by hydrolysis to energize formation of the amide crosslink (41, 42). The crosslinking of actin then causes depolymerization of the actin cytoskeleton both through the removal of G-actin as a substrate for actin assembly and by addition of actin dimers to growing F-actin blocking further filament assembly (40, 41).
The next domain in the V. cholerae MARTX is the Rho inactivation domain (RID). This effector is a member of the circular permuted thiol peptidase family, although the RID is predicted to have a distinct arrangement of its catalytic site compared to other proteins in this family (43, 44). After delivery to the cytosol by processing from CPD, the presence of a four helical bundle type membrane localization domain targets the RID to the plasma membrane (45, 46). At the membrane, the cellular protein modified or proteolyzed by the RID is unknown, but the enzymatic modification causes a change in cellular signaling resulting in loss of all active GTP-bound Rho. As Rho is a master regulator of stress fiber assembly, cells treated with RID round due to lack of cytoskeletal assembly (47). Interestingly, although both ACD and RID independently affect polymerized actin, the domains are not synergistic, and thus, neither actin crosslinking nor Rho inactivation is accelerated by the presence of the other domain in the same toxin (26, 44).
The effector located between RID and CPD is the alpha/beta hydrolase (ABH) effector. Sequence analysis indicates this effector is a member of the expansive structural protein family known as the alpha/beta hydrolases that includes proteases, lipases, and esterases (48, 49). This domain has been shown to indirectly activate the small GTPase CDC42. Interestingly, this activity is antagonistic with RID, which inactivates CDC42, such that a toxin with both domains shows little overall change in CDC42 activation. Thus, the biological consequence of the ABH domain remains currently unknown (26).
Role of V. cholerae MARTX in Cholera Pathogenesis
The loss of cell structure due to actin depolymerization by V. cholerae has been linked both with loss of tight junction integrity of polarized intestinal epithelial cell and with inhibition of phagocytosis (26, 50). It unclear if affects on epithelial cells are important in vivo as only minimal damage occurs to the intestine during cholera disease (51, 52). By contrast, consistent with a primary role for inhibition of phagocytosis during infection, the MARTX toxin of V. cholerae is linked to evasion of innate immune defenses during intestinal infection of mice. The toxin works in concert with another secreted toxin, the cytolysin/hemolysin HlyA, such that strains deleted of both hlyA and rtxA genes fail to colonize the small intestine of mice and are avirulent (52, 53). The contribution of these toxins in vivo is, in part, to inhibit clearance of the infecting bacteria from the small intestine by neutrophils, accounting for their requirement for bacterial colonization (54).
Given the role of the MARTX toxin in innate immune evasion rather than diarrheal disease, and its apparent redundancy with HlyA, it is not surprising that some strains would eliminate the toxin entirely. Both classical strains that caused cholera epidemics during the 19th century and the currently circulating altered El Tor strains have either a large deletion or an introduction of an opal stop codon in the rtxA gene that renders these strains rtxA null mutants (8, 11). These strains also have sequence differences in the cholera toxin B (CtxB) sub-unit of cholera toxin, the primary toxin associated with diarrhea (55). It has been postulated that CtxB may control immunity during infection just as it does when functioning as a vaccine adjuvant (56). Thus, it is possible that the MARTX likely became redundant in both classical and altered El Tor strains that carry the modified CtxB and it was thus eliminated by spontaneous mutation (8, 11).
Rare V. cholerae MARTX Toxin Variants among Environmental Isolates
Environmental strains of V. cholerae are not commonly associated with disease but can cause sporadic outbreaks or food-borne infection (57, 58, 59, 60, 61). Although less conserved at the amino acid level than the El Tor strains, the MARTX toxins of the environmental strains with genomes tightly linked with the V. cholerae human isolates do predominantly retain the ACD-RID-ABH effector domain structure (11). However, among the extensive database of sequenced environmental isolates are strains that have a distinct effector domain structure. The strain RC385 isolated from Chesapeake Bay has a unique domain structure that includes an adenylate cyclase (AC) similar to the T3S effector ExoY from Pseudomonas aeruginosa (11, 62, 63). This AC domain is flanked by duplicated copies of a novel domain of unknown function that has been designated Domain X (DmX) (63). This domain is present also in one of four MARTX toxins of the insect pathogen Photorhabdus asymbiotica and in V. vulnificus biotype 3 strains (see below) (11, 63, 64).
The non-O1, non-O139 strain HE-09 isolated from the environment in Haiti (11, 37) and strain PS-15 isolated from Puget Sound in Washington State (65) also have unique toxin structures. For both strains, the toxins have in the first position, DUF1, a domain of unknown function that has no homology to any non-MARTX proteins. This domain is present in the first position of a number of other MARTX toxins including those from V. vulnificus (34, 35) and the nematode symbionts Xenorhabdus bovienii and Xenorhabdus nematophila (29, 66). In the second position is a RID domain similar to the common arrangement of V. cholerae MARTX toxins. The two strains vary however in the third position. For PS-15, the domain of unknown function in the third position (known as DUF5) shares limited homology with the C1 and C2 domains of Pasteurella multocida toxin (PMT) and has been partially characterized in V. vulnificus as a cytotoxin (see below). In HE-09, the third domain is a novel domain denoted as VIP2, as it is similar to the catalytic domain of ADP-ribosyltransferase toxins that ADP-ribosylate G-actin, including Clostridium botulinum C2 toxin (67), Aeromonas hydrophila VgrG1 (68), and Bacillus cereus VIP2 (69).
Thus, with the increasing monitoring of V. cholerae in the environment by genome sequencing, there is the potential greater diversity of this toxin will be revealed, suggesting V. cholerae is altering its MARTX toxin as part of its niche adaptation. However, both clinical and environmental isolates that are closely linked to human pathogenesis all share a highly conserved three domain (ACD-RID-ABH) structure that does not vary, suggesting that this particular arrangement of the toxin is potentially linked to its role in human infection.
MARTX TOXINS OF V. VULNIFICUS
Extensive Diversity among V. vulnificus MARTX Toxins
V. vulnificus is a pathogen that causes wound and food-borne intestinal diseases, both of which can spread from the site of initial infection to cause sepsis with high mortality rates (70). In contrast to the conservation of the MARTX toxin domain structure among most of the V. cholerae isolates, the V. vulnificus MARTX toxins are strikingly diverse (Fig. 3) (34, 35, 63). Interestingly, variation does not seem to track with multi-locus sequence typing-based evolutionary trees, indicating that variation arises by homologous recombination between strains rather that clonal expansion of strains after the rtxA1 gene has recombined (34, 35).
Among the biotype 1 V. vulnificus isolates most commonly associated with clinical infections, there are two prominent MARTX toxin arrangements. The first is the five effector arrangement represented by sequenced clinical isolates, CMCP6 (71) and YJ016 (72), and also identified in sequenced environmental isolate, JY1305 (73). This variant has been independently termed MARTX/C (35) or type I (34). These strains have DUF1 in the first position, RID in the second position, ABH in the third position. The domain in the fourth position has homology to a portion of the Makes caterpillars floppy (Mcf) toxins of Photorhabdus luminescens (74, 75). This domain has recently been shown to be an autoprocessing cysteine protease associated with a cytopathic cell rounding (76).
The DUF5 effector domain in the fifth position has homology to PMT as noted above, but this does not inform about its function, since the aligned domain in PMT has not been characterized. Indeed, expression of DUF5 in both mammalian cell and yeast is cytotoxic, although expression of the aligned domain from PMT does not apparently affect the cells (77). The cellular target of DUF5 is currently unknown but is likely to be a membrane protein. DUF5, similar to V. cholerae RID, has a functional four helical bundle type membrane localization domain and DUF5 has been found to tightly localize to cellular junctions (45, 46). Surprisingly, within the conserved membrane localization domain for V. vulnificus RID, many strains have a point mutation in a critical residue that affects protein localization; therefore, RID from V. vulnificus is not predicted to localize to eukaryotic cell membranes and it is unclear if it will be functional after translocation to cells (46).
The second prominent MARTX toxin arrangement of V. vulnificus biotype 1 strains of the clinical clade is thought to have arisen by homologous recombination in the rtxA1 gene, apparently with rtxA DNA acquired from V. anguillarum. Independently, the same arrangement arose in biotype 1 strains within the environmental clade by recombination with rtxA DNA, possibly from a V. vulnificus plasmid. Both recombination events resulted in rtxA1 genes that encode for MARTX toxins with only four effectors and missing the DUF5 effector domain (35). This variant is termed MARTX/M (35) or type II (34) and is encoded by many clinical isolates including sequenced strains MO6–24/O, ATCC 27562, and B2 (34, 35, 78, 79, 80).
Other variants of the MARTX toxin have arisen among clinical and environmental biotype 1 isolates and these include a mosaic of effector domains, including those found in CMCP6 and M06–24/O, but also toxins with actin crosslinking activity due to the ACD (35), predicted actin ADP-ribosylating toxins due to the VIP2 domain, and predicted AC toxins due to the acquisition of the ExoY-like AC domain (63). The promiscuous nature of the recombination is evident in environmental isolate E64MW, which has complete AC and VIP2 domains, but also incomplete remnants of ABH and MCF domains from the progenitor toxins (Fig. 3) (63, 73).
Cell Biological Effects of Intoxication by V. vulnificus MARTX
The most prominent activity of both MARTX/C-type (or type I) and MARTX/M-type (or type II) biotype 1 MARTX toxins is the ability to induce cytolysis of a broad range of cell types including erythrocytes, epithelial cells, and macrophages (21, 22, 24, 27, 81). The lysis had been proposed to occur by calcium-dependent necrosis in epithelial cells dependent on the carboxy-terminal repeat region plus the CPD (18). However, the relevance of this finding to programmed necrotic epithelial cell death is as yet unclear as CPD proteins alone are known to induce cell lysis when transfected (31), but not when presented from the bacterium (44). In addition, more recent studies show that calcium in the media is required for secretion of the toxin from the bacterium, not for necrosis, and that cell lysis depends on both the amino and carboxy repeat regions of the toxin (28). Moreover, protein corresponding to the C-terminus does not induce necrosis when added to cell culture media (82).
At lower multiplicity of infection or short incubation times such that epithelial cells are not lysed, V. vulnificus with the common MARTX/C (type I) and MARTX/M (type II) arrangements have been linked to activities other than cytolysis. These include induction of caspase 3-dependent apoptosis (83), cytoskeleton disassembly (84), and activation of Rac2 and Nox for increased generation of reactive oxygen species (ROS) (85). When the bacteria are added to cultured macrophages, the action of the MARTX toxin of V. vulnificus inhibits bacterial killing (27). At low multiplicity of infection, macrophages are also known to undergo NLRP3-dependent inflammasome activation to cleave caspase-1 and to release the proinflammatory cytokine IL-1β (86). How each of these activities are linked to the various domains of the large toxin has not, as yet, been determined.
Role of Biotype 1 V. vulnificus MARTX Toxin during Mouse Infection
The MARTX toxin of V. vulnificus has been shown to be essential for pathogenesis of several mouse models. In models of septicemia where bacteria are delivered intraperitoneally, mutants disrupted in the gene rtxA1 show a 250- to 450-fold increase in LD50 in ironoverloaded mice (21, 22) and a 100- to 150-fold increase in normal mice (24, 27). These data show that the MARTX toxin is important for septicemic disease.
During subcutaneous infection to mimic wound infection, rtxA1 exerted a 72-fold effect on virulence in normal mice, but only a 13-fold defect in virulence in neutropenic mice. In addition to decreased virulence, rtxA1 mutants are defective for colonization of the wound site, and thereby, do not successfully spread from the initial site of infection and are cleared. All together, these data support a role for the MARTX toxin in innate immune evasion and suggest that the MARTX toxin has a shared role with other secreted factors during wound infection (27).
For food-borne infection, the MARTX toxin has been shown to have a more significant impact on infection than either septicemic or wound-induced models. Initial studies with MARTX/M (or type II) variant strain M06–24/O indicated that rtxA1 mutants are ~100-fold less virulent than wild type in intragastrically inoculated infant mice (24), and these data are similar to findings in intragastrically inoculated adult mice where an rtxA1 mutant in M06–24/O is 180-fold less virulent (35). Yet, a similar mutant in the MARTX/C-type (or type I) MARTX variant strain CMCP6 exerted a 2,600-fold effect on virulence. Conversion of the MARTX/C-type toxin in CMCP6 to the MARTX/M type toxin by recombination decreased virulence by 54-fold indicating that, for the food-borne route of infection, the MARTX/ C-type toxin is more potent than the MARTX/M-type toxin (35). These data indicate that there may be selection in the environment for decreased virulence of V. vulnificus during intestinal infection of aquatic animals, perhaps to avoid killing fish or other animals that become colonized with V. vulnificus.
During intestinal infection, similar to wound infection, the toxin also has a role in promoting growth of the bacterium within the small intestine such that mice infected with mutants without the toxin have increased survival (81). However, apparently in contrast to wound infection where other secreted factors contribute to tissue destruction (27), necrosis of the intestinal villi is directly caused by the MARTX toxin with a secondary additive effect with the secreted cytolysin/hemolysin VvhA. The combination of cytotoxins destroys the intestinal epithelium, providing a route for bacterial dissemination to the bloodstream and other organs (81). The more significant role of the MARTX toxin in this process makes it the most important virulence factor contributing to V. vulnificus food-borne disease and accounts for the more significant impact of the toxin during LD50 studies of CMCP6 MARTX on intestinal infection (35) compared to wound and septicemic models (22, 27).
Biotype 2 MARTX Toxins and their Role in Infection
V. vulnificus biotype 2 strains are associated with vibriosis in fish and sporadic infections in humans (87). Two prevalent serovars have been described with both serovar A and serovar E able to cause disease in eels, sea bass, tilapia, and trout, although strains of serovar E are more virulent and able to cause disease in both sea bass and trout at lower inocula levels. Colonization studies indicate that serovar A strains predominantly colonize the anus and mouth, while serovar E strains colonize the gills (88, 89).
All biotype 2 strains, including the representative strain CECT4999, carry a plasmid that encodes an entire rtx locus (90, 91). This plasmid is essential for virulence of eels due not to the rtxA gene, but to a different RTX-family toxin encoded by vep07, which enhances bacterial survival in eel serum (91). The reason for the lack of essentiality of the plasmid-encoded MARTX toxin is that the rtxA gene on the plasmid is a duplicate of a toxin encoded on the chromosome (34). All biotype 2 chromosomal- and plasmid-encoded MARTX toxins share the same domain structure, known as MARTX type III (34, 91). They all have an ACD in the first position, suggesting these toxins have actin crosslinking activity similar to V. cholerae and the rare variants of the biotype 1 toxins that have the same overall structure (35). The ACD is followed by an MCF domain, an ABH domain, and then a second copy of MCF (91). The implication of the duplication of MCF is unknown as the function of this domain has not, as yet, been elucidated (92).
Deletion of either the plasmid or the chromosomal rtxA gene copy has minimal impact on virulence of either mice or eels. However, loss of both copies rendered the bacterium essentially avirulent for eels both by intraperitoneal infection or by immersion inoculation (23). The same strain showed a 100-fold decreased virulence in mice (23) similar to the impact of the rtxA1 toxin genes of V. vulnificus biotype 1 strains during in-traperitoneal infection of noniron overloaded mice (24, 27). Thus, the function of this toxin is absolutely essential for virulence of eels and likely for sporadic infection of humans. However, a strain with deletions in both copies of rtxA was able to colonize the gills of eels, invade to the bloodstream, and colonize other organs, in some cases better than wild type bacteria. Yet, the eels do not succumb to infection. The difference in infection appears to be that the toxins enhance survival in eel blood, kidneys, and liver, indicating that growth of bacteria in these organs and presence of the toxin itself may be directly linked to animal death from septic shock (23). Further, the rtxA-encoded toxins appear to be essential for protection from phagocytic cells and from predation by amoeba that colonize the gills of eels (23). How both copies of the genes and whether there are individual differences in the contribution of the toxins to virulence remains to be investigated.
Biotype 3 MARTX Toxins and their Role in Infection
Biotype 3 strains are thought to have emerged from biotype 1 and are hybrids of the two major clades of the biotype 1 strains (93, 94). The clonal biotype 3 strain is notable as it was responsible for a major outbreak of V. vulnificus wound infections linked to tilapia ponds and aquaculture during the late 1990s (95). Although the outbreak was abated by changes in fish marketing practices, infections with V. vulnificus biotype 3 still occur in Israel associated with tilapia and wild carp (96). Two genomes of biotype 3 strains are available and analysis of these reveals that they have a unique structure of the MARTX toxin (63, 97, 98). These toxins, similar to the common biotype 1 arrangements, have DUF1-RID-ABH in the first three positions. However, the last two positions have been swapped for the Exo Y-like AC domain and DmX (63). In the strain ATCC BAA87, isolated from a wound during the original Israel outbreak, the rtxA1 gene that encodes the MARTX toxin was found to be essential for wound-induced virulence in mice. In tissue culture, the acquisition of the ExoY-like AC domain was correlated with an rtxA1-dependent acquisition of the adenylate cyclase activity and this activity was directly due to newly acquired AC domain (63). How gain of the AC domain contributed to the virulence of this strain is not yet known, but it is intriguing to consider that this exchange contributed to the local emergence of this pathogen in Israel.
Indeed, as a global concept, given the extensive variation of the V. vulnificus MARTX toxins, it will be interesting to consider how this extensive swapping of domains between strains and species could contribute in the future to emergence of sudden outbreaks and epidemics.
MARTX TOXINS OF V. ANGUILLARUM AND V. ORDALLI
V. anguillarum (occasionally known as Listonella anguillarum) and the closely related species V. ordalli (formerly V. anguillarum biotype 2) are marine pathogens that cause hemorrhagic septicemia in cultured and wild fish, mollusks, and crustaceans (99, 100).
The representative isolate V. anguillarum M93 (serotype J-O-1) originally isolated from an ayu fish in Japan (101) produces a MARTX toxin that has been shown to be the major virulence factor for vibriosis. Using juvenile Atlantic salmon as a model, an isogenic rtxA mutant is avirulent at a dose that causes 100% fatality from the wild type strain. The toxin produced by M93 is hemolytic and induces rounding of salmon kidney cells (19). As encoded in M93, the MARTX toxin has three domains: RID, ABH, and MCF with the presence of a RID domain consistent with studies demonstrating V. cholerae RID induces rounding of cells (19, 47).
The MARTX toxin of the V. anguillarum O2β serotype strain RV22 isolated from a turbot fish in Spain (102) is identical to the toxin of M93 except for three conservative amino acid changes arising from only four single nucleotide polymorphisms across the 13,200 nt gene (7). Thus, despite having been isolated from drastically different geographic zones, the toxins are virtually unchanged.
By contrast, the O1 strain 775 isolated from Coho salmon in Washington state (7, 103) and the isolate NB10 from rainbow trout in Sweden (104) (GenBank LK021130.1) have a different MARTX toxin organization, although the amino acid sequences are identical despite the disparate geographical locations. This toxin has a remnant of a RID, but the toxin has undergone a recombination to acquire VIP2 and AC domains in place of the ABH and MCF domains. The toxin undergoes a further change in strain 96F (isolated from striped bass in Chesapeake Bay) where the AC domain is retained in the last position, but acquires ACD and MCF in the first and second positions (7, 105). Finally, the closely related species V. ordalli 33509 isolated from Coho salmon in the Puget Sound has another distinct arrangement consisting solely of VIP2 and AC domains (7, 106). How all this heterogeneity accounts for colonization of different species, different organs within species, or clinical outcome of disease is currently not known but merits further investigation given the economic importance of this disease to the aquaculture industry (99).
PLASMID-ASSOCIATED MARTX TOXIN OF V. NIGRIPULCHRITUDOM
The shrimp pathogen, V. nigripulchritudo, has gene sequences for a MARTX toxin that has four domains comprised of DmX, DUF1, RID, and AC domains. The rtxA toxin gene and the necessary maturation and secretion genes are carried on a large 247-kilobase plasmid that also carries sequences for a siderophore and a metalloprotease. This plasmid has been identified in highly pathogenic and moderately pathogenic strains, but has a modified structure in nonpathogenic strains including absence of the rtx locus. During infection studies, the plasmid is essential for virulence in shrimp injected intramuscularly. Future studies will be necessary to determine if the requirement for the plasmid in pathogenesis is due to the gene for the MARTX toxin directly or due to other genes (107).
MARTX TOXIN VARIATION IN V. SPLENDIDUS
V. splendidus is a heterogeneous bacterium found in seawater. For studies of genetic diversity and evolution, many strains have been subjected to whole genome sequencing and five of these have assembled contiguous sequences that carry genes for full-length MARTX toxins. Strains 12F01 and 12B01 are closely related by hsp60 sequencing and pulse-field gel electrophoresis genotyping (108), and they encode an identical MARTX toxin. Other isolates, FF-6, and possibly, FF-500, also encode an identical toxin (109). In the first position of this toxin is a domain of unknown function from MARTX toxins of the insect pathogens, P. luminescens and P. asymbiotica (PasyHD), that has otherwise not been identified among the Vibrio genus (1). This domain includes a subdomain that in the Bordetella pertussis type III secretion effector BteA has been shown to direct the effector to ezrin-rich lipid rafts (110). Other domains of the V. splendidus toxin are the ABH, MCF, and DUF5.
Two other V. splendidus genomes, 5S-101 and ZF-90, encode a different toxin in which the PasyHD domain has been swapped with an ACD, but otherwise is highly conserved (111). No investigations with these toxins have been performed; therefore, the role of these toxins in the lifestyle of V. splendidus is currently unknown.
MARTX TOXIN OF MARINE SPONGE-ASSOCIATED V. CARIBBEANICUS
The final Vibrio species that genome analysis reveals carries a gene for a MARTX toxin is V. caribbeanicus. This is a recently recognized Vibrio species isolated from a marine sponge in the Caribbean Sea off the coast of Curaçao (112). The type strain N384(T) (ATCC BAA-2122) has been sequenced and the genome deposited as V. caribbenthicus (GenBank AEIU00000000). Sequence analysis indicates this strain produces a MARTX toxin with RID, ABH, and AC domains, but no other information is known about this species or how the toxin might be advantageous to its interaction with the sponge.
SUMMARY
Overall, the vibrios are a rich source of MARTX toxins that are likely to have a range of functional activities. In addition to variation across the effector domains, it is likely that sequence variations within the repeat regions will contribute to host selection or tissue distribution within a host. It is interesting to speculate that each Vibrio isolate is exquisitely selected to colonize or infect its specific host and has achieved this by evolving its MARTX toxin to carry a unique combination of conserved region modifications for receptor recognition coupled to an optimal effector domain repertoire.
In some cases, it is possible that a species is still sampling the optimal configuration to occupy a certain niche, accounting for the many different variants found within some species such as V. vulnificus. By contrast, the conservation of the toxin arrangement, for example in V. cholerae, would suggest that these strains are already optimally suited to their particular niche, and thus, a specific arrangement has already been evolu-tionarily selected. In the future, additional studies regarding how the conserved regions contribute to toxin specificity needs to be addressed along with how different effector domain arrangements contribute to host-bacterial interactions. Thus far, investigations have focused on connection of the holotoxins to pathogenesis with limited consideration for the function of any one domain or how domain changes can alter the patho-genesis program. In addition, for other Vibrio species, future studies could determine if the MARTX toxin can contribute to the development of a commensal relationship or if they are solely related to induction of disease in a variety of animal hosts.
ACKNOWLEDGMENT
I declare no conflicts of interest with regard to the manuscript.
Footnotes
ADDENDUM IN PROOF
The molecular mechanism of DUF5 has been recently published. Antic I, Biancucci M, Zhu Y, Gius DR, Satchell KJ. 2015. Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain. Nature Communication. Accepted 5/1/2015.
REFERENCES
- 1.Satchell KJ. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- 2.Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egerer M, Satchell KJ. Inositol hexakisphosphate-induced auto-processing of large bacterial protein toxins. PLoS Pathog. 2010;6:e1000942. doi: 10.1371/journal.ppat.1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 5.Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, Alves N, Jr, Ussery DW, Iida T, Thompson FL. Genomic taxonomy of vibrios. BMC Evol Biol. 2009;9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72:6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, Crosa JH. Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii . Infect Immun. 2011;79:2889–2900. doi: 10.1128/IAI.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann U, Wu S, Flaherty KM, McKay DB. Three-dimensional structure of the alkaline protease of Pseudomonas aeru-ginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolores J, Satchell KJ. Analysis of Vibrio cholerae genome sequences reveals unique rtxA variants in environmental strains and an rtxA-null mutation in recent altered El Tor isolates. mBio. 2013;4:e00624–e00612. doi: 10.1128/mBio.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boardman BK, Satchell KJ. Vibrio cholerae strains with mutations in an atypical type I secretion system accumulate RTX toxin intracellularly. J Bacteriol. 2004;186:8137–8143. doi: 10.1128/JB.186.23.8137-8143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BC, Lee JH, Kim MW, Kim BS, Oh MH, Kim KS, Kim TS, Choi SH. Vibrio vulnificus rtxE is important for virulence, and its expression is induced by exposure to host cells. Infect Immun. 2008;76:1509–1517. doi: 10.1128/IAI.01503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KE, Bang JS, Baek CH, Park DK, Hwang W, Choi SH, Kim KS. IVET-based identification of virulence factors in Vibrio vulnificus MO6–24/O. J Microbiol Biotechnol. 2007;17:234–243. [PubMed] [Google Scholar]
- 16.Fullner KJ, Mekalanos JJ. In vivo covalent crosslinking of actin by the RTX toxin of Vibrio cholerae . EMBO J. 2000;19:5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen A, Lupardus PJ, Albrow VE, Guzzetta A, Powers JC, Garcia KC, Bogyo M. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat Chem Biol. 2009;5:469–478. doi: 10.1038/nchembio.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YR, Lee SE, Kang IC, Nam KI, Choy HE, Rhee JH. A bacterial RTX toxin causes programmed necrotic cell death through calcium-mediated mitochondrial dysfunction. J Infect Dis. 2013;207:1406–1415. doi: 10.1093/infdis/jis746. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Rock JL, Nelson DR. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect Immun. 2008;76:2620–2632. doi: 10.1128/IAI.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YR, Kim BU, Kim SY, Kim CM, Na HS, Koh JT, Choy HE, Rhee JH, Lee SE. Outer membrane vesicles of Vibrio vulnificus deliver cytolysin-hemolysin VvhA into epithelial cells to induce cytotoxicity. Biochem Biophys Res Commun. 2010;399:607–612. doi: 10.1016/j.bbrc.2010.07.122. erratum in 403: 491–492. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Kim MW, Kim BS, Kim SM, Lee BC, Kim TS, Choi SH. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J Microbiol. 2007;45:146–152. [PubMed] [Google Scholar]
- 22.Liu M, Alice AF, Naka H, Crosa JH. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus . Infect Immun. 2007;75:3282–3289. doi: 10.1128/IAI.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CT, Pajuelo D, Llorens A, Chen YH, Leiro JM, Padros F, Hor LI, Amaro C. MARTX of Vibrio vulnificus biotype 2 is a virulence and survival factor. Environ Microbiol. 2013;15:419–432. doi: 10.1111/j.1462-2920.2012.02854.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim YR, Lee SE, Kook H, Yeom JA, Na HS, Kim SY, Chung SS, Choy HE, Rhee JH. Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell Microbiol. 2008;10:848–862. doi: 10.1111/j.1462-5822.2007.01088.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen YC, Chung YT. A conserved GTPase YchF of Vibrio vulnificus is involved in macrophage cytotoxicity, iron acquisition, and mouse virulence. Int J Med Microbiol. 2011;301:469–474. doi: 10.1016/j.ijmm.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Dolores JS, Agarwal S, Egerer M, Satchell KJ. Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol Microbiol. 2015;95:590–604. doi: 10.1111/mmi.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo HR, Lin JH, Chen YH, Chen CL, Shao CP, Lai YC, Hor LI. RTX Toxin enhances the survival of Vibrio vulnificus during infection by protecting the organism from phagocytosis. J Infect Dis. 2011;203:1866–1874. doi: 10.1093/infdis/jir070. [DOI] [PubMed] [Google Scholar]
- 28.Kim BS, Gavin HE, Satchell KJ. Distinct Roles of the Repeat-Containing Regions and Effector Domains of the Vibrio vulnificus Mul-tifunctional-Autoprocessing Repeats-in-Toxin (MARTX) Toxin. mBio. 2015;6:e00324–e00315. doi: 10.1128/mBio.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satchell KJ. MARTX: multifunctional-autoprocessing RTX toxins. Infect Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudryashova E, Heisler D, Zywiec A, Kudryashov DS. Ther-modynamic properties of the effector domains of MARTX toxins suggest their unfolding for translocation across the host membrane. Mol Microbiol. 2014;92:1056–1071. doi: 10.1111/mmi.12615. [DOI] [PubMed] [Google Scholar]
- 31.Sheahan KL, Cordero CL, Satchell KJ. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 2007;26:2552–2561. doi: 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prochazkova K, Satchell KJ. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J Biol Chem. 2008;283:23656–23664. doi: 10.1074/jbc.M803334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJ. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J Biol Chem. 2009;284:26557–26568. doi: 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roig FJ, Gonzalez-Candelas F, Amaro C. Domain organization and evolution of multifunctional autoprocessing repeats-in-toxin (MARTX) toxin in Vibrio vulnificus . Appl Environ Microbiol. 2011;77:657–668. doi: 10.1128/AEM.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwak JS, Jeong HG, Satchell KJ. Vibrio vulnificus rtxA1 gene recombination generates toxin variants with altered potency during intestinal infection. Proc Natl Acad Sci USA. 2011;108:1645–1650. doi: 10.1073/pnas.1014339108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004;363:223–233. doi: 10.1016/s0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 37.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci USA. 2012;109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudryashov DS, Durer ZA, Ytterberg AJ, Sawaya MR, Pashkov I, Prochazkova K, Yeates TO, Loo RR, Loo JA, Satchell KJ, Reisler E. Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin. Proc Natl Acad Sci USA. 2008;105:18537–18542. doi: 10.1073/pnas.0808082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geissler B, Bonebrake A, Sheahan KL, Walker ME, Satchell KJ. Genetic determination of essential residues of the Vibrio cholerae actin cross-linking domain reveals functional similarity with glutamine synthetases. Mol Microbiol. 2009;73:858–868. doi: 10.1111/j.1365-2958.2009.06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordero CL, Kudryashov DS, Reisler E, Satchell KJ. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J Biol Chem. 2006;281:32366–32374. doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudryashov DS, Cordero CL, Reisler E, Satchell KJ. Characterization of the enzymatic activity of the actin cross-linking domain from the Vibrio cholerae MARTXVc toxin. J Biol Chem. 2008;283:445–452. doi: 10.1074/jbc.M703910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudryashova E, Kalda C, Kudryashov DS. Glutamyl phosphate is an activated intermediate in actin crosslinking by actin crosslinking domain (ACD) toxin. PLoS One. 2012;7:e45721. doi: 10.1371/journal.pone.0045721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei J, Grishin NV. The Rho GTPase inactivation domain in Vibrio cholerae MARTX toxin has a circularly permuted papain-like thiol protease fold. Proteins. 2009;77:413–419. doi: 10.1002/prot.22447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahrens S, Geissler B, Satchell KJ. Identification of a His-Asp-Cys catalytic triad essential for function of the Rho inactivation domain (RID) of Vibrio cholerae MARTX toxin. J Biol Chem. 2013;288:1397–1408. doi: 10.1074/jbc.M112.396309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geissler B, Tungekar R, Satchell KJ. Identification of a conserved membrane localization domain within numerous large bacterial protein toxins. Proc Natl Acad Sci USA. 2010;107:5581–5586. doi: 10.1073/pnas.0908700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geissler B, Ahrens S, Satchell KJ. Plasma membrane association of three classes of bacterial toxins is mediated by a basic-hydrophobic motif. Cell Microbiol. 2012;14:286–298. doi: 10.1111/j.1462-5822.2011.01718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheahan KL, Satchell KJ. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae . Cell Microbiol. 2007;9:1324–1335. doi: 10.1111/j.1462-5822.2006.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ollis DL, Cygler CE, Dijkstra B, Frolow F, Franken SM, Remington HM, Silman I, Schrag J. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 49.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 50.Fullner KJ, Lencer WI, Mekalanos JJ. Vibrio cholerae-induced cellular responses of polarized T84 intestinal epithelial cells dependent of production of cholera toxin and the RTX toxin. Infect Immun. 2001;69:6310–6317. doi: 10.1128/IAI.69.10.6310-6317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathan MM, Chandy G, Mathan VI. Ultrastructural changes in the upper small intestinal mucosa in patients with cholera. Gastroenter-ology. 1995;109:422–430. doi: 10.1016/0016-5085(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 52.Olivier V, Haines GK, 3rd, Tan Y, Satchell KJ. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivier V, Queen J, Satchell KJ. Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PLoS One. 2009;4:e7352. doi: 10.1371/journal.pone.0007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Queen J, Satchell KJ. Neutrophils Are essential for containment of Vibrio cholerae to the intestine during the proinflammatory phase of infection. Infect Immun. 2012;80:2905–2913. doi: 10.1128/IAI.00356-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Satchell KJF. Activation and suppression of the proinflammatory immune response by Vibrio cholerae toxins . Microbes Infect. 2003;5:1241–1247. doi: 10.1016/j.micinf.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Lee YL, Hung PP, Tsai CA, Lin YH, Liu CE, Shi ZY. Clinical characteristics of non-O1/non-O139 Vibrio cholerae isolates and poly-merase chain reaction analysis of their virulence factors. J Microbiol Immunol Infect. 2007;40:474–480. [PubMed] [Google Scholar]
- 58.Chandrasekhar MR, Krishna BV, Patil AB. Changing characteristics of Vibrio cholerae: emergence of multidrug resistance and non-O1, non-O139 serogroups. Southeast Asian J Trop Med Public Health. 2008;39:1092–1097. [PubMed] [Google Scholar]
- 59.Thomas A, Straif-Bourgeois S, Sokol TM, Ratard RC. Vibrio infections in Louisiana: twenty-five years of surveillance 1980–2005. J La State Med Soc. 2007;159:205–208. 210–201. [PubMed] [Google Scholar]
- 60.Dalsgaard A, Forslund A, Bodhidatta L, Serichantalergs O, Pitarangsi C, Pang L, Shimada T, Echeverria P. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol Infect. 1999;122:217–226. doi: 10.1017/s0950268899002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altekruse SF, Bishop RD, Baldy LM, Thompson SG, Wilson SA, Ray BJ, Griffin PM. Vibrio gastroenteritis in the US Gulf of Mexico region: the role of raw oysters. Epidemiol Infect. 2000;124:489–495. doi: 10.1017/s0950268899003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziolo KJ, Jeong HG, Kwak JS, Yang S, Lavker RM, Satchell KJ. Vibrio vulnificus Biotype 3 MARTX toxin is an adenylate cyclase toxin essential for virulence in mice. Infect Immun. 2014;82:2148–2157. doi: 10.1128/IAI.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, Vlisidou I, Barron A, Bignell A, Clark L, Ormond D, Mayho M, Bason N, Smith F, Simmonds M, Churcher C, Harris D, Thompson NR, Quail M, Parkhill J, Ffrench-Constant RH. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens . BMC Genomics. 2009;10:302. doi: 10.1186/1471-2164-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Lindquist IE, Sundararajan A, Rajanna C, Floyd JT, Smith KP, Andersen JL, He G, Ayers RM, Johnson JA, Werdann JJ, Sandoval AA, Mojica NM, Schilkey FD, Mudge J, Varela MF. Genome sequence of non-O1 Vibrio cholerae PS15. Genome Announc. 2013;1:e00227–e00212. doi: 10.1128/genomeA.00227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE, Cowles KN, Darby C, de Léon L, Drace K, Du Z, Givaudan A, Herbert Tran EE, Jewell KA, Knack JJ, Krasomil-Osterfeld KC, Kukor R, Lanois A, Latreille P, Leimgruber NK, Lipke CM, Liu R, Lu X, Martens EC, Marri PR, Médigue C, Menard ML, Miller NM, Morales-Soto N, Norton S, Ogier JC, Orchard SS, Park D, Park Y, Qurollo BA, Sugar DR, Richards GR, Rouy Z, Slominski B, Slominski K, Snyder H, Tjaden BC, van der Hoeven R, Welch RD, Wheeler C, Xiang B, Barbazuk B, Gaudriault S, Goodner B, Slater SC, Forst S, Goldman BS, Goodrich-Blair H. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One. 2011;6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aktories K, Barmann M, Ohishi I, Tsuyama S, Jakobs KH, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 68.Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol. 2010;192:155–168. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 70.Gulig PA, Bourdage KL, Starks AM. Molecular Pathogenesis of Vibrio vulnificus . J Microbiol. 2005;43:118–131. [PubMed] [Google Scholar]
- 71.Kim HU, Kim SY, Jeong H, Kim TY, Kim JJ, Choy HE, Yi KY, Rhee JH, Lee SY. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol Syst Biol. 2011;7:460. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, Liu YM, Chen HJ, Shen AB, Li JC, Su TL, Shao CP, Lee CT, Hor LI, Tsai SF. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 2003;13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One. 2012;7:e37553. doi: 10.1371/journal.pone.0037553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waterfield NR, Daborn PJ, Dowling AJ, Yang G, Hares M, Ffrench-Constant RH. The insecticidal toxin makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol Lett. 2003;229:265–270. doi: 10.1016/S0378-1097(03)00846-2. [DOI] [PubMed] [Google Scholar]
- 75.Dowling AJ, Daborn PJ, Waterfield NR, Wang P, Streuli CH, Ffrench-Constant RH. The insecticidal toxin Makes caterpillars floppy (Mcf) promotes apoptosis in mammalian cells. Cell Microbiol. 2004;6:345–353. doi: 10.1046/j.1462-5822.2003.00357.x. [DOI] [PubMed] [Google Scholar]
- 76.Agarwal S, Agarwal S, Biancucci M, Satchell KJ. Induced autoprocessing of the cytopathic Makes Caterpillars Floppy-like effector domain of the Vibrio vulnificus MARTX toxin. Cell Microbiol. 2015 Apr 23; doi: 10.1111/cmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antic I, Biancucci M, Satchell KJ. Cytotoxicity of the Vibrio vulnificus MARTX toxin Effector DUF5 is linked to the C2A Subdomain. Proteins. 2014;82:2643–2656. doi: 10.1002/prot.24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park JH, Cho YJ, Chun J, Seok YJ, Lee JK, Kim KS, Lee KH, Park SJ, Choi SH. Complete genome sequence of Vibrio vulnificus MO6–24/O. J Bacteriol. 2011;193:2062–2063. doi: 10.1128/JB.00110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z, Chen H, Chen X, Zhou T, Zhao L, Zhang C, Jin W. Genome sequence of the human-pathogenic bacterium Vibrio vulnificus type strain ATCC 27562. J Bacteriol. 2012;194:6954–6955. doi: 10.1128/JB.01890-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang ZG, Wu Z, Xu SL, Zha J. Genome sequence of the human-pathogenetic bacterium Vibrio vulnificus B2. J Bacteriol. 2012;194:7019. doi: 10.1128/JB.01955-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeong HG, Satchell KJ. Additive function of Vibrio vulnificus MARTXVv and VvhA cytolysins promotes rapid growth and epithelial tissue necrosis during intestinal infection. PLoS Pathog. 2012;8:e1002581. doi: 10.1371/journal.ppat.1002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee TH, Kim MH, Lee CS, Lee JH, Rhee JH, Chung KM. Protection against Vibrio vulnificus infection by active and passive immunization with the C-terminal region of the RtxA1/MARTXVv protein. Vaccine. 2014;32:271–276. doi: 10.1016/j.vaccine.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 83.Lee BC, Choi SH, Kim TS. Vibrio vulnificus RTX toxin plays an important role in the apoptotic death of human intestinal epithelial cells exposed to Vibrio vulnificus . Microbes Infect. 2008;10:1504–1513. doi: 10.1016/j.micinf.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 84.Dhakal BK, Lee W, Kim YR, Choy HE, Ahnn J, Rhee JH. Caenorhabditis elegans as a simple model host for Vibrio vulnificus infection. Biochem Biophys Res Commun. 2006;346:751–757. doi: 10.1016/j.bbrc.2006.05.168. [DOI] [PubMed] [Google Scholar]
- 85.Chung KJ, Cho EJ, Kim MK, Kim YR, Kim SH, Yang HY, Chung KC, Lee SE, Rhee JH, Choy HE, Lee TH. RtxA1-induced expression of the small GTPase Rac2 plays a key role in the pathogenicity of Vibrio vulnificus . J Infect Dis. 2010;201:97–105. doi: 10.1086/648612. [DOI] [PubMed] [Google Scholar]
- 86.Toma C, Higa N, Koizumi Y, Nakasone N, Ogura Y, McCoy AJ, Franchi L, Uematsu S, Sagara J, Taniguchi S, Tsutsui H, Akira S, Tschopp J, Núñez G, Suzuki T. Pathogenic Vibrio activate NLRP3 inflammasome via cytotoxins and TLR/nucleotide-binding oligomeriza-tion domain-mediated NF-κB signaling. J Immunol. 2010;184:5287–5297. doi: 10.4049/jimmunol.0903536. [DOI] [PubMed] [Google Scholar]
- 87.Amaro C, Biosca EG. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl Environ Microbiol. 1996;62:1454–1457. doi: 10.1128/aem.62.4.1454-1457.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fouz B, Llorens A, Valiente E, Amaro C. A comparative epi-zootiologic study of the two fish-pathogenic serovars of Vibrio vulnificus biotype 2. J Fish Dis. 2010;33:383–390. doi: 10.1111/j.1365-2761.2009.01130.x. [DOI] [PubMed] [Google Scholar]
- 89.Fouz B, Roig FJ, Amaro C. Phenotypic and genotypic characterization of a new fish-virulent Vibrio vulnificus serovar that lacks potential to infect humans. Microbiology. 2007;153:1926–1934. doi: 10.1099/mic.0.2006/005405-0. [DOI] [PubMed] [Google Scholar]
- 90.Roig FJ, Amaro C. Plasmid diversity in Vibrio vulnificus biotypes. Microbiology. 2009;155:489–497. doi: 10.1099/mic.0.023424-0. [DOI] [PubMed] [Google Scholar]
- 91.Lee CT, Amaro C, Wu KM, Valiente E, Chang YF, Tsai SF, Chang CH, Hor LI. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J Bacteriol. 2008;190:1638–1648. doi: 10.1128/JB.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daborn PJ, Waterfield N, Silva CP, Au CP, Sharma S, Ffrench-Constant RH. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci USA. 2002;99:10742–10747. doi: 10.1073/pnas.102068099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bisharat N, Cohen DI, Harding RM, Falush D, Crook DW, Peto T, Maiden MC. Hybrid Vibrio vulnificus . Emerg Infect Dis. 2005;11:30–35. doi: 10.3201/eid1101.040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Efimov V, Danin-Poleg Y, Raz N, Elgavish S, Linetsky A, Kashi Y. Insight into the evolution of Vibrio vulnificus biotype 3’s genome. Front Microbiol. 2013;4:393. doi: 10.3389/fmicb.2013.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bisharat N1, Agmon V, Finkelstein R, Raz R, Ben-Dror G, Lerner L, Soboh S, Colodner R, Cameron DN, Wykstra DL, Swerdlow DL, Farmer JJ., 3rd Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Israel Vibrio Study Group. Lancet. 1999;354:1421–1424. doi: 10.1016/s0140-6736(99)02471-x. [DOI] [PubMed] [Google Scholar]
- 96.Zaidenstein R, Sadik C, Lerner L, Valinsky L, Kopelowitz J, Yishai R, Agmon V, Parsons M, Bopp C, Weinberger M. Clinical characteristics and molecular subtyping of Vibrio vulnificus illnesses, Israel. Emerg Infect Dis. 2008;14:1875–1882. doi: 10.3201/eid1412.080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Danin-Poleg Y, Elgavish S, Raz N, Efimov V, Kashi Y. Genome Sequence of the Pathogenic Bacterium Vibrio vulnificus Biotype 3. Genome Announc. 2013;1:e0013613. doi: 10.1128/genomeA.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Phillips KE, Schipma MJ, Satchell KJ. Draft genome sequence of Israeli outbreak-associated Vibrio vulnificus biotype 3 clinical isolate BAA87. Genome Announc. 2014;2:e00031–e00014. doi: 10.1128/genomeA.00032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Actis LA, Tolmasky ME, Crosa JH. Vibriosis. In: Woo PTK, Bruno BW, editors. Fish Diseases and Disorders: Viral, Bacterial, and Fungal Infections. 2nd ed. Vol. 3. Oxfordshire, UK: CABI International; 2010. pp. 570–605. [Google Scholar]
- 100.Thompson FL, Thompson CC, Dias GM, Naka H, Dubay C, Crosa JH. The genus Listonella MacDonell and Colwell 1986 is a later heterotypic synonym of the genus Vibrio Pacini 1854 (Approved Lists 1980)-a taxonomic opinion. Int J Syst Evol Microbiol. 2011;61:3023–3027. doi: 10.1099/ijs.0.030015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Denkin SM, Nelson DR. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl Environ Microbiol. 1999;65:3555–3560. doi: 10.1128/aem.65.8.3555-3560.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemos ML, Salinas P, Toranzo AE, Barja JL, Crosa JH. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum . J Bacteriol. 1988;170:1920–1925. doi: 10.1128/jb.170.4.1920-1925.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crosa JH, Schiewe MH, Falkow S. Evidence for plasmid contribution to the virulence of fish pathogen Vibrio anguillarum . Infect Immun. 1977;18:509–513. doi: 10.1128/iai.18.2.509-513.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Milton DL, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum . J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toranzo AE, Barja JL, Potter SA, Colwell RR, Hetrick FM, Crosa JH. Molecular factors associated with virulence of marine vibrios isolated from striped bass in Chesapeake Bay. Infect Immun. 1983;39:1220–1227. doi: 10.1128/iai.39.3.1220-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schiewe M, Crosa JH. Vibrio ordalii sp. nov.: a causative agent of vibriosis in fish. Curr Microbiol. 1981;6:343–348. [Google Scholar]
- 107.Le Roux F, Labreuche Y, Davis BM, Iqbal N, Mangenot S, Goarant C, Mazel D, Waldor MK. Virulence of an emerging pathogenic lineage of Vibrio nigripulchritudo is dependent on two plasmids. Environ Microbiol. 2011;13:296–306. doi: 10.1111/j.1462-2920.2010.02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thompson JR1, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 109.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, Polz MF, Alm EJ. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.French CT, Panina EM, Yeh SH, Griffith N, Arambula DG, Miller JF. The Bordetella type III secretion system effector BteA contains a conserved N-terminal motif that guides bacterial virulence factors to lipid rafts. Cell Microbiol. 2009;11:1735–1749. doi: 10.1111/j.1462-5822.2009.01361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 112.Hoffmann M, Monday SR, Allard MW, Strain EA, Whittaker P, Naum M, McCarthy PJ, Lopez JV, Fischer M, Brown EW. Vibrio caribbeanicus sp. nov., isolated from the marine sponge Scleritoderma cyanea . Int J Syst Evol Microbiol. 2012;62:1736–1743. doi: 10.1099/ijs.0.032375-0. [DOI] [PubMed] [Google Scholar]
- 113.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]