Abstract
Monomeric clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated 9 (Cas9) nucleases have been widely adopted for simple and robust targeted genome editing but also have the potential to induce high-frequency off-target mutations. In principle, two orthogonal strategies for reducing off-target cleavage, truncated guide RNAs (tru-gRNAs) and dimerization-dependent RNA-guided FokI-dCas9 nucleases (RFNs), could be combined as tru-RFNs to further improve genome editing specificity. Here we identify a robust tru-RFN architecture that shows high activity in human cancer cell lines and embryonic stem cells. Additionally, we demonstrate that tru-gRNAs reduce the undesirable mutagenic effects of monomeric FokI-dCas9. Tru-RFNs combine the advantages of two orthogonal strategies for improving the specificity of CRISPR/Cas nucleases and therefore provide a highly specific platform for performing genome editing.
Introduction
Programmable RNA-guided nucleases, engineered from the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) bacterial adaptive immune system, are robust and versatile tools that have substantially simplified targeted genome editing in living cells and organisms.1–3 Nuclease-induced double-stranded breaks (DSBs) can be repaired by nonhomologous end-joining (NHEJ) often resulting in variable-length insertions and deletions (indels) or by homology-directed repair with an exogenous “donor” template. Streptococcus pyogenes Cas9 nuclease can be programmed to recognize and cleave DNA target sites adjacent to a 5′-NGG-3′ protospacer adjacent motif (PAM). The specificity of Cas9-mediated cleavage for different genomic sites can be easily modified by varying a 17–20 nt region of target-site complementarity at the 5′ end of a ∼100 nt single guide RNA (gRNA) that complexes with Cas9.4
Observations of high-frequency off-target effects induced by CRISPR/Cas9 nucleases5–9 have spurred the development of novel strategies to increase its specificity. One strategy for improving Cas9 cleavage specificity has been to utilize truncated gRNAs (tru-gRNAs), in which the region of target-site complementarity at the 5′ end of the gRNA has been shortened by as many as 3 nt. The use of tru-gRNAs substantially reduces CRISPR/Cas9 off-target effects while generally maintaining full on-target activity.10 One hypothesis for how tru-gRNAs improve specificity is that they reduce excess binding energy of the CRISPR/Cas9:gRNA complex to target DNA, thereby conferring higher sensitivity to mismatches at the gRNA:DNA target-site interface.
Another strategy developed by our group and others11,12 is to fuse the dimerization-dependent nonspecific FokI cleavage domain to the amino-terminus of a catalytically inactive Cas9 protein (dCas9), thereby creating a dimeric RNA-guided FokI-dCas9 nuclease (RFN). Because FokI is only active as a dimer,13 this system is dependent on recruitment of two FokI-dCas9 monomers to adjacent “half-sites” with their PAM sites facing outward away from an intervening spacer sequence to enable FokI dimerization and efficient DNA cleavage of the spacer region. Therefore, an RFN target site is essentially double the length of a conventional Cas9 target site.
In principle, the tru-gRNA and RFN strategies could be combined to create CRISPR/Cas9-based nucleases with the desirable properties of improved sensitivity to mismatches, an extended double-length target site, and dimerization-dependent activity. Here we show that the on-target activities of RFNs can be directed by tru-gRNAs and that the activities of these tru-RFNs can be comparable to the activities of RFNs directed by full-length gRNAs. Additionally, the use of tru-gRNAs can reduce the residual undesired mutagenic activity of monomeric RFNs. Our findings suggest the utility of tru-RFNs for applications requiring the highest possible genome editing specificity.
Methods
Experimental design to test activity of dimeric RNA-guided FokI nucleases directed by tru-gRNAs
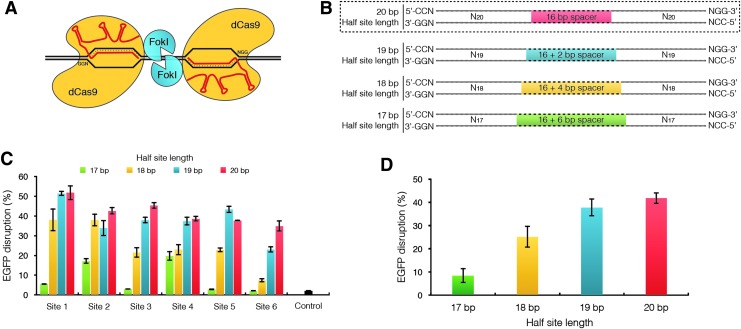
To determine whether pairs of gRNAs truncated at the 5′ end could efficiently direct activity of dimeric RFNs (Fig. 1A), we designed a series of gRNA pairs with variable-length half-sites (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hum). We reasoned that as the FokI-dCas9 fusion protein is anchored by contacts to NGG PAMs, truncating the gRNA at the 5′ end should not change the effective position of the protein with respect to a bound target site. As previously described, we used the ribonuclease Csy4 to process multiple gRNAs flanked by Csy4 recognition sites on a single transcript into individual gRNAs bearing any 5′ nucleotide,11 enabling us to test varying lengths of target-site complementarity without being limited by the 5′ G requirement for efficient RNA Pol III transcription initiation at the human U6 promoter.11
Figure 1.
Optimization of dimeric tru-RFN architecture. (A) Schematic of dimeric RNA-guided FokI-dCas9 nucleases (RFNs) binding to DNA. Two FokI-dCas9 proteins are targeted to adjacent PAM-out DNA half-site sequences by two gRNAs enabling FokI dimerization and cleavage of the intervening spacer sequence. (B) Standard RFN and tru-RFN-binding sites. The overall length of the dimeric target site is 62 bp with the 5′-NGG-3′ PAMs facing outward. The spacer length is dependent on the gRNA half-site length: 20, 19, 18, and 17 bp half-sites correspond to 16, 18, 20, and 22 bp spacers, respectively. (C) Mutagenic activity of standard RFNs or tru-RFNs with 17–20 bp half-site lengths. All RFNs are targeted to the same six target sites within the EGFP gene (Supplementary Table S1). Control is U2OS.EGFP nucleofected with ptdTomato-N1 plasmid. EGFP disruption in U2OS.EGFP cells was measured by flow cytometry. Error bars represent s.e.m., n=3. (D) Mean EGFP disruption values of (C) grouped according by gRNA half-site length. Error bars represent s.e.m., n=6. gRNAs, guide RNAs; RFN, RNA-guided FokI-dCas9 nucleases.
Single and multiplex gRNA expression plasmids
Plasmids encoding single or multiplex gRNAs were assembled by cloning target-site oligoduplexes (Integrated DNA Technologies) and a constant region oligoduplex (for multiplex gRNAs) into pSQT1313 (Addgene plasmid #53370). Cloning of single or multiplex gRNAs was performed as previously described.11
Tissue culture and transfections
Cell culture experiments were carried out in HEK293 cells, feeder-free HUES9 cells, wild-type U2OS cells, or in U2OS cells harboring a stably integrated, single-copy, destabilized EGFP gene (U2OS.EGFP cells). HEK293 and U2OS cells were cultured in Advanced DMEM (Life Technologies) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies), and penicillin/streptomycin at 37°C with 5% CO2. In addition, U2OS.EGFP medium was supplemented with 400 μg/ml of G418. HUES9 cells were plated on Geltrex-coated tissue culture plates and cultured in mTeSR 1 medium (Stemcell Technologies). Two hours before transfection until harvesting of genomic DNA, mTeSR 1 medium was supplemented with 10 μM of Rho-associated kinase inhibitor Y-27632 (Sigma).
HEK293 cells were transfected with TransIT-LT1 (Mirus Bio) according to the manufacturer's instructions as described previously.11 U2OS cells and U2OS.EGFP cells were transfected using the DN-100 program of a Lonza 4D-Nucleofector according to the manufacturer's instructions as described previously.11 HUES9 cells were transfected using the CA-137 program and the P3 primary cell solution (Lonza). An amount of 975 ng of human codon-optimized pCAG-Csy4-T2A-nls-FokI-dCas9-nls (pSQT1601; Addgene plasmid #53369) or pCAG-Csy4-T2A-Cas9n-nls (pNW3; Addgene plasmid #53372) was transfected along with 325 ng of gRNA or tru-gRNA vector and 10 ng of ptdTomato-N1 expression plasmid (Clontech). All transfections were performed in triplicate.
Mutation rate quantification by EGFP disruption and T7EI assay
The EGFP disruption assay to quantify nuclease-induced mutation frequencies was performed as previously described.11 EGFP and tdTomato fluorescence was measured three days posttransfection using a BD Biosciences LSR II or Fortessa FACS analyzer.
T7 endonuclease I (T7E1) assays to estimate mutagenesis frequencies were performed as previously described.14 Three days posttransfection, genomic DNA was isolated using the Agencourt DNAdvance Genomic DNA Isolation kit (Beckman Coulter Genomics) according to the manufacturer's instructions. To amplify genomic loci, Phusion Hot-start Flex DNA polymerase (NEB) and primers listed in Supplementary Table S2 were used in a touchdown PCR protocol (98°C, 3 min; [98°C, 10 s; 72–62°C, −1°C/cycle, 15 s; 72°C, 30 s]×10 cycles, [98°C, 10 s; 62°C, 15 s; 72°C, 30 s]×25 cycles; 72°C, 3 min). An amount of 200 ng of purified PCR amplicons was denatured, re-annealed, and digested with T7 Endonuclease I (NEB) as previously described. Mutation rates were quantified using a Qiaxcel capillary electrophoresis instrument (Qiagen) as previously described.14
Illumina library preparation and analysis
Genomic sites were amplified for deep sequencing with Phusion Hot-start FLEX DNA polymerase (NEB) and primers listed in Supplementary Table S2 and the touchdown PCR protocol as described above. Amplicons of 200–350 bp were purified using Ampure XP beads (Beckman Coulter Genomics) according to manufacturer's instructions. Dual-indexed TruSeq Illumina deep sequencing libraries were prepared using a high-throughput “with bead” protocol (KAPA Biosystems) on a Sciclone G3 liquid-handling workstation. Adapter-ligated libraries were quantified using a Qiaxcel capillary electrophoresis instrument (Qiagen) and sequenced on an Illumina MiSeq Sequencer by the Dana-Farber Cancer Institute Molecular Biology Core. Paired-end reads of 150 bp were mapped to human genome reference GChr37 using BWA-MEM.15 Indel mutations or point mutations were quantified as previously described.14 Integrative Genomics Viewer16 and Python were used for mutation analyses.
Results
Optimizing the length of tru-gRNAs for use with dimeric RFNs
To test whether tru-gRNAs can be used with FokI-dCas9 fusions, we measured the targeting efficiency of FokI-dCas9 fusions with tru-gRNAs bearing various lengths of complementarity to the target DNA site at six different target sites in an EGFP reporter gene. To quantify mutagenesis frequencies, we used a well-established human U2OS cell-based EGFP disruption assay in which cleavage of an EGFP gene sequence followed by mutagenic NHEJ-mediated repair results in loss of EGFP expression (see Methods). For each of these six EGFP target sites, we designed four gRNA pairs with half-site complementarity ranging from 17 to 20 bp. In each pair, the two gRNAs were designed to be equal length and separated by intervening spacer sequences that maintain the original distances between the two PAM sites bound by each monomer of FokI-dCas9 (Fig. 1B and Supplementary Table S1). gRNA pairs were coexpressed with FokI-dCas9 in our EGFP reporter cell line and the percentage of EGFP-disrupted cells was quantified three days posttransfection (Fig. 1C). We found that EGFP disruption rates with gRNA pairs bearing 19 bp of half-site complementarity maintained on average ∼90% of the mutagenic activities of matched gRNA pairs bearing 20 bp of half-site complementarity. Pairs of gRNAs bearing 17 or 18 bp of half-site complementarity showed more dramatic average decreases in activity of 40% or 80%, respectively, across the 6 different target sites (Fig. 1D). Given these results, we chose to use tru-gRNAs with 19 bp half-site complementarity lengths (and 18 bp spacers between the 2 monomeric target sites) for all subsequent experiments.
Activities of dimeric tru-RFNs at endogenous human genes
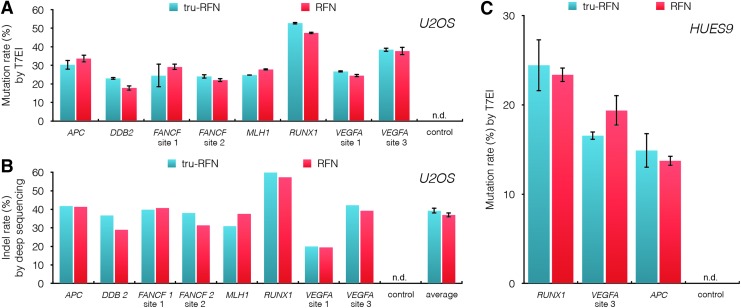
To assess whether tru-RFNs could also efficiently edit endogenous human genes, we targeted matched standard RFNs or tru-RFNs to eight different sites in human U2OS cells (Supplementary Table S2) and measured mutation frequencies by T7EI assay (Fig. 2A). Tru-RFNs efficiently mutagenized all 8 sites (range of 23–53%; mean of 31%) as did standard RFNs (range of 18–47%; mean of 30%). No statistically significant difference was detected between mutagenesis rates for conventional RFNs versus tru-RFNs (p=0.527, two-way ANOVA). Deep sequencing of the same genomic DNA used for the T7EI assay confirmed the similar activity rates of matched standard and tru-RFNs (Fig. 2B).
Figure 2.
Standard RFN and tru-RFN activity at endogenous target sites in human U2OS and HUES9 cells. (A) Comparison of standard RFN and tru-RFN mutagenesis frequencies at eight endogenous dimeric target sites in human U2OS cells. Mutation rates were quantified by T7EI assay. p=0.527; two-way ANOVA; n.d., not detected; error bars represent s.e.m., n=3. (B) Orthogonal verification of mutation rates from (A) by deep sequencing. Each value was determined from a single deep-sequencing library prepared from genomic DNA that had been pooled from three independent transfections. p=0.2352; paired t-test; n.d., not detected. Error bar of average mutation rates represents s.e.m., n=8. (C) Genome editing efficiencies in human embryonic stem cells using RFNs and tru-RFNs. The RUNX1, VEGFA site 3, and APC loci were targeted by the same pairs of standard or tru-gRNAs as in Fig. 2A. Mutation rates were quantified using the T7EI assay. p=0.880; two-way ANOVA; n.d. not detected. Error bars represent s.e.m., n=3. APC, adenomatous polyposis coli; DDB2, damage-specific DNA binding protein 2; FANCF, Fanconi anemia, complementation group F; MLH1, Mutl homolog 1; RUNX1, Runt-related transcription factor 1; VEGFA, human vascular endothelial growth factor A.
We next sought to determine whether RFNs and tru-RFNs could also function robustly in a more clinically relevant cell type. Using an optimized transfection protocol for the human embryonic stem cell line HUES9, we targeted three genes that have been associated with human pancreatic cancer, RUNX1, VEGFA, and APC, with matched tru-RFNs and standard RFNs (Fig. 2C).17–19 As we found with U2OS cells, both standard and tru-RFNs robustly mutagenized their target sites in HUES9 cells. In addition, we did not detect a significant difference in mutagenesis frequencies between full-length RFNs and tru-RFNs (p=0.880, two-way ANOVA).
tru-RFN monomers induce fewer indels and point mutations than conventional RFN monomers at their target sites
Previously, we observed that coexpression of FokI-dCas9 with a single gRNA could induce low-frequency mutagenesis at the gRNA target site.11 These included two classes of mutations: variable-length indels, presumably induced by low-frequency cleavage of the target site, and hypermutation of certain base positions near the end of the half-site closest to the spacer (i.e., the part of the half-site that interacts with the 5′ end of the gRNA). We hypothesized that use of tru-gRNAs might reduce both types of unwanted mutagenesis by reducing the binding stability of the FokI-dCas9 complex, which could reduce both FokI dimerization and cleavage as well as decrease the length of unwound target-site DNA that might be exposed to cellular deaminases that could induce hypermutation.
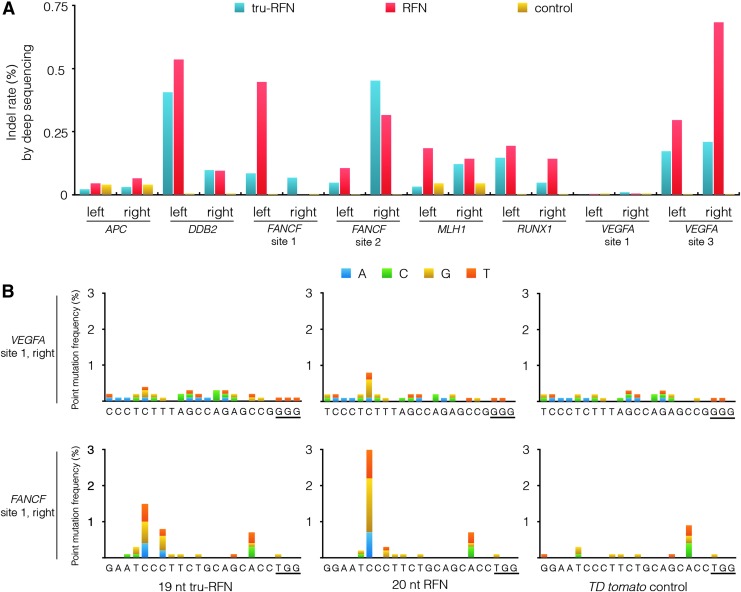
To quantify the residual mutagenic activity of standard RFN and tru-RFN monomers, we expressed FokI-dCas9 with single gRNAs targeted to either the “left” or “right” half-site of the eight dimeric target sites analyzed in this study and assessed indel frequencies by deep sequencing. On average, coexpression of FokI-dCas9 with tru-gRNAs as compared with conventional full-length gRNAs reduces unwanted indel mutation frequencies by 40% (p=0.047, Fig. 3A). We also analyzed the deep sequencing data to assess whether tru-gRNAs decreased the frequencies of point mutations introduced by RFNs. Unlike low-level indel mutations that seem to occur at many different target sites of monomeric RFNs, we previously observed point mutations only at the VEGFA site 1 right and FANCF site 1 right target sites.11 Similarly, we found that among the various tru-gRNAs we tested, only the VEGFA site 1 right and FANCF site 1 right gRNAs induce point mutations with FokI-dCas9; however, at those two sites, tru-RFNs induce lower frequencies of point mutations relative to matched standard RFNs (Fig. 3B). As we observed previously with standard RFNs, tru-RFNs induced point mutations primarily at hot spots between positions 13 and 15 of the target site (base positions numbered by increasing distance from the PAM sequence as previously described, where position 1 is closest to the PAM11).
Figure 3.
Indel mutation and point mutation frequencies induced by standard RFN or tru-RFN monomers. (A) Mutation rates of RFN and tru-RFN monomers targeted to left or right half-site of the eight dimeric target sites of Fig. 2A. For each locus, we assessed indel frequencies with individual full-length gRNAs or tru-gRNAs targeting either the left or right half-site. Mutation frequencies were determined by deep sequencing, and the numerical values for this data are presented in Supplementary Table S3. Each indel frequency value was determined from a single deep-sequencing library prepared from genomic DNA that had been pooled from three independent transfection experiments. p=0.047; paired t-test. (B) Comparison of point mutation frequencies in the VEGFA site 1 and FANCF site 1 target sites of FokI-dCas9 coexpressed with single tru-gRNAs or single conventional gRNAs. As a negative control, deep sequencing was performed on genomic DNA of U2OS cells that have only been transfected with a ptdTomato-N1 plasmid. Each point mutation frequency value was determined from a single deep-sequencing library prepared from genomic DNA that had been pooled from three independent transfection experiments. Note that, for point mutation analysis, the same raw deep sequencing data as in Fig. 3A were used.
Discussion
Here we show that truncated gRNAs can be used with the dimeric RFN platform to create tru-RFNs for highly specific genome editing in human cells. We identified an optimal gRNA 5′ truncation length that maintains robust on-target activity similar to that of conventional RFNs. Interestingly, while Cas9 shows full activity with most gRNAs that are truncated by up to three nucleotides, tru-RFNs induce efficient mutagenesis only with two gRNAs each truncated by one nucleotide. Use of gRNAs bearing 5′ truncations of two or three nucleotides leads to greater reductions or loss of detectable activity by FokI-dCas9. One potential explanation for the reduced tolerance of dimeric RFNs for 5′ gRNA truncation is that greater monomeric binding affinity by dCas9 may be required to stabilize FokI dimerization and cleavage (even with the advantages of potentially cooperative binding) compared with wild-type Cas9 binding and cleavage. This reduced tolerance to gRNA truncation suggests that dimeric RFNs may also be less tolerant of mismatches at the gRNA:DNA interface, a hypothesis that would need to be confirmed by future studies.
An important consideration for designing active RFNs is picking dimeric target sites with the appropriate spacing between gRNA half-sites. Tru-RFNs have the same requirement for half-site positioning as standard RFNs, as measured by the distance between the PAMs of adjacent RFN half-sites. To facilitate the use of the tru-RFN technology, we have also modified our recently developed web-based CasBLASTR tool20 so that it can find properly spaced standard and tru-RFN dimeric target sites in a DNA sequence of choice. CasBLASTR also offers instructions for cloning of multiplex gRNA expression vectors.
One potential limitation of the RFN technology is that FokI-dCas9 guided to a target locus by a single gRNA might induce indel mutations, possibly by recruitment of a second FokI-dCas9 monomer from solution. Our earlier study supported this idea because low-frequency on-target mutagenesis is observed when FokI-dCas9 is expressed with just one gRNA. Presumably, if this phenomenon can be observed at the intended target site, then indels might also be induced at other monomeric off-target sites elsewhere in the genome. Our study suggests that the use of tru-gRNAs to direct FokI-dCas9 can reduce the frequencies of indel mutations caused by RFN monomers, presumably by decreasing the energy of this interaction.
Previously, we observed that RFN monomers can also introduce undesired point mutations at their target sites, although at a frequency lower than that for Cas9 nickase (Cas9n) monomers.11 While the mechanism for this activity is unknown, we speculated that it might be the result of interactions between endogenous cellular deaminases and unwound DNA exposed as ssDNA during Cas9n or FokI-dCas9 target-site recognition. Tru-RFNs may help reduce the frequencies of these point mutations, perhaps because of effects of reduced binding affinity resulting in decreased residence time of the FokI-dCas9 protein bound to unwound target-site DNA. Alternatively, the structure and nature of the R-loop might be different in this hypermutation region when the gRNA is truncated (i.e., less extensively unwound), which could change its effective solvent-exposed profile. Further studies are required to answer remaining questions such as why those point mutations have only been detected at certain positions within certain target sites, why point mutation rates induced by Cas9 nickases can be up to 10-fold higher than point mutation rates of monomeric RFNs (Supplementary Fig. S1), and what the practical significance of these point mutations is for research or clinical applications of the CRISPR/Cas technology.
It is challenging to quantify the specificity improvement achieved by tru-RFNs without a method for identifying bona fide off-target cleavage sites. However, it has become increasingly clear that in silico off-target prediction algorithms21,22 and dCas9 ChIP seq23–25 methodologies have substantial limitations for comprehensive profiling off-target mutagenesis induced by CRISPR/Cas9 nucleases.26 We conducted preliminary experiments to test whether our recently developed genome-wide unbiased identification of DSBs enabled by sequencing or GUIDE-seq method26 could be directly applied to dimeric RFNs, and found that the first step of end-protected double-stranded oligodeoxynucleotide tag integration was relatively inefficient in comparison to wild-type Cas9 (data not shown). In future studies, we will seek to optimize genome-wide specificity profiling methods such as GUIDE-seq26 to directly measure the off-target effects of dimeric RFNs and tru-RFNs.
The dimeric RFN platform enables high-efficiency genome editing with reduced off-target effects relative to standard monomeric Cas9. Our work demonstrates that the use of tru-gRNAs likely further reduces potential low-frequency indels induced by RFNs, generally without sacrificing on-target editing frequencies in both human transformed somatic and embryonic stem cells. Therefore, our data show that tru-RFNs provide a useful and further improved tool for high-precision genome editing applications in human cells.
Supplementary Material
Acknowledgments
This work was funded by a National Institutes of Health (NIH) Director's Pioneer Award (DP1 GM105378), NIH P50 HG005550, NIH R01 AR063070, and the Jim and Ann Orr Massachusetts General Hospital Research Scholar Award. S.Q.T. was supported by NIH F32 GM105189.
Author Disclosure
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. S.Q.T. and J.K.J. have filed a patent application covering the RFN technology.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, Chylinski K, Fonfara I, et al. . A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y, Foden JA, Khayter C, et al. . High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattanayak V, Lin S, Guilinger JP, et al. . High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013;31:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu PD, Scott DA, Weinstein JA, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Aach J, Stranges PB, et al. . CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013;31:833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SW, Kim S, Kim Y, et al. . Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014;24:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Sander JD, Reyon D, et al. . Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 2014;32:279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai SQ, Wyvekens N, Khayter C, et al. . Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 2014;32:569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 2014;32:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bitinaite J, Wah DA, Aggarwal AK, et al. . FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA 1998;95:10570–10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyon D, Tsai SQ, Khayter C, et al. . FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012;30:460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010;26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. . Integrative genomics viewer. Nat Biotechnol 2011;29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birnbaum DJ, Adélaïde J, Mamessier E, et al. . Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer 2011;50:456–465 [DOI] [PubMed] [Google Scholar]
- 18.Inoue M, Hager JH, Ferrara N, et al. . VEGF-A has a critical, nonredundant role in angiogenic switching and pancreatic β cell carcinogenesis. Cancer Cell 2002;1:193–202 [DOI] [PubMed] [Google Scholar]
- 19.Horii A, Nakatsuru S, Miyoshi Y, et al. . Frequent somatic mutations of the APC gene in human pancreatic cancer. Cancer Res 1993;52:6696–8 [PubMed] [Google Scholar]
- 20.Kleinstiver BP, Prew MS, Tsai SQ, et al. . Engineered CRISPR-Cas9 nucleases with altered and improved PAM specificities. Nature 2015. DOI: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran FA, Hsu PD, Wright J, et al. . Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods 2014;11:122–123 [DOI] [PubMed] [Google Scholar]
- 23.Kuscu C, Arslan S, Singh R, et al. . Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 2014;32:677–683 [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Scott DA, Kriz AJ, et al. . Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 2014;32:670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan J, Lu G, Xie Z, et al. . Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res 2014;24:1009–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai SQ, Zheng Z, Nguyen NT, et al. . GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.