Highlights
-
•
Noroviruses are now recognized as the most common cause of viral gastroenteritis.
-
•
The 5′ and 3′ ends of caliciviruses genome fold into characteristic structures conserved within the family.
-
•
The tirmini of calicivirus genome is involved in recruiting host factors to the replication complex.
-
•
The 5′ and 3′ ends of the MNV genome have been shown to interact with host proteins and further stabilize this interaction.
Keywords: Caliciviruses, RNA secondary structures, RNA–protein interactions, Norovirus
Abstract
The Caliciviridae family of small positive sense RNA viruses contains a diverse range of pathogens of both man and animals. The molecular mechanisms of calicivirus genome replication and translation have not been as widely studied as many other RNA viruses. With the relatively recent development of robust cell culture and reverse genetics systems for several members of the Caliciviridae family, a more in-depth analysis of the finer detail of the viral life cycle has now been obtained. As a result, the identification and characterization of the role of RNA structures in the calicivirus life cycle has also been possible. This review aims to summarize the current state of knowledge with respect to the role of RNA structures at the termini of calicivirus genomes.
1. Introduction
The Caliciviridae family of small positive-strand RNA viruses contains pathogens of various animals including humans, dolphins, reptiles, sheep, dogs, cattle, chickens and amphibians. The family is currently divided into 5 genera; Vesivirus, Lagovirus, Norovirus, Sapovirus and Nebovirus. A number of other genera have been proposed but have yet to be approved. The most widely studied genera within the Caliciviridae are the Norovirus genus as noroviruses are now recognized as the most common cause of viral gastroenteritis (Glass et al., 2009). Our knowledge of the calicivirus life cycle lags behind that of the majority of other RNA viruses due to the inability of many members of the family to replicate efficiently in immortalized cells. Despite this, there have been numerous advances in the study of calicivirus biology over the past 10 years; these include the recent development of a mouse model for human norovirus infection (Taube et al., 2013), the use of Tulane virus as a model for calicivirus pathogenesis (Farkas et al., 2008, Wei et al., 2008) and the recent observation that human noroviruses posses a potential tropism for B-cells (Jones et al., 2014). The majority of our knowledge of the intracellular life of caliciviruses has been established using animal virus that replicate efficiently in cell culture, including (but not limited to), feline calicivirus (FCV) from the Vesivirus genus, murine norovirus (MNV) from the Norovirus genus and porcine sapovirus (PSaV) from the Sapovirus genus. Recent review articles have summarized the current state of knowledge on the norovirus life cycle (Thorne and Goodfellow, 2014), norovirus immune responses (Karst et al., 2014a), and the pathological mechanism underlying norovirus infection (Karst et al., 2014a, Karst et al., 2014b, Thorne and Goodfellow, 2014). This review aims to summarize the current state of knowledge on the role of RNA structures at the 5′ and 3′ extremities of the calicivirus genomes and the role they play in the viral life cycle.
The genomes of caliciviruses share a highly conserved overall structure (Fig. 1 ); typically ∼7.3–7.7 kb in length, polyadenylated at the 3′ end and covalently linked to a small virus-encoded protein (VPg) at the 5′ end. A shorter than genome length, subgenomic RNA (sgRNA) is also produced during replication, which is 3′ co-terminal to the genomic RNA. Calicivirus genomes typically contain three open reading frames; ORF1 encodes a large polyprotein that is post and co-transnationally cleaved by the virus-encoded protease (3C or NS6), producing the non-structural proteins. ORF2 encodes the major capsid protein VP1 and ORF3 encodes the minor capsid protein VP2. A detailed description of the function of the viral proteins is beyond the scope of this review but has recently been described (Thorne and Goodfellow, 2014). While most caliciviruses have three ORFs, the main exception to this is MNV that encodes a fourth overlapping ORF, which produces a newly identified protein virulence factor 1 (VF1) within the VP1 coding region (McFadden et al., 2011, Thackray et al., 2007). In addition, in the case of rabbit hemorrhagic disease virus (RHDV) and some sapoviruses, the capsid-coding region is in frame with ORF1 providing two mechanisms for the synthesis of VP1, i.e. as part of the large polyprotein from ORF1 and as a single protein from the sgRNA (Fig. 1) (Wirblich et al., 1996).
Fig. 1.
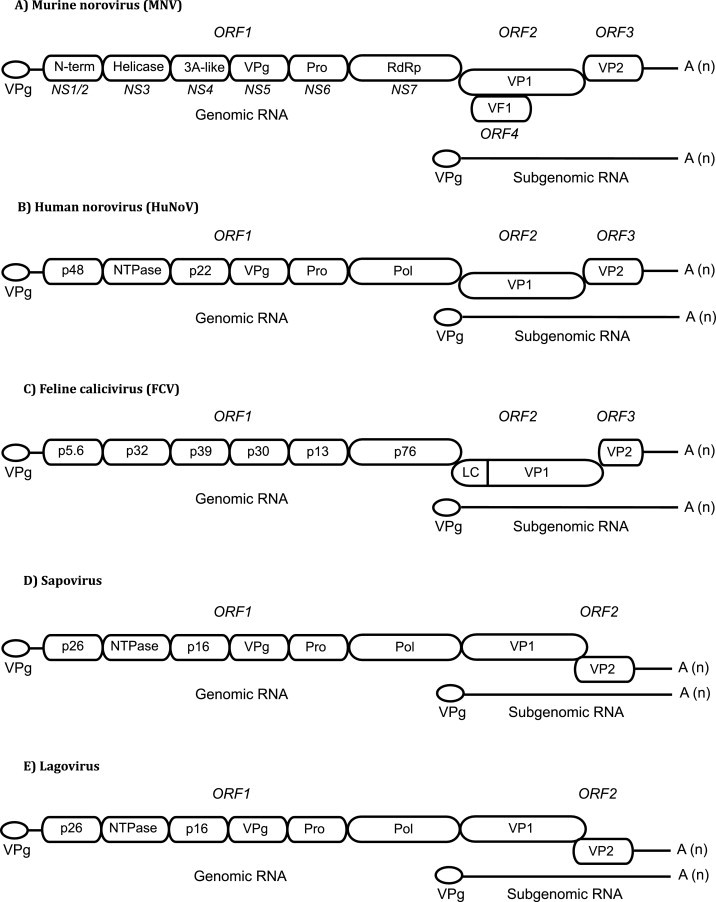
Calicivirus genome organization. The full-length genome is organized into 2–4 ORFs. A subgenomic RNA (sgRNA) consisting of ORF2 and ORF3 is also produced during replication. The viral protein, VPg, is covalently attached to the 5′ end of genomic and sgRNAs and the 3′ end is polyadenylated. ORF1 encodes a polyprotein which is post-translationally cleaved into non-structural proteins, whereas ORF2 and ORF3 encode the structural proteins VP1 and VP2, respectively. The genome layout of (A) MNV, (B) HuNoV, (C) FCV, (D) sapovirus, and (E) lagovirus are shown. The genome of MNV encodes a unique virulence factor (VF1) translated from a recently discovered ORF4. The FCV genome encodes for a leader capsid peptide (LC) at the 5′ end of VP1.
Calicivirus genomes are not capped but instead have the VPg covalently linked to the 5′ end of the viral RNA (Rowlands et al., 1978). The calicivirus VPg protein is typically 13–15 kDa in size, much larger than the ∼22 amino acid VPg seen in picornaviruses and it is essential for the translation of viral RNA (Herbert et al., 1997). The viral genome is translated via the direct interaction of the calicivirus VPg protein with the canonical translation initiation factors (Goodfellow, 2011). The initiation factors involved in calicivirus VPg-dependent translation vary between caliciviruses, but interactions have been described with eIF3 (Daughenbaugh et al., 2003), eIF4E (Chaudhry et al., 2006, Goodfellow et al., 2005) and more recently eIF4G (Chung et al., 2014). As described in more detail below, it is highly likely that the cellular proteins interact with the 5′ and 3′ extremities of the calicivirus genomes also contribute to the regulation of viral genome translation.
2. The identification of RNA secondary structures in calicivirus genomes
Similar to other positive-strand RNA viruses, the 5′ and 3′ extremities of calicivirus genomes contain highly conserved RNA secondary structures that play many roles during viral life cycle, including viral RNA replication, translation, and encapsidation (Bailey et al., 2010, Simmonds et al., 2008, Vashist et al., 2012). In the majority of RNA viruses, these cis-acting RNA elements are typically found within the untranslated regions (UTRs) of the viral genome (Liu et al., 2009). However, in some cases where the UTRs are short, these structures can extend into the neighboring coding sequences. Caliciviruses contain short UTRs at both ends of their genomic and sgRNAs. For example, the 5′ UTRs of the MNV genome are only 5 nucleotides (ntds), while the 3′ UTR is 78 ntds (Karst et al., 2003). The presence of RNA secondary structures in the genomes of caliciviruses was first noted in the Norwalk virus (NV) genome (Jiang et al., 1993), where a double stem-loop at the 5′ end of the NV genome was described. However, the biological function of this stem-loop has never been described. The presence of hairpin stem-loop structures at the 3′ end of animal caliciviruses, including RHDV and FCV was also noted in 1994 (Seal et al., 1994).
More recently, the availability of full-length calicivirus genome sequences has enabled an unbiased approach to be used to identify and characterize RNA structures within calicivirus genomes. Computational analysis has been used to identify regions of within caliciviruses that demonstrate significant synonymous codon suppression and/or regions of extensive RNA secondary structure that is evolutionarily conserved between diverse isolates. This analysis has shown that the 5′ and 3′ extremities of the genomic and subgenomic viral RNAs contain the majority of conserved RNA structures, although structures are also present within other regions of the genome of some caliciviruses (Simmonds et al., 2008, Vashist et al., 2012). In almost all cases, the suppression of synonymous codon usage extends 200–250 bases into the coding regions at the 5′ and 3′ ends of the viral genome. To date, biochemical evidence of the existence of RNA structures in these regions is limited but some data exists for FCV and MNV (Karakasiliotis et al., 2010, Vashist et al., 2012).
3. The 5′ ends of the genomic and subgenomic RNAs
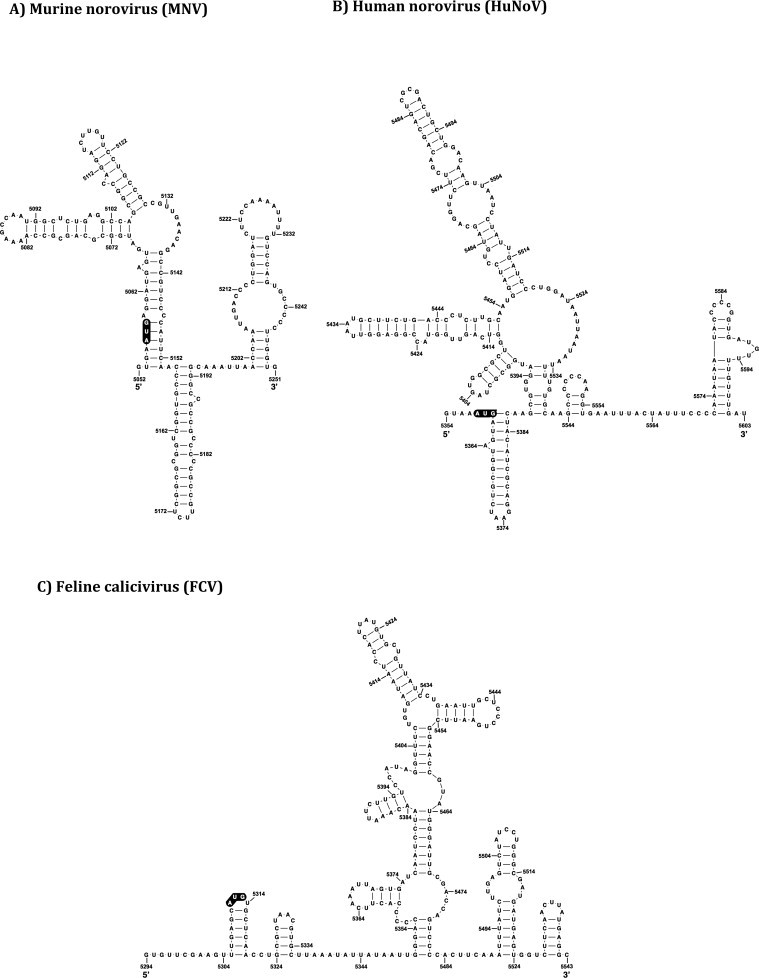
As described above, calicivirus genomes invariably have short UTRs at the 5′ ends of the genomic and sgRNA. Therefore, in all cases, evolutionarily conserved RNA secondary structures at the 5′ ends of the viral genome extend into the flanking coding regions; ORF1 in the case of the viral genomic RNA and ORF2 in the case of the sgRNA (Simmonds et al., 2008). Fig. 2 illustrates the predicted secondary structures that are found at the 5′ end of a number of representative calicivirus genomes. There is limited information with respect to the role that these RNA structures play in the viral life cycle, however mutational analysis of MNV has revealed that the stem-loop structures located at the 5′ end of the genome are important for viral replication; synonymous mutations that destabilized the stem-loop structures starting at positions 8 and 29 (referred as SL8 and SL29) caused a significant decrease in virus titer and in the case of SL29, a small plaque phenotype (Simmonds et al., 2008).
Fig. 2.

Calicivirus 5′ genomic RNA structure. The RNA secondary structures of various caliciviruses, predicted using mfold (Zuker, 2003), and biochemically confirmed for MNV and FCV (Karakasiliotis et al., 2010, Vashist et al., 2012), are shown. The ORF1 initiation codon for each virus is shown in black. The predicted RNA secondary structure are (A) (MNV, GenBank accession number DQ285629), (B) (HuNoVs, GenBank accession number NC_001959), and (C) (FCV, GenBank accession number L40021) 5′ ends. Four potential PTB binding sites (BS1-4) in the FCV genome are highlighted in gray. Note that these are potential binding sites, and only BS2-3 was identified to bind to PTB.
The 5′ end of the calicivirus genomic RNA is known to interact with host cellular proteins, and in a limited number of cases these interactions have also been shown to play important roles in the virus life cycle (Gutiérrez-Escolano et al., 2000, Karakasiliotis et al., 2010, Simmonds et al., 2008, Vashist et al., 2012). These studies have unfortunately been limited in nature and have focused almost exclusively on three caliciviruses, namely MNV (strain MNV-1 CW1), NV (strain NC1959) and FCV (strain Urbana). Table 1 summarizes the host factors that are known to interact with the 5′ and 3′ extremities of MNV, NV, and FCV genomes.
Table 1.
Host factors that interact with the 5′ and 3′ extremities of caliciviruses genome.
| Region | Virus | Host factors | References |
|---|---|---|---|
| 5′ | MNVa | La, PCBP1, PCBP2, hnRNP A1, Nucleolin, DDX3 | López-Manríquez et al. (2013) and Vashist et al. (2012) |
| NV | La, PCBP2, PTB, hnRNPL | Gutiérrez-Escolano et al. (2000) | |
| FCV | PTB | Karakasiliotis et al. (2006) | |
| 3′ | MNVa | La, PCBP1, PCBP2, hnRNP A1, DDX3, YBX-1 | Vashist et al. (2012) |
| NV | La, PABP, PTB, Nucleolin | Cancio-Lonches et al. (2011) and Gutiérrez-Escolano et al. (2003) | |
| FCV | Nucleolin | Cancio-Lonches et al. (2011) | |
Numerous other proteins were identified binding to the 5′ and 3′ of MNV RNA (Vashist et al., 2012), but only those which are functionally important are shown in the table.
Mobility shift and cross-linking assays were used to identify cellular proteins that interact with RNA structures at the 5′ end of the NV genome (Gutiérrez-Escolano et al., 2000). The cellular proteins lupus autoantigen (La), poly(rC)-binding protein (PCBP-2), polypirimidine tract-binding protein (PTB) and heterogeneous nuclear ribonucleoprotein L (hnRNPL), were found to be components of an ribonucleoprotein (RNP) complex formed on the 5′ end of the NV genome, although the function of these interactions has yet to be determined (Gutiérrez-Escolano et al., 2000). La, PCBP-2 and PTB have all previously been implicated in the life cycles of many other RNA viruses (Blyn et al., 1997, Kim and Jang, 1999, Svitkin et al., 1994, Witherell et al., 1993). In the case of picornaviruses, they are thought to stimulate internal ribosome entry site (IRES)-mediated translation. The precise mechanism by which they stimulate IRES-mediated translation is unknown but it has been suggested they function as RNA chaperones to promote the correct folding of the RNA structure facilitating the recruitment of translation initiation factors (Li and Nagy, 2011). As highlighted above, calicivirus translation relies on the interaction of cellular translation initiation factors with the VPg protein covalently linked to the 5′ end of the viral genome. Given the proximity of VPg to the RNA structures at the 5′ ends, it is possible that the direct recruitment of cellular RNA-binding proteins also contribute to stimulate or regulate viral VPg-mediated translation. Evidence for this exists in FCV and is described in more detail below.
More recently, unbiased approaches that use RNA affinity chromatography combined with mass spectrometry have been used to identify host cellular proteins that interact specifically with the RNA structures at both the 5′ and 3′ termini of the MNV genome (Vashist et al., 2012). Using this approach, a total of 99 cellular proteins were identified as interacting with the 5′ end of the MNV genomic RNA (ntds 1–250) and 33 cellular proteins interacting with the 3′ end of the MNV genomic RNA (ntds 7141–7397). As noted above, the RNA structures within these regions are primarily in the coding regions of ORF1 and ORF3. These regions were selected as baits due to the fact that they contain the majority of the RNA structures that are evolutionarily conserved between MNV isolates and are therefore likely to contain the major cis-acting replication signals. The roles of a small number of the identified proteins were also studied using RNA interference to reduce the intracellular levels of three identified host factors, namely La, PTB and the cellular RNA helicase DDX3. siRNA-mediated knockdown of the three host factors significantly reduced MNV replication in immortalized cells, suggesting they contribute to some aspect of the norovirus life cycle (Vashist et al., 2012). Clearly further detailed studies will be required to analyze the precise roles of the identified factors in the norovirus life cycle but given their reported roles in the translation of other viral RNAs (Blyn et al., 1997, De Nova-Ocampo et al., 2002, Hahm et al., 1998) it is tempting to speculate that they may also function as translational enhancers.
The function of PTB in the FCV life cycle has been characterized in much more detail. PTB is considered one of the most well characterized cellular RNA-binding proteins as it plays a role in the life cycle of many RNA viruses including coronaviruses (Sola et al., 2011), picornaviruses (Chang et al., 1993, Lin et al., 2009a) and flaviviruses (Agis-Juárez et al., 2009, De Nova-Ocampo et al., 2002, Roby et al., 2014). PTB interacts with the 5′ ends of the FCV genomic and sgRNAs, binding to at least two sites at the 5′ end of the genomic RNA (Fig. 2) (Karakasiliotis et al., 2010). Based on the observation that recombinant PTB was able to specifically suppress FCV VPg-dependent translation in vitro, it was hypothesized that PTB functioned as a negative regulator of FCV translation (Karakasiliotis et al., 2006, Karakasiliotis et al., 2010). RNAi-mediated reduction in PTB levels resulted in an increase in the translation of replication incompetent VPg-linked FCV RNA but lead to a decrease in overall virus production after infection. PTB was also seen to redistribute from a primarily nuclear localization to the cytoplasm during the course of FCV replication (Karakasiliotis et al., 2010). Based on these data, it was proposed that PTB functions to regulate whether FCV RNA is translated or replicated. All RNA viruses must regulate their mechanisms of translation and replication, as the two processes cannot occur on the same viral RNA template at the same time due to their opposing polarities, i.e. the ribosome moves in a 5′–3′ direction along the template whereas the viral RNA-dependent RNA polymerase moves in a 3′–5′ direction. The regulation of this process has been most notably characterized in poliovirus where it is thought that it is primarily mediated by the cleavage of PCBP2, which occurs as a result of the accumulation of the viral 3CD protease (Perera et al., 2007). Some reports also describe that other translation initiation factors are cleaved during poliovirus replication and may also contribute to the regulation of viral translation (Etchison et al., 1982, Gamarnik and Andino, 1998, Lloyd, 2006). Based on the data obtained with FCV, a model has been proposed where a gradual, but specific, recruitment of PTB from the nucleus to the cytoplasmic viral replication complex leads to the repression of viral translation in the later stages of the viral life cycle. The net effect is to clear viral RNA of translating ribosomes, contributing to the stimulation of viral RNA replication or enabling the packaging of viral RNA into viral capsids (Karakasiliotis et al., 2010). The mechanisms involved in the control of PTB relocation are not as yet known, however previous reports have suggested that phosphorylation of key residues within PTB may affect its cellular localization (Ma et al., 2007, Xie et al., 2003).
The 5′ end of the calicivirus sgRNA also contains regions of extensive RNA secondary structures (Fig. 3 ), which based on the observation they are conserved between isolates, would suggest they play a role in the viral life cycle. Unfortunately, very limited information is available with respect to the interaction of these RNA structures with viral or cellular proteins. A limited mutagenesis study has been used to study the potential role of cis-acting elements in the 5′ end of the sgRNA of NV (Chang et al., 2008). Data suggests that the sequence of the first base, at the start of the sgRNA, invariably a G in all caliciviruses, is important for virus replication, as mutating this to A, U, and C resulted in no replication of a GFP-containing Norwalk virus replicon (Chang et al., 2008). RNA structures at the 5′ end of the sgRNA of FCV were found to interact directly with PTB (Karakasiliotis et al., 2006). As noted above, PTB has been shown to function as a negative regulator of FCV VPg-dependent translation of both the genomic and sgRNAs. The identification of other cellular and viral factors that interact with the 5′ ends of the calicivirus sgRNA will require further study but may be greatly facilitated by the recent description of potential cell culture (Jones et al., 2014) and reverse genetics systems for human norovirus (Katayama et al., 2014).
Fig. 3.
Predicted 5′ RNA secondary structures of calicivirus sgRNA. Schematic illustration of the predicted RNA secondary structures of the 5′ sgRNA of (A) MNV, (B) HuNoV, and (C) FCV predicted by mfold (Zuker, 2003). The initiation codon of ORF2 is highlighted in black.
4. The 3′ extremity
Unlike the very short 5′ ends of calicivirus genomes, the 3′ extremities typically contain un-translated regions upstream of the poly A tail that vary in size from 46 to 78 ntds in length. Computational analysis of all the available calicivirus genomes confirmed the presence of RNA structures at the 3′ ends of the viral genome (Simmonds et al., 2008). A terminal stem-loop (SL) was predicted to exist in the 3′ UTR of all caliciviruses, although the size and sequence composition varies greatly. In addition, bioinformatic approaches have invariably identified extensive regions of RNA secondary structure with the VP2 coding region, which were also confirmed using biochemical probing (Bailey et al., 2010, Simmonds et al., 2008). Mutational analyses of these regions in the FCV and human norovirus genomes have shown that they are essential for virus replication. In the case of HuNoV, a number of deletion mutants were introduced at the VP2 and 3′ UTR, and it was found that the 66 bases of 3′ UTR as well as the last part of the RNA nucleotides that encode ORF3 are important for viral replication (Chang et al., 2008). This also appears to be true for FCV; the ORF3 coding region contains a cis-acting RNA sequence that is essential for viral replication in cell culture (Sosnovtsev et al., 2005). The function of this RNA has as yet to be determined.
The terminal 3′ stem-loop structure of NV has been shown to interact with at least 10 host proteins using a UV crosslinking assay (Gutiérrez-Escolano et al., 2003). Subsequent immunoprecipitations identified La protein, PTB and PABP as components of the RNP complex on the 3′ end of the NV RNA. Furthermore, the 3′ UTRs of NV and FCV were found to interact with the host factor nucleolin and the potential binding sites have been predicted (Fig. 4 ) (Cancio-Lonches et al., 2011). Nucleolin was also found to interact with recombinant forms of the viral protease-RNA-dependent RNA polymerase (Pro-Pol, NS6/7) precursor protein of both NV and FCV. This interaction was confirmed by co-immunoprecipitation and co-localization studies in FCV infected cells, indicating that nucleolin and the viral replicase components interact. RNAi-mediated reduction in cellular nucleolin levels slowed FCV replication and the appearance of cytopathic effect (Cancio-Lonches et al., 2011). While these data confirm a potential role for nucleolin in the viral life cycle, the precise role remains unknown, however given the direct interaction with the viral RNA-dependent RNA polymerase containing precursor protein, it is likely to contribute directly to RNA synthesis. Due to a lack of an efficient culture system for human noroviruses, the functional role for nucleolin in the human norovirus life cycle is as yet undetermined and will require additional study. However, it has been shown that nucleolin binds to other positive-strand RNA viruses, and plays essential roles during their viral life cycle. Nucleolin was found to bind to the 3′ UTR of tombusvirus RNA, and by doing so reduces viral replication during the early stages of replication (Jiang et al., 2010). In addition, nucleolin interacts directly with the nonstructural protein NS5B of hepatitis C virus, and this interaction results in inhibition of RNA-dependent RNA polymerase activity in vitro (Hirano et al., 2003). More recently, it has been suggested that the dengue capsid protein (C) interacts with nucleolin, and that blocking this interaction with an aptamer or by reducing intracellular nucleolin levels by siRNAs, reduced the production of infectious virus, without any obvious effect on viral replication (Balinsky et al., 2013).
Fig. 4.
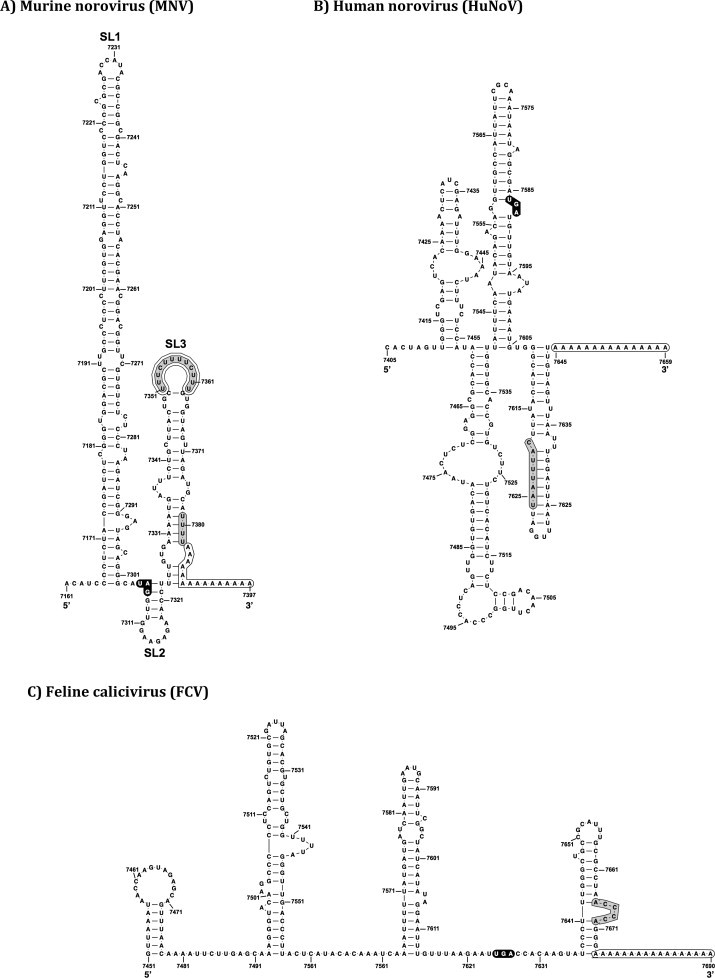
RNA structure at the calicivirus 3′ extremity. Schematic representation of the mfold predicted (Zuker, 2003) and biochemically confirmed RNA secondary structures (Bailey et al., 2010) at the 3′ end of calicivirus genomes. The stop codon of the ORF3 is shown in black circles, while the poly A tail is highlighted for each virus at the end of the RNA sequence of the genome. (A) MNV contains three predicted stem-loop structures (SL1-3). The variable p(Y) tract in SL3 is highlighted in gray, and the nucleotides (UUUU) at position 7380 are highlighted in black prior to the poly A tail. In (B) HuNoV and (C) FCV the predicted binding site of the host factor nucleolin is highlighted in gray.
The role of 3′ end RNA structures in the norovirus life cycle has been characterized using the MNV model system. The MNV 3′ end contains three predicted stem-loop structures SL1-3 (Fig. 4). SL1 is the largest, typically 138 ntds, and is within the VP2 coding region. SL2 is the smallest, 18 ntds and largely non-coding whereas SL3 is entirely non-coding. SL2 contains a single-stranded GA rich sequence whereas SL3 contains a polypyrimidine p(Y) tract (Bailey et al., 2010). Complete deletion of the GA-rich tract of SL2 or deletion of SL3 or both SL2 and 3 rendered the cDNA clones of MNV non-infectious, confirming that the integrity of the entire 3′ UTR is critical for function. Mutations that disrupt the terminal stem-loop structure, shown as SL3 in Fig. 4, are lethal in the context of an infectious cDNA clone (Simmonds et al., 2008). Furthermore, the sequence of this stem-loop was also shown to be important as the introduction of compensatory mutations that repaired the base-pairing of the stem-loop structure were also debilitating for virus replication. This observation fits with the previous discovery that the last nucleotide in the 3′ UTR prior to the poly A tail (U7380) is essential for MNV viability as changing this to C prevented recovery of infectious virus from cDNA (Chaudhry et al., 2007). Further studies suggested that the highly conserved p(Y) tract within loop region of the terminal stem loop (Fig. 4) contributed to MNV virulence in the natural host (Bailey et al., 2010). The single-stranded p(Y) tract in SL3 is present in all MNV isolates, but varies in length between isolates from 9 to 18 (Thackray et al., 2007). The cellular RNA-binding proteins PTB and PCBP2 interact with the p(Y) tract however replacement of the p(Y) tract with a GNRA tetraloop (GUAA) or a tetra-A sequence (AAAA) had no effect on viral replication in immortalized murine macrophage cells (Bailey et al., 2010). Importantly however viral fitness in cell culture was compromised; in mixed infections where wild-type virus and a virus containing a GNRA tetraloop in place of the p(Y) tract (SL3 GNRA), were mixed at 1:1 or 1:10 ratio, the wild type virus dominated after just a few passages in cell culture. Infection of mice deficient in STAT1 with MNV-1 typically results in systemic, lethal infections whereby mice will typically succumb to infection within 5–7 days. Infection results in significant clinical signs including weight loss, piloerection, diarrhea and eye discharge concomitant with high levels of infectious virus present in all tissues and shed into feces. In mice infected with the MNV mutant SL3 GNRA a delayed onset of clinical signs were observed, however all mice eventually succumbed to infection, identifying the first RNA-structure based virulence determinant for any calicivirus. Importantly, these data add to the well-established body of literature indicating a critical role for RNA–protein interactions in viral pathogenesis.
5. Genome circularisation
A significant body of evidence has shown that the 5′ and the 3′ ends of positive-strand RNA virus genomes interact with each other to coordinate essential functions of virus replication (Gamarnik and Andino, 1998, Alvarez et al., 2008, Liu et al., 2009). These interactions are often mediated by long-range RNA–RNA interaction between the 5′ and 3′ ends of the genome. For example, in the case of dengue virus, disruption of sequence complementarity leads to virus attenuation (Alvarez et al., 2005). In many cases the 5′–3′ end interactions are further stabilized by the associate of host proteins. For example, in the case of bovine viral diarrhea virus (BVDV), sequence complementarity alone is not sufficient to maintain the 5′–3′ end interaction and cellular proteins stabilize these interactions by acting as facilitators (Isken et al., 2003). To date, only two reports exist on the studies of 5′–3′ end interactions in caliciviruses (López-Manríquez et al., 2013, Sandoval-Jaime and Gutiérrez-Escolano, 2009). In silico analyses of NV genome identified possible complementary sequences, which were promoted or stabilized by interaction with cellular proteins. The observation was further supported by the mutational analysis of the NV sequences, demonstrating that the complementarity of the sequence was essential to maintain the 5′–3′ interaction, even in the presence of cellular proteins. Although, the identity of cellular proteins involved in this 5′–3′ end interaction of NV is not known, recent work has identified a similar 5′–3′ end interaction within the MNV genome and provided evidence that the cellular factors heterogeneous ribo-nucleoprotein (hnRNP) A1 and PCBP2 played essential role in stabilizing these interactions. The physical interaction between the 5′ and 3′ end is stimulated by the hnRNP A1 and PCBP2 proteins. Their role in genome circularization was confirmed by electron microscopy. In addition RNAi-mediated reduction in the intracellular levels of each protein confirmed a function in the MNV life cycle. However, it should be noted that proteins of the hnRNP A/B family can often replace each other functionally, and that inherent over-expression of these proteins is often observed in immortalized cells. This can often lead to difficulty in observing their correct functionality in common cell lines (Lin et al., 2009b). Clearly, further studies in either primary cells or animal models will aid in the identification of molecular factors essential for genome circularization of caliciviruses.
6. Conclusions
Overall, whilst there is increasing data on the presence and function of RNA structural elements in calicivirus genomes, until recently, the lack tractable experimental systems for many of the family members have significantly impaired their characterization. The last 10 years have seen a dramatic increase in the development of such experimental systems and this has lead to an inevitable increase in our knowledge of the calicivirus life cycle. Concomitant with this has been the ability to use reverse genetics, cell culture systems and small animal models with which to characterize the role of viral RNA structures. Despite a significant increase in the available data, there are noteworthy areas that have to date not been studied. For example, no data exists on the functional role of RNA structures in many members of the family, and the potential role of RNA structures in viral encapsidation have yet to be studied in any great detail. With the development of experimental systems for HuNoV, including reverse genetics (Katayama et al., 2014), a possible cell culture system (Jones et al., 2014), and a small animal model (Taube et al., 2013), the functional characterization of RNA structures and RNA–protein interactions in the life cycle of HuNoV may now be possible. Such studies may contribute to the development of novel antiviral strategies for the control of noroviruses and more widely to other members of the Caliciviridae. More importantly however, they provide new insights into the life cycle of a diverse range of previously poorly characterized pathogens.
Acknowledgments
We acknowledge support from the Wellcome Trust (Ref: WT097997MA) and Ministry of Higher Education in Saudi Arabia (BA). IG is a Wellcome Senior Fellow.
Contributor Information
Bader Alhatlani, Email: bysa2@cam.ac.uk.
Ian Goodfellow, Email: ig299@cam.ac.uk.
References
- Agis-Juárez R.A., Galván I., Medina F., Daikoku T., Padmanabhan R., Ludert J.E., del Angel R.M. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J. Gen. Virol. 2009;90:2893–2901. doi: 10.1099/vir.0.013433-0. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Filomatori C.V., Gamarnik A.V. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′ UTRs. Virology. 2008;375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Ludueña S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., Karakasiliotis I., Vashist S., Chung L.M.W., Rees J., Reese J., McFadden N., Benson A., Yarovinsky F., Simmonds P., Goodfellow I. Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J. Virol. 2010;84:2859–2870. doi: 10.1128/JVI.02053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn L.B., Towner J.S., Semler B.L., Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancio-Lonches C., Yocupicio-Monroy M., Sandoval-Jaime C., Galvan-Mendoza I., Ureña L., Vashist S., Goodfellow I., Salas-Benito J., Gutiérrez-Escolano A.L. Nucleolin interacts with the feline calicivirus 3′ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication. J. Virol. 2011;85:8056–8068. doi: 10.1128/JVI.01878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.-O., George D.W., Patton J.B., Green K.Y., Sosnovtsev S.V. Leader of the capsid protein in feline calicivirus promotes replication of Norwalk virus in cell culture. J. Virol. 2008;82:9306–9317. doi: 10.1128/JVI.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.I.H.A., Brown E.A., Lemon S.M. Cell type-specific proteins which interact with the 5′ nontranslated region of hepatitis A virus. RNA. 1993;67:6716–6725. doi: 10.1128/jvi.67.11.6716-6725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y., Nayak A., Bordeleau M.-E., Tanaka J., Pelletier J., Belsham G.J., Roberts L.O., Goodfellow I.G. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006;281:25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y., Skinner M.A., Goodfellow I.G. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007;88:2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L., Bailey D., Leen E.N., Emmott E.P., Chaudhry Y., Roberts L.O., Curry S., Locker N., Goodfellow I.G. Norovirus translation requires an interaction between the C terminus of the genome-linked viral protein VPg and eukaryotic translation initiation factor 4G. J. Biol. Chem. 2014;289:21738–21750. doi: 10.1074/jbc.M114.550657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh K.F., Fraser C.S., Hershey J.W.B., Hardy M.E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M., Villegas-Sepúlveda N., del Angel R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Etchison D., Milburn S.C., Ederys I., Sonenberggl N., Hershey J.W.B. Inhibition of HeLa cell protein. J. Biol. Chem. 1982:14806–14810. [PubMed] [Google Scholar]
- Farkas T., Sestak K., Wei C., Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. http://dx.doi.org/10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I. The genome-linked protein VPg of vertebrate viruses – a multifaceted protein. Curr. Opin. Virol. 2011 doi: 10.1016/j.coviro.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberté J.-F., Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Escolano A.L., Brito Z.U., del Angel R.M., Jiang X. Interaction of cellular proteins with the 5′ end of Norwalk virus genomic RNA. J. Virol. 2000;74:8558–8562. doi: 10.1128/jvi.74.18.8558-8562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Escolano A.L., Vázquez-Ochoa M., Escobar-Herrera J., Hernández-Acosta J. La, PTB, and PAB proteins bind to the 3′ untranslated region of Norwalk virus genomic RNA. Biochem. Biophys. Res. Commun. 2003;311:759–766. doi: 10.1016/j.bbrc.2003.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B., Kim Y.K., Kim J.H., Kim T.Y., Jang S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert T.P., Brierley I., Brown T.D.K. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- Hirano M., Kaneko S., Yamashita T., Luo H., Qin W., Shirota Y., Nomura T., Kobayashi K., Murakami S. Direct interaction between nucleolin and hepatitis C virus NS5B. J. Biol. Chem. 2003;278:5109–5115. doi: 10.1074/jbc.M207629200. [DOI] [PubMed] [Google Scholar]
- Isken O., Grassmann C.W., Sarisky R.T., Kann M., Zhang S., Grosse F., Kao P.N., Behrens S. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22 doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Wang M., Wang K., Estes M.K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Li Z., Nagy P.D. Nucleolin/Nsr1p binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology. 2010;396:10–20. doi: 10.1016/j.virol.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.K., Watanabe M., Zhu S., Graves C.L., Keyes L.R., Grau K.R., Gonzalez-Hernandez M.B., Iovine N.M., Wobus C.E., Vinje J., Tibbetts S.A., Wallet S.M., Karst S.M. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasiliotis I., Chaudhry Y., Roberts L.O., Goodfellow I.G. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 2006;87:3339–3347. doi: 10.1099/vir.0.82153-0. [DOI] [PubMed] [Google Scholar]
- Karakasiliotis I., Vashist S., Bailey D., Abente E.J., Green K.Y., Roberts L.O., Sosnovtsev S.V., Goodfellow I.G. Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PLoS ONE. 2010;5:e9562. doi: 10.1371/journal.pone.0009562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Goodfellow I.G., Green K.Y., Virgin H.W. Advances in norovirus biology. Cell Host Microbe. 2014 doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Lay M., Davidson J., Virgin H.W. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Karst S.M., Zhu S., Goodfellow I.G. The molecular pathology of noroviruses. J. Pathol. 2014 doi: 10.1002/path.4463. [DOI] [PubMed] [Google Scholar]
- Katayama K., Murakami K., Sharp T.M., Guix S., Oka T., Takai-Todaka R., Nakanishi A., Crawford S.E., Atmar R.L., Estes M.K. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4043–E4052. doi: 10.1073/pnas.1415096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Jang S.K. La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J. Gen. Virol. 1999;80:3159–3166. doi: 10.1099/0022-1317-80-12-3159. [DOI] [PubMed] [Google Scholar]
- Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011 doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-Y., Chen T.-C., Weng K.-F., Chang S.-C., Chen L.-L., Shih S.-R. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-Y., Shih S.-R., Pan M., Li C., Lue C.-F., Stollar V., Li M.-L. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009;83:6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wimmer E., Paul A.V. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Manríquez E., Vashist S., Ureña L., Goodfellow I., Chavez P., Mora-Heredia J.E., Cancio-Lonches C., Garrido E., Gutiérrez-Escolano A.L. Norovirus genome circularisation and efficient replication is facilitated by binding of PCBP2 and hnRNP A1. J. Virol. 2013 doi: 10.1128/JVI.03433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Liu G., Sun Y., Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim. Biophys. Acta Mol. Cell Res. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- McFadden N., Bailey D., Carrara G., Benson A., Chaudhry Y., Shortland A., Heeney J., Yarovinsky F., Simmonds P., Macdonald A., Goodfellow I. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 2011;7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R., Daijogo S., Walter B.L., Nguyen J.H.C., Semler B.L. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby J.A., Pijlman G.P., Wilusz J., Khromykh A.A. Noncoding subgenomic flavivirus RNA: multiple functions in west Nile virus pathogenesis and modulation of host responses. Viruses. 2014 doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands D.J., Harris T.J., Brown F. More precise location of the polycytidylic acid tract in foot and mouth disease virus RNA. J. Virol. 1978;26:335–343. doi: 10.1128/jvi.26.2.335-343.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Jaime C., Gutiérrez-Escolano A.L. Cellular proteins mediate 5′–3′ end contacts of Norwalk virus genomic RNA. Virology. 2009;387:322–330. doi: 10.1016/j.virol.2009.02.041. [DOI] [PubMed] [Google Scholar]
- Seal B.S., Neill J.D., Ridpath J.F. Predicted stem-loop structures and variation in nucleotide sequence of 3′ noncoding regions among animal calicivirus genomes. Virus Genes. 1994;8:243–247. doi: 10.1007/BF01704518. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Karakasiliotis I., Bailey D., Chaudhry Y., Evans D.J., Goodfellow I.G. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008;36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Galán C., Mateos-Gómez P.a., Palacio L., Zúñiga S., Cruz J.L., Almazán F., Enjuanes L. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication–transcription sites. J. Virol. 2011;85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.-O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Meerovitch K., Lee H.S., Dholakia J.N., Kenan D.J., Agol V.I., Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube S., Kolawole A.O., Höhne M., Wilkinson J.E., Handley S.A., Perry J.W., Thackray L.B., Akkina R., Wobus C.E. A mouse model for human norovirus. MBio. 2013;4 doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray L.B., Wobus C.E., Chachu K.A., Liu B., Alegre E.R., Henderson K.S., Kelley S.T., Virgin H.W. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L.G., Goodfellow I.G. Norovirus gene expression and replication. J. Gen. Virol. 2014;95:278–291. doi: 10.1099/vir.0.059634-0. [DOI] [PubMed] [Google Scholar]
- Vashist S., Urena L., Chaudhry Y., Goodfellow I. Identification of RNA–protein interaction networks involved in the norovirus life cycle. J. Virol. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Farkas T., Sestak K., Jiang X. Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J. Virol. 2008;82:11429–11436. doi: 10.1128/JVI.00696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirblich C., Thiel H.J., Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell G.W., Gil A., Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- Xie J., Lee J.-A., Kress T.L., Mowry K.L., Black D.L. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


