Abstract
CRISPR/Cas9 derived from the bacterial adaptive immunity pathway is a powerful tool for genome editing, but the safety profiles of in vivo delivered Cas9 (including host immune responses to the bacterial Cas9 protein) have not been comprehensively investigated in model organisms. Nonalcoholic steatohepatitis (NASH) is a prevalent human liver disease characterized by excessive fat accumulation in the liver. In this study, we used adenovirus (Ad) vector to deliver a Streptococcus pyogenes–derived Cas9 system (SpCas9) targeting Pten, a gene involved in NASH and a negative regulator of the PI3K-AKT pathway, in mouse liver. We found that the Ad vector mediated efficient Pten gene editing even in the presence of typical Ad vector-associated immunotoxicity in the liver. Four months after vector infusion, mice receiving the Pten gene-editing Ad vector showed massive hepatomegaly and features of NASH, consistent with the phenotypes following Cre-loxP-induced Pten deficiency in mouse liver. We also detected induction of humoral immunity against SpCas9 and the potential presence of an SpCas9-specific cellular immune response. Our findings provide a strategy to model human liver diseases in mice and highlight the importance considering Cas9-specific immune responses in future translational studies involving in vivo delivery of CRISPR/Cas9.
Introduction
Rapidly advancing genome-editing technologies hold great promise for realizing therapeutic gene manipulation at the DNA level, and have triggered much excitement in the gene therapy field.1,2 Particularly with the advent of the CRISPR/Cas9 platform adopted from the bacterial adaptive immune system, genome editing in mammalian cells and in a range of model organisms has become increasingly straightforward.3–5 One important application of genome editing is to rapidly generate somatic mouse models of certain human diseases,6–8 thus bypassing the time- and resource-consuming genetic manipulation from embryo transgenesis to breeding in multiple rounds. For example, we have previously showed that hydrodynamic injection of plasmid DNA expressing an SpCas9 system to target cancer genes in wild-type mice could yield cancer-related phenotypes in liver.9–11 Despite being convenient, hydrodynamic injection can deliver plasmid DNA to only about 20% of hepatocytes in mice,12 which limits gene editing efficiency. To enhance delivery, several groups have used viral vectors, especially the adenovirus (Ad) vector with large cargo capacity, to systemically deliver SpCas9 into mouse liver for either therapeutic purpose or disease modeling.13,14 In these two studies, expected phenotypic outcomes following targeted gene editing were described 3 and 14 days after Ad vector injection, respectively. However, it has been well documented that systemic Ad vector infection in immunocompetent mice triggers strong immune responses to both viral gene and transgene products in the liver that eventually clear transduced hepatocytes.15 Therefore, information about the sustainability of Ad vector-mediated gene editing and the resulting phenotypic changes in the liver remains elusive.
Another poorly characterized aspect of in vivo CRISPR/Cas9 genome editing in mammals concerns host immune responses mounted against the bacteria-derived Cas9 protein. Theoretically, only a single, transient delivery of CRISPR/Cas9 system is required to make permanent genetic modifications, and such transient function may reduce off-target effect.16,17 However, many of the Cas9 constructs used in vivo may express for a long time, such as following recombinant adeno-associated virus (rAAV)-mediated delivery.18 Under these conditions, Cas9-specific immune responses, especially cytotoxic cellular responses, have the potential to destroy targeted cells and limit long-term therapeutic efficacy.
Phosphatase and tensin homolog (PTEN) is frequently mutated in many human sporadic cancers, as well as in patients with cancer predisposition syndromes such as Cowden disease.19 About half of the patients with primary hepatoma show decreased or absent PTEN protein expression, although PTEN does not mutate frequently in hepatocytes. Moreover, reduced PTEN expression is well correlated with increased tumor grade, advanced disease stage, and poor prognosis in patients with hepatocellular carcinoma (HCC). In a mouse model generated by the Cre-loxP system, hepatocyte-specific Pten deficiency led to massive hepatomegaly (liver swelling) and steatohepatitis (fatty liver disease), as well as adenomas or HCC at older ages.20,21 In light of these health risk associations, it is clear that Pten plays an important role in lipid metabolism, hepatocyte homeostasis, and tumorigenesis in the liver.19
In this study, we used an Ad vector to deliver an SpCas9 system targeting Pten in mouse liver, hypothesizing that Ad vector-mediated somatic gene editing can produce sustained liver phenotypes despite potential confounding effects of Ad vector-associated immune responses to transduced hepatocytes. We purposefully controlled the experimental conditions closely to resemble those of the previously published study of hydrodynamic injection of plasmid DNA carrying the same sgRNA and SpCas9 expression cassettes,9 allowing us to compare gene editing efficiency between these two methods. We found the Ad vector-mediated delivery to be more efficient for Pten gene editing than hydrodynamic injection two weeks after treatment, although typical Ad vector-associated immune responses and liver damage were detected during this period of time. Four months after infection, mice receiving the Pten gene-editing Ad vector showed massive hepatomegaly and steatohepatitis, consistent with the phenotype following Cre-loxP-induced Pten deficiency in mouse liver. In addition, evidence of both humoral and cellular immunity against SpCas9 following systemic Ad vector-mediated CRISPR/Cas9 delivery in mice was observed. To our knowledge, this is the first study reporting SpCas9-specific immune responses in mammals. These findings indicate that delivery methodology can affect Cas9-specific immune responses, and argue for close monitoring of Cas9-specific immune responses, as well as engineering efforts to limit Cas9 immunogenicity, in future translational studies involving in vivo delivery of CRISPR/Cas9 system to model organisms and human subjects.
Materials and Methods
Recombinant Ad genome construction and viral vector production
The SpCas9 cassette in Ad.sgPten was derived from pX330,4 where a hybrid chicken beta-actin promoter drives the expression of a codon-optimized S. pyogenes Cas9 cDNA with 3× FLAG tag and 2× SV40 nuclear localization signal. U6 promoter-driven sgRNA targeting Pten was PCR-amplified and cloned with the SpCas9 cassette into an Ad backbone plasmid22 using the Gibson cloning method. The guide RNA sequence is 5′-gagaucguuagcagaaacaaa-3′. EGFP driven by the hybrid chicken beta-actin promoter was cloned into a separate Ad backbone plasmid. E1/E3-deleted Ad type 5 vectors were produced using standard methods by either ViraQuest, Inc. (Ad.sgPten), or University of Massachusetts Medical School Viral Vector Core (Ad.GFP).
Animal use
Eight-week-old FVB/NJ female mice were initially used to study Pten gene editing efficiency and liver steatosis. In later studies characterizing immune responses, 8–10-week-old FVB/NJ female mice and 8-week-old C57BL/6 female mice were used. All FVB/NJ mice were purchased from Jackson Laboratory, and all C57BL/6 mice were obtained by in-house breeding. An amount of 2.5×1010 viral particles (5×108 plaque-forming units, PFU) diluted in 150 μl of phosphate-buffered saline (PBS) or equal volume of PBS without viral particles was delivered through tail vein injection. All animal study protocols were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Cell culture and infection with Ad vectors
Human embryonic kidney 293-T antigen transformed (HEK293T) cells and a mouse cell line carrying the oncogenic KrasG12D allele and inactivated p53 (KP cells)23 were maintained under standard cell culture conditions. Cells were seeded in 24-well plates 1 day before infection, and infected with Ad.GFP or Ad.sgPten. Seventy-two hours later, cells were harvested and subjected to the Surveyor nuclease assay as described below. The gene-editing reporter cell line was created by infecting HEK293 cells with lentiviral vectors to induce stable gene integration. The GFP and mCherry fluorescence was imaged 72 hr after Ad.sgPten infection.
Surveyor nuclease assay
Genomic DNA was extracted from cell culture using QE buffer (Epicentre) or from mouse liver tissue using High Pure PCR Template Preparation Kit (Roche). PCR was performed using Herculase II High-Fidelity Polymerase (Agilent) to amplify the DNA region of interest. PCR product was purified with QIAquick Gel Extraction Kit (Qiagen), and subjected to a Surveyor nuclease assay or to deep sequencing (see description below). The Surveyor nuclease assay was performed using the SURVEYOR Mutation Detection Kit (Transgenomic) as previously described,9 followed by electrophoresis using a 4–20% Novex TBE Gel (Life Technologies) with ethidium bromide staining.
Deep sequencing
Libraries were made from the purified PCR products described in the Surveyor nuclease assay according to the Nextera XT protocol, and sequenced on the Illumina Nextseq platform (75 bp paired-end). Standard Illumina sequencing analysis procedures were adopted to process the data.9 Reads were mapped to the Pten reference sequence, and insertion/deletion mutations were determined using VarScan2.
Histological analysis
Mice were humanely euthanized by CO2 asphyxiation followed by cervical dislocation. A portion of liver was fixed in 10% formalin solution overnight and embedded in paraffin, and 4-μm-thick sections were stained with hematoxylin and eosin. Three fields were imaged from each mouse liver by an investigator blinded to the treatment information.
Fluorescence microscopy of liver sections
Upon harvesting, a portion of liver tissue was directly embedded in O.C.T. Compound (Tissue-Tek) and stored under −80°C until sectioning, and 8-μm-thick cryosections were collected and mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). Native GFP signal and DAPI nuclear staining were captured by a fluorescence microscope (Leica). Three fields were imaged from each mouse liver by an investigator blinded to the treatment information. GFP+ cells were counted using the Imaris 8.1.1 image processing program (Bitplane). Briefly, using the 3D View option in Imaris, total DAPI was counted based on threshold intensity and cell diameter parameters. Afterward, a GFP threshold was applied across all images. DAPI and GFP signals above their respective thresholds that showed overlap were counted as GFP+ cells. To calculate the percentage of GFP+ cells, the number of GFP+ cells was divided by total DAPI. During automated analysis, cells on the edge of each image were excluded.
Serum ALT and AST assays
Blood was collected by facial vein bleeding, and serum was separated using a Microtainer tube with serum separator (BD). ALT and AST levels were assayed in duplicate using the ALT Color Endpoint Reagent Kit and AST Color Endpoint Reagent Kit (TECO Diagnostics), respectively, according to the manufacturer's instructions.
Reverse transcription-quantitative PCR
A portion of mouse liver was flash-frozen in liquid nitrogen immediately following harvesting, and stored under −80°C until RNA extraction. Total RNA was extracted using Direct-zol RNA MiniPrep Kit with DNase I treatment (ZYMO Research). About 1.5 μg of RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Multiplexed qPCR was performed in triplicates using TaqMan reagents that detect mouse Tnf-α (Life Technologies; Assay ID: Mm00443258_m1) and Gapdh (Life Technologies; cat # 4352339E), or mouse Ifn-γ (Life Technologies; Assay ID: Mm01168134_m1) and Gapdh. Tnf-α or Ifn-γ expression level was normalized to Gapdh.
Immunocytochemistry
Formalin-fixed paraffin-embedded liver sections as described above (see Histological analysis section) was immunostained using primary antibodies against Pten (Cell Signaling; cat # 9559, 1:200), CD4 (eBioscience; cat # 14-9766-82, 1:100), or CD8a (eBioscience; cat # 14-0808-82, 1:100). Proper secondary antibodies were chosen according to the host species of each primary antibody. Three fields were imaged from each mouse liver by an investigator blinded to the treatment information.
ELISA for SpCas9-specific antibody
Ninety-six-well Nunc MaxiSorp Plates (Thermo Scientific) were coated with the SpCas9 protein (0.5 μg/well, PNA Bio, cat # CP01) in 1× coating buffer diluted from Coating Solution Concentrate Kit (KPL) overnight at 4°C. The plates were washed with 1× wash buffer diluted from 20× Wash Solution (KPL) and blocked with 1% BSA Blocking Solution (KPL) for 1 hour at room temperature. Mouse sera were diluted 1000-fold with 1% BSA Diluent Solution (KPL), added to the wells, and incubated for 1 hr at room temperature with shaking (200 rpm). The mouse monoclonal antibody against SpCas9 (Epigentek; clone 7A9, cat # A-9000-100) was serially diluted and used as a standard to quantify IgG1. After washing, each well was incubated with 100 μl of HRP-labeled goat anti-mouse IgG1, IgG2a, or IgG2b (Santa Cruz; diluted 1:5000) for 1 hour at room temperature. The wells were washed four times and incubated with 100 μl of ABTS ELISA HRP Substrate (KPL). Optical density at 410 nm was measured using a Synergy HT microplate reader (BioTek). The IgG1 standard curve was generated using the four-parameter logistic regression equation with the Gen5 software (BioTek).
Cytokine release assay
Splenocytes were isolated from mice by mashing freshly harvested spleen and passing through a 70 μm cell strainer (BD). Red blood cells were lysed using Red Blood Cell Lysis Buffer (Roche Diagnostics). Cell number and viability were determined by Guava ViaCount assay (Guava Technologies). For the cytokine release assay, 200,000 live cells per well were seeded into a 96-well plate in 200 μl complete RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin (Life Technologies). Cells were incubated with or without the SpCas9 protein (5 μg/ml, PNA Bio) at 37°C/5% CO2. Three days later, culture medium was collected after centrifugation, and the levels of IL-1β, IL-2, IL-6, IL-10, IL-12 (p70), Ifn-γ, and Tnf-α were measured by the Bio-Plex Mouse Cytokine Assay (BioRad) according to the manufacturer's instructions, using a Bio-Plex MAGPIX Multiplex Reader (Luminex).
Statistical analysis
Comparison between two groups was analyzed by Student's t-test. Comparison among three groups was analyzed by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test to compare between two groups. All analysis was performed using Prism 6 software (GraphPad Software).
Results
Ad-mediated Pten gene editing in vitro
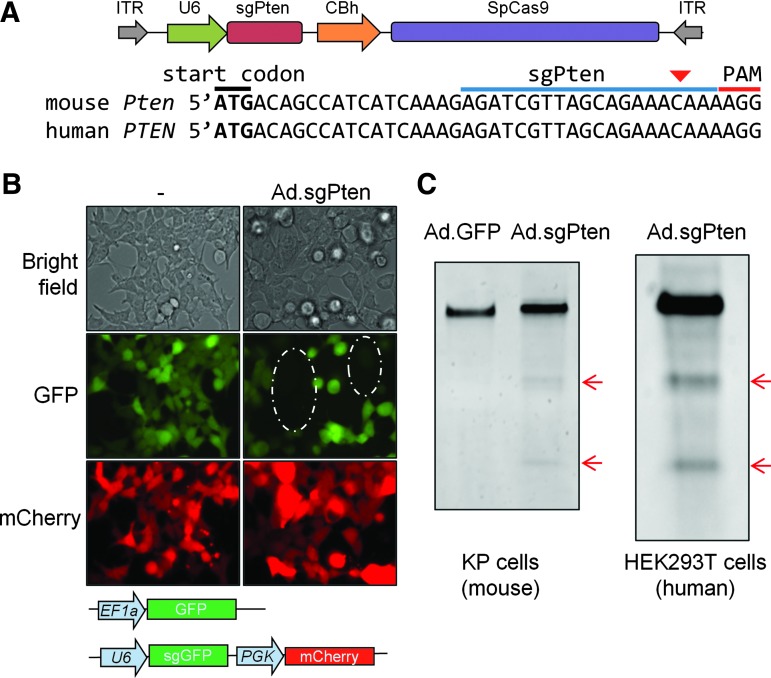
We packaged an E1/E3-deleted Ad vector to co-express SpCas9 and an sgRNA targeting the mouse Pten gene (Ad.sgPten hereafter) (Fig. 1A). To validate SpCas9 expression and function from this Ad vector, we used a gene-editing reporter cell line that stably expresses GFP, mCherry, and an sgRNA targeting the GFP gene (sgGFP). In the absence of Ad.sgPten infection, almost all cells expressed both GFP and mCherry (Fig. 1B). In contrast, Ad.sgPten infection generated a cell population showing only mCherry but not GFP, suggesting that SpCas9 expression from Ad.sgPten together with sgGFP reconstituted a functional CRISPR gene-editing system, and introduced disruptive insertion/deletion (indel) mutations in GFP as a result of error-prone nonhomologous end-joining repair (Fig. 1B). The sgPten target sequence is conserved between the mouse Pten and human PTEN genes (Fig. 1A), allowing us to test Pten gene editing using Ad.sgPten in both mouse (KP) and human (HEK293T) cell lines. In both cell lines, Ad.sgPten infection resulted in indel mutations in Pten as evidenced by Surveyor assay (Fig. 1C). These results validated the Pten gene-editing function of Ad.sgPten.
Figure 1.
Ad.sgPten functions in Pten gene editing in vitro. (A) Viral genome of Ad.sgPten. Also shown is the conserved target sequence of sgPten in mouse and human genomes. The positions of start codon, sgPten binding site, and protospacer adjacent motif (PAM) are labeled. Predicted SpCas9 cleavage site is indicated by red arrowhead. CBh, hybrid chicken beta-actin promoter; ITR, inverted terminal repeat. (B) Representative bright-field and fluorescence images of cells harboring a gene-editing reporter system with or without Ad.sgPten infection. Integrated expression cassettes of GFP, sgGFP, and mCherry are depicted. EF1a, elongation factor 1a promoter; PGK, phosphoglycerate kinase promoter. Circled areas indicate depletion of GFP in a subset of cells. (C) Surveyor assay to detect mutations within targeted genomic region following Ad.sgPten infection of KP cell and HEK293T cells. Ad.GFP infection serves as control. Red arrows indicate positive bands after mismatch-specific DNA nuclease digestion of PCR products.
In vivo Ad-mediated Pten gene editing models steatosis in mouse liver
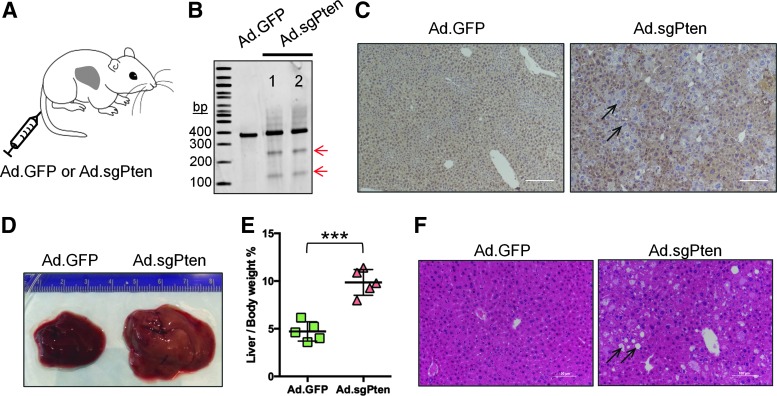
We then delivered Ad.sgPten into mouse liver to examine Pten gene editing efficiency and associated phenotypic outcome in vivo. Eight-week-old female FVB/NJ mice received 2.5×1010 particles/animal through tail vein injection. We injected Ad.GFP into another group of strain-, age-, and gender-matched mice as a control (Fig. 2A). Mice were sacrificed at different time points to examine gene editing efficiency and phenotypic consequences following Pten gene disruption in the liver. Two weeks postinfection (p.i.), Surveyor assay using liver genomic DNA revealed indel mutations (Fig. 2B). Deep sequencing confirmed the presence of indels surrounding the predicted SpCas9 cleavage site (Table 1). At one month after treatment, loss of Pten immunostaining in liver sections was readily observed (Fig. 2C), demonstrating successful Pten gene disruption by error-prone indel mutations. We have previously employed hydrodynamic injection to deliver plasmid harboring the same sgRNA and SpCas9 expression cassettes into strain-, age-, and gender-matched mice, and indel frequency in liver was estimated to be only about 3% two weeks postinjection.9 With Ad.sgPten, we achieved total indel frequencies of 14.8% and 22.8% in the two mice examined (Table 1 and Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hum), 5–8-fold greater than that obtained with hydrodynamic injection of plasmid.
Figure 2.
In vivo adenovirus-mediated Pten gene editing models steatosis in mouse liver. (A) Cartoon showing tail vein injection of Ad vector to an adult mouse. (B) Surveyor assay to detect Pten gene mutation in the liver two weeks after Ad vector infection. 1 and 2 indicate samples from two mice. Arrows indicate positive bands. (C) Immunohistochemistry of liver sections showing loss of Pten staining in a subset of cells (arrows) one month after Ad vector infection. The scale bars represent 100 μm, and 29±6% hepatocytes are Pten negative (n=3). (D) Representative photographs of whole mouse liver harvested four months after infection. A ruler with numbered unit in centimeter was included to gauge size. (E) Dot plot showing liver weight per body weight ratios of five mice per treatment group four months after infection. Each dot represents one mouse. Mean values and standard deviations are plotted. ***p<0.001 by t-test. (F) Representative images of H&E staining of liver sections showing steatosis (lipid accumulation) in Ad.sgPten-infected mice (arrows) four months after infection.
Table 1.
Deep sequencing revealed insertion/deletion (indel) mutations surrounding the predicted SpCas9 cleavage site in mouse Pten gene
| Mouse | Position in reference sequencea | Reference | Indelb | Reads supporting reference | Reads supporting indel | Indel % |
|---|---|---|---|---|---|---|
| #1 | 124 | A | */+C | 5888 | 372 | 5.87 |
| 121 | G | */−A | 6925 | 193 | 2.65 | |
| Indels <1% | 6.28 | |||||
| Total | 14.8 | |||||
| #2 | 124 | A | */+C | 5514 | 534 | 8.68 |
| 121 | G | */−A | 6670 | 206 | 2.91 | |
| 124 | A | */−C | 5514 | 77 | 1.25 | |
| 121 | G | */−AA | 6670 | 74 | 1.04 | |
| Indels <1% | 8.92 | |||||
| Total | 22.8 |
The predicted SpCas9 cleavage site in reference is between positions 125 and 126, or three nucleotides upstream of the PAM sequence (Fig. 1A).
Only indels of a frequency >1% are shown. See Supplementary Table S1 for all indels. Indels are denoted as the changes downstream of the reference nucleotide.
Importantly, mice receiving Ad.sgPten showed liver phenotypes recapitulating Pten genetic deletion. Four months p.i., pale liver appearance and dramatic hepatomegaly were observed in the mice infected with Ad.sgPten (Fig. 2D). The ratio of liver weight to body weight was increased from about 5% in Ad.GFP-infected mice to about 10% in Ad.sgPten-infected mice (Fig. 2E). H&E staining revealed massive liver steatosis (accumulation of lipid shown by arrows) in the mice infected by Ad.sgPten, but not in the mice infected by Ad.GFP (Fig. 2F). These data demonstrated that Ad-mediated Pten gene editing is a viable means to rapidly generate somatic mouse model of liver steatosis, and that Ad infection outperformed plasmid hydrodynamic injection in inducing liver Pten gene editing.
Ad.sgPten induced typical Ad vector-associated immune responses and liver damage
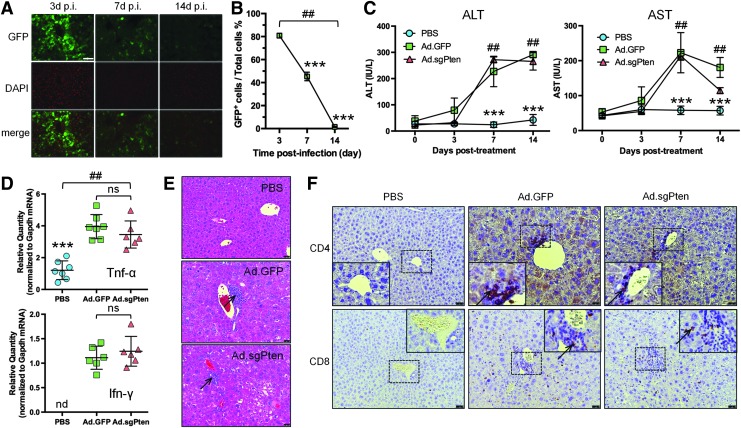
It is well known that immune responses associated with systemic delivery of Ad vectors can clear infected hepatocytes and induce transaminitis that indicates liver damage.15,24–26 Such a destructive process might have eliminated some hepatocytes that had undergone successful Pten gene editing, and thus complicated our study. To estimate the efficiency and kinetics of liver transduction over time under our experimental setting (including our specific mouse strain, gender, age, vector dose, and route of administration [see Materials and Methods for details]), we infected mice with an Ad vector expressing GFP (Ad.GFP), and counted GFP-positive cells in liver sections at 3, 7, and 14 days p.i. by fluorescence microscopy (Fig. 3A). This time-course experiment revealed an expectedly high but transient transduction rate of 80.8% of liver cells 3 days p.i., which rapidly diminished to 45.3% seven days p.i. and to 1.4% fourteen days p.i. (Fig. 3B). Correspondingly, serum liver transaminases ALT and AST were significantly elevated in Ad.GFP- and Ad.sgPten-infected groups at 7 and 14 days p.i. (Fig. 3C), indicating substantial acute liver damage.
Figure 3.
Ad vector-associated immune responses and liver damage. (A) Representative images of liver sections showing EGFP native fluorescence (green-colored), nuclear staining by DAPI (red-colored), and merged signals at 3, 7, and 14 days postinfection (p.i.). The scale bar represents 100 μm. (B) Quantification of percent of GFP-positive (GFP+) cells in liver at 3, 7, and 14 days postinfection. Three fields of each liver section from 4 to 5 mice per time point were analyzed. Data are plotted as mean and standard deviation. ##p<0.01 (among three time points) by one-way ANOVA. ***p<0.001 (versus 3d p.i.) by Tukey's test. (C) Serum levels of alanine transaminase (ALT) and aspartate transaminase (AST) at pretreatment and 3, 7, and 14 days posttreatment. Data are plotted as mean and standard deviation of 6–7 mice per treatment group in international unit per liter (IU/liter). ##p<0.01 (among three treatment groups of the same time point) by one-way ANOVA. ***p<0.001 (versus Ad.GFP or Ad.sgPten) by Tukey's test. (D) mRNA expression of Tnf-α (upper panel) and Ifn-γ (lower panel) normalized to Gapdh expression in mouse liver two weeks after treatment as determined by RT-qPCR. Each symbol represents one mouse. Mean values and standard deviations are plotted. ##p<0.01 (among three treatment groups) by one-way ANOVA. ***p<0.001 (versus Ad.GFP or Ad.sgPten). nd, not detectable; ns, not significant. (E) Representative H&E staining images of liver sections showing cell infiltration in Ad.GFP- and Ad.sgPten-infected mice 14 days p.i. (arrows). The scale bars represent 50 μm. (F) Representative images by immunohistochemistry showing positive staining of CD4 (upper panels) and CD8 (lower panels) in Ad.GFP- and Ad.sgPten-infected mice 14 days p.i. (arrows). Enlarged images of boxed areas are embedded. The scale bars represent 50 μm.
We sacrificed a group of mice at 14 days to further characterize the immunotoxicity in the liver. Compared with PBS treatment, both Ad.GFP and Ad.sgPten infections elicited similarly elevated expression of inflammatory cytokines including Tnf-α and Ifn-γ in the liver (Fig. 3D), indicative of innate and/or T helper type 1 (Th1)-related immune response. H&E staining of liver sections showed inflammatory cell infiltration following Ad.GFP and Ad.sgPten infection (Fig. 3E). Immunocytochemistry analysis identified CD4+ and CD8+ T-cells in the liver sections of both Ad.GFP and Ad.sgPten groups (Fig. 3F). Together, these data may represent well-studied immune responses caused by Ad vector-mediated gene transfer in the liver, which might negatively affect the sustainability of Ad.sgPten-mediated gene editing. However, it is possible that as Pten-edited cells lost the inhibitory effect of Pten on cell cycle and proliferation control, and rapidly proliferated through clonal expansion during liver damage and regeneration, the nonintegrating and nonreplicating Ad vector genome was gradually diluted and eventually lost during hepatocyte division, leaving the Pten-edited daughter cells spared from immune attack. Studies are under the way to define the nature (i.e., viral- and/or transgene-directed) and consequences (e.g., loss of transduced cells and vector genomes and/or promoter shutdown) of such immune responses.
Immune responses triggered by Ad vector infection in the liver consist of two branches, namely, the responses toward viral gene and transgene products, respectively. These immune responses can influence one another. Nonetheless, the robust immunogenic setting following Ad.sgPten infection provided us with a unique opportunity to study SpCas9-specific immunity in vivo.
Detection of SpCas9-specific immune responses
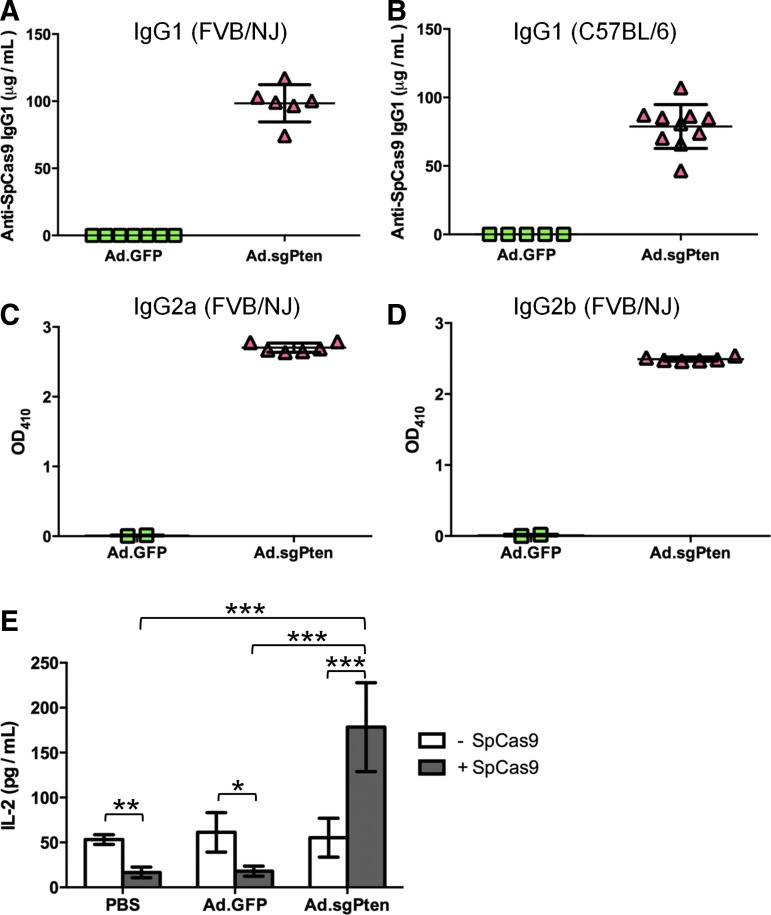
We further sought to examine the SpCas9-specific immune responses, considering that they may be relevant not only to disease modeling via somatic mutagenesis but also to in vivo therapeutic applications. We developed an ELISA assay to quantify the antibody against SpCas9 in the sera collected from Ad.GFP- and Ad.sgPten-infected mice. Fourteen days p.i., robust IgG1 antibody formation in all Ad.sgPten-infected mice was detected, whereas the mice infected with Ad.GFP showed nondetectable SpCas9-specific IgG1 (Fig. 4A). To rule out a mouse strain-specific effect, we infected age- and gender-matched C57BL/6 mice with the same dose of either Ad.GFP or Ad.sgPten, and detected SpCas9-specific IgG1 in only the Ad.sgPten group (Fig. 4B). In addition, we also detected SpCas9-specific IgG2a and IgG2b in Ad.sgPten-infected mice (Fig. 4C, D).
Figure 4.
SpCas9-specific immune responses following Ad.sgPten infection in mice 14 days p.i. Detection of anti-SpCas9 IgG1 levels in FVB/NJ mouse sera (A) and C57BL/6 mouse sera (B), anti-SpCas9 IgG2a (C), and IgG2b (D) levels in FVB/NJ mouse sera as determined by ELISA. Each symbol represents one mouse. Mean values and standard deviations are also plotted. (E) IL-2 induction in cultured splenocytes isolated from PBS-, Ad.GFP-, or Ad.sgPten-infected mice with (gray bars) or without (white bars) SpCas9 protein stimulation. Mean values from 6 to 7 mice per group are plotted with standard deviation. *p<0.05; **p<0.01; ***p<0.001.
To characterize the cellular response against SpCas9, we isolated and cultured splenocytes from PBS-, Ad.GFP-, and Ad.sgPten-treated mice, and these splenocytes were subjected to a cytokine release assay with or without SpCas9 protein stimulation. SpCas9 stimulation of splenocytes for three days in vitro increased the levels of a range of cytokines secreted in culture medium, including IL-1β, IL-6, IL-10, IL-12, Tnf-α, and Ifn-γ. However, the increase in these cytokines was not specific to splenocytes recovered from the Ad.sgPten-infected mice (Supplementary Fig. S1). Interestingly, IL-2 secretion from Ad.sgPten-primed splenocytes was significantly increased from 50 to 175 pg/ml upon SpCas9 stimulation, whereas the PBS and Ad.GFP-primed control splenocytes showed decreased IL-2 (Fig. 4E). Increased IL-2 secretion from Th1 cells could be associated with the development of CD4+ and/or CD8+ T-cell responses. Here we did not intend to define immune epitopes for B- and/or T-cell responses in SpCas9 and role of SpCas9-specific cytotoxic T lymphocyte (CTL) in the liver damage, which will be addressed in our follow-up study. Nonetheless, the data presented here indeed demonstrated that SpCas9, as a foreign bacterial protein, could induce humoral immunity.
Discussion
Ad vectors for human gene therapy have been hampered by their destructive immunotoxicity.27 However, they may be well-suited to deliver the CRISPR/Cas9 gene-editing system to model organisms such as mouse. First, the large cargo size of Ad vector allows for packaging the entire CRISPR/Cas9 system in one vector even with multiplexed targeting capacity. Second, Ad vector can transduce a broad range of cell types, including dividing and quiescent cells, in vivo and with high efficiency. Third, although Ad vector-mediated gene expression is transient, it is believed that only transient expression of CRISPR/Cas9 system is required to induce permanent genome editing. However, Ad vector-associated immune responses are capable of clearing transduced hepatocytes under many experimental conditions. For example, Cheng et al. delivered a CRISPR/Cas9-nickase Ad vector targeting Cebpa to mouse liver through tail vein injection, and they observed about 50% loss of both Ad vector genome and Cas9 expression in liver 7 days p.i., and nearly complete loss 3 months p.i., suggesting efficient clearance of Ad vector-transduced liver cells.14 This and another proof-of-concept study13 employing an Ad vector for CRISPR/Cas9 genome editing in mouse liver showed short-term phenotypic outcome up to 7 days p.i., meanwhile raising the question of whether the gene-editing effects would be preserved over a longer period of time after clearance of initially transduced liver cells. This is important for disease modeling since a stable phenotype may be required to study disease mechanism or to evaluate therapeutic intervention.
Here, we demonstrated that Pten gene editing in mouse liver by Ad vector-mediated CRISPR/Cas9 somatic gene editing could induce steatosis at 4 months p.i., although typical Ad vector-associated immune responses and liver damage were readily detected at 14 days p.i. and earlier. Our results support using Ad vectors to deliver a genome-editing system in mouse liver for long-term human disease modeling, turning the shortcoming of transient in vivo transduction by Ad vector into an advantage. However, we cannot rule out the possibility that silencing Pten, a known tumor suppressor gene, might provide edited liver cells with a growth advantage during liver damage and regeneration, which might contribute to shaping the liver phenotype at 4 months p.i. In addition, using the same experimental setting as the previous report utilizing hydrodynamic injection to deliver Cas9 and sgPten, we achieved up to 22.8% Pten gene editing efficiency with Ad.sgPten compared with only 3% with hydrodynamic injection 14 days posttreatment,9 further supporting the advantage of Ad vectors in inducing gene editing in mouse liver.
In recent nonhuman primate studies and gene therapy clinical trials using rAAV vector as a gene delivery platform, investigators found that various host immune responses directed against the transgene product or rAAV capsid can compromise therapeutic efficacy.28–30 As CRISPR/Cas9 genome editing is an appealing approach to directly repairing disease-related gene mutations in situ,10,31 characterizing Cas9-specific host immune responses following gene delivery, and the development of needed precautions or workarounds will be necessary features in future translational studies. We demonstrated that systemic Ad.sgPten delivery in mice elicited SpCas9-specific immune responses, including the generation of antibodies against SpCas9 as well as increased IL-2 secretion from Ad-sgPten-primed splenocytes upon SpCas9 protein stimulation. Notably, although our current study provided no direct evidence of a SpCas9-specific CTL response, we are in the process of constructing an SpCas9 short peptide library and defining immune epitopes for further study of SpCas9-specific immunity. It should be noted that the SpCas9-specific immune responses characterized in this study were generated following systemic Ad vector delivery. The strong immune adjuvant effect of Ad vector infection32 and the steatohepatitis induced by Pten genome editing may contribute to or escalate the formation of adaptive immune responses against SpCas9. Therefore, SpCas9-specfic immune responses should be further studied during the use of other in vivo gene delivery platforms and in other species. Furthermore, a proper delivery method should be carefully chosen and optimized to avoid any negative impact of such immune responses.
To minimize off-target effect, transient in vivo delivery of Cas9 (e.g., by direct delivery of protein) was attempted.16 Provided that Cas9-specific immunotoxicity could negatively affect therapeutic efficacy, a short-term immune suppression may be sufficient to ensure gene editing during the transient presence of Cas9. However, we envision several cases where Cas9 expression is sustained. For example, rAAV-delivered Cas9 expression in vivo has the potential to last for years and even an entire lifetime in recipients. In addition, the catalytically dead Cas9 (dCas9) fused with effector protein domains has been shown to either activate or silence specific gene expression at the transcriptional level, representing a versatile approach to regulating gene expression.33–35 This usually requires the continuing presence of dCas9 effector to exert a stable gene regulatory effect. Potential immune responses against Cas9, such as a cytotoxic T-cell response attacking transduced and edited cells marked by an MHC I-Cas9 epitope, can limit therapeutic efficacy. Under these circumstances, developing novel methods of controlling Cas9 expression in vivo will be valuable.16,36–38 In addition, by using proper effector domains to fuse with dCas9, permanent transcriptional regulation through epigenetic modulation may be possible.
In summary, we successfully employed an Ad vector to deliver a CRISPR/Cas9-mediated Pten gene-editing system to mouse liver, and achieved a liver phenotype consistent with genetic Pten deficiency at 4 months p.i., thus supporting the use of Ad vector-mediated genome editing for somatic disease modeling in mice. The SpCas9-specific immune responses detected following Ad.sgPten infection in mice warrant further investigation into the nature of the immunogenicity, the optimal method of Cas9 delivery and expression, and its potential impact on gene editing efficiency in future translational studies.
Supplementary Material
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 2.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nature medicine 2015;21:121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mou H, Kennedy Z, Anderson DG, et al. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med 2015;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 2015;15:387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014;516:428–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014;514:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Park A, Mou H, et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol 2015;16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther 1999;6:1258–1266 [DOI] [PubMed] [Google Scholar]
- 13.Ding Q, Strong A, Patel KM, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res 2014;115:488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng R, Peng J, Yan Y, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett 2014;588:3954–3958 [DOI] [PubMed] [Google Scholar]
- 15.Schagen FH, Ossevoort M, Toes RE, Hoeben RC. Immune responses against adenoviral vectors and their transgene products: a review of strategies for evasion. Crit Rev Oncol Hematol 2004;50:51–70 [DOI] [PubMed] [Google Scholar]
- 16.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 2014;33:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014;24:1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2012;13:283–296 [DOI] [PubMed] [Google Scholar]
- 20.Horie Y, Suzuki A, Kataoka E, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest 2004;113:1774–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiles B, Wang Y, Stahl A, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected]. Proc Natl Acad Sci USA 2004;101:2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014;516:423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 2009;4:1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Zajac AJ, McPherson SA, et al. Primary adenovirus-specific cytotoxic T lymphocyte response occurs after viral clearance and liver enzyme elevation. Gene Ther 2005;12:1079–1088 [DOI] [PubMed] [Google Scholar]
- 25.Michou AI, Santoro L, Christ M, et al. Adenovirus-mediated gene transfer: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther 1997;4:473–482 [DOI] [PubMed] [Google Scholar]
- 26.Tao N, Gao GP, Parr M, et al. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol Ther 2001;3:28–35 [DOI] [PubMed] [Google Scholar]
- 27.Lehrman S. Virus treatment questioned after gene therapy death. Nature 1999;401:517–518 [DOI] [PubMed] [Google Scholar]
- 28.Nathwani AC, Tuddenham EG, Rangarajan S, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 2011;365:2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mingozzi F, Meulenberg JJ, Hui DJ, et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood 2009;114:2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson PR, Schnepp BC, Zhang J, et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat Med 2009;15:901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin H, Kanasty RL, Eltoukhy AA, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet 2014;15:541–555 [DOI] [PubMed] [Google Scholar]
- 32.Geutskens SB, van der Eb MM, Plomp AC, et al. Recombinant adenoviral vectors have adjuvant activity and stimulate T cell responses against tumor cells. Gene Ther 2000;7:1410–1416 [DOI] [PubMed] [Google Scholar]
- 33.Agne M, Blank I, Emhardt AJ, et al. Modularized CRISPR/dCas9 effector toolkit for target-specific gene regulation. ACS Synth Biol 2014;3:986–989 [DOI] [PubMed] [Google Scholar]
- 34.Maeder ML, Linder SJ, Cascio VM, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013;10:977–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014;159:647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 2015;11:198–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis KM, Pattanayak V, Thompson DB, et al. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 2015;11:316–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dow LE, Fisher J, O'Rourke KP, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 2015;33:390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.