SUMMARY
Subsets of long-lived, tumor-initiating stem cells often escape cancer therapies. However, sources and mechanisms that generate tumor heterogeneity and drug-resistant cell population are still unfolding. Here, we devise a functional reporter system to lineage trace and/or genetic ablate signaling in TGF-β-activated squamous cell carcinoma stem cells (SCC-SCs). Dissecting TGF-β’s impact on malignant progression, we demonstrate that TGF-β concentrating near tumor-vasculature generates heterogeneity in TGF-β signaling at tumor-stroma interface and bestows slower-cycling properties to neighboring SCC-SCs. While non-responding progenies proliferate faster and accelerate tumor growth, TGF-β-responding progenies invade, aberrantly differentiate, and affect gene expression. Intriguingly, TGF-β-responding SCC-SCs show increased protection against anti-cancer drugs, but slower-cycling alone does not confer survival. Rather, TGF-β transcriptionally activates p21, which stabilizes NRF2, thereby markedly enhancing glutathione metabolism and diminishing effectiveness of anti-cancer therapeutics. Together, these findings establish a surprising non-genetic paradigm for TGF-β signaling in fueling heterogeneity in SCC-SCs, tumor characteristics, and drug resistance.
INTRODUCTION
Most tumors are of a clonal origin but often exhibit heterogeneity in phenotypic and functional properties including proliferation, morphology, motility, and differentiation. Such heterogeneity has also been implicated in the ability to survive therapy and seed metastases (Hanahan and Weinberg, 2011). Cumulative mutations resulting from genomic instability certainly produce heterogeneity (Greaves and Maley, 2012). However, developmental diversity of cell types is accomplished without genetic alterations, raising the possibility that cellular diversity within tumors may also arise from non-genetic factors. Contributing variations might come from the tumor microenvironment, which can transmit gradients of signaling factors, oxygen, and metabolites to tumor cells depending upon their proximity to the local sources (Meacham and Morrison, 2013; Kreso and Dick, 2014). While the hypothesis is attractive, experimental evidence is lacking, and non-genetic mechanisms that drive tumor heterogeneity remain largely unknown.
Irrespective of the basis for tumor heterogeneity, the long-lived capacity of tumor-initiating stem cells (SCs) to self-renew, initiate, and propagate cancers place these cells at the roots of diversity. Furthermore, SCs are often few in number and can exist in slow-cycling states, which has led to speculation that cancer SCs may be the source of recurrence following anti-cancer therapy (Hope et al., 2004; Berns, 2005; Notta et al., 2011; Visvader and Stingl, 2014). Another potentially intertwined factor is the need for long-lived SCs to adjust their metabolism in order to withstand stress and reactive oxygen species (ROS) (Diehn et al., 2009). In turn, such metabolic reprogramming can alter cellular behavior and lead to cancer progression (Bigarella et al., 2014). To this end, variations in cycling rates and/or local microenvironments could generate metabolic heterogeneities in cancer SCs, which could ultimately affect tumor heterogeneity and drug resistance.
An excellent tumor model for addressing these issues is squamous cell carcinoma (SCC). Among the most common and life-threatening cancers world-wide, SCCs exhibit high rates of tumor recurrence following anti-cancer therapy. Both functionally and molecularly, populations enriched for SCC-SCs have been identified, purified, and characterized. These SCC-SC-enriched populations represent ~1%–5% of the tumor and reside at the tumor-stroma interface. They are typified by elevated integrins, and other markers, e.g. CD34, CD44, and SOX2 (Malanchi et al., 2008; Schober and Fuchs, 2011; Lapouge et al., 2012). They also express VEGFA, suggestive of enrichment at the vasculature (Beck et al., 2011). Interestingly, heterogeneity, particularly in proliferative rates, exists within SCC-SC-enriched populations (Schober and Fuchs, 2011). Whether a slow-cycling property allows some SCs to escape chemotherapy and contribute to cancer recurrence has not been explored.
Notably, SCC-SC numbers increase by ~10-fold when TβRII, an essential component of the transforming growth factor β (TGF-β) transmembrane receptor, is abrogated (Schober and Fuchs, 2011). TGF-β is a well-established inhibitor of normal epithelial cell proliferation, and conditional ablation of Tgfbr2 predisposes epithelial tissues to cancer (Lu et al., 2006; Ijichi et al., 2006; Muñoz et al., 2006; Guasch et al., 2007). Paradoxically, although elevated TGF-β signaling in skin prevents chemical induction of benign papillomas, TGF-β enhances their malignant conversion to SCCs and promotes metastasis (Cui et al., 1996; Massagué, 2012).
Researchers often attribute these seemingly distinct effects of TGF-β to cumulative genetic changes during tumorigenesis. However, since cycling rates of SCs are heterogeneous within an SCC and since SC numbers increase in the absence of TGF-β signaling, we posited that heterogeneity in TGF-β-responsiveness might exist within SCC progenitors, and might simultaneously restrict their proliferation and promote invasion and malignant transformation. If so, TGF-β-mediated differences in cycling rates of SCC-SCs could contribute to metabolic heterogeneity, as well as ultimately, heterogeneity in response to anti-cancer therapies. Elucidating how TGF-β functions in cancer progression and metastasis is a prerequisite for ascertaining whether disrupting this pathway is prudent for metastatic therapeutics when its tumor suppressive features might co-exist.
The TGF-β signaling pathway has been extensively studied. When latent TGF-β ligands are processed and activated, they can bind to TβRII, which phosphorylates TβRI, the other essential component of this bipartite transmembrane receptor. Activated TβRI propagates the signal by phosphorylating intracellular downstream effectors, SMAD2 and SMAD3 (SMAD2/3), which complex with SMAD4, translocate to the nucleus and bind to specific DNA sequence motifs called SMAD-binding elements (SBEs). Upon binding, pSMAD2/3-SMAD4 complexes interact with additional transcriptional regulators to transactivate TGF-β-responsive target genes (Massagué, et al., 2005; Mullen et al., 2011).
The ability of TGF-β signaling to activate target genes enables the pathway to impact diverse cellular processes including not only proliferation but also differentiation, migration, apoptosis, and ECM remodeling (Derynck and Miyazono, 2008; Massagué, 2012; Oshimori and Fuchs, 2012a). Important questions now emerge regarding TGF-β’s ability to unleash its varied and temporal effects on tumor progression. Do TGF-β’s seemingly opposing actions on proliferation and invasion act sequentially or do they act simultaneously in tumor progression? Are these dissimilar events dependent upon progressively distinct genetic states that emerge during malignancy? Does TGF-β contribute to heterogeneity in the tumor microenvironment and, if so, how? Can heterogeneity in TGF-β signaling impact SCC-SCs differentially and might this allow some cancer SCs to escape anti-cancer drugs? If so, is it because of its ability to impact proliferation rates, affect metabolic states and/or alter the expression of key target genes?
To tackle these issues, we devised a strategy to monitor, track, and modify TGF-β signaling in mouse skin during malignant progression. In so doing, we’ve been able to delineate the temporal functions of TGF-β in SCCs as they develop and progress. Combined with transcriptional profiling, molecular, biochemical, and genetic studies, we unearth important functions for TGF-β signaling during the process and unveil its impact not only on cancer SC proliferation but also on the emergence of tumor heterogeneity and anti-cancer drug resistance. Moreover, we show that metabolic reprogramming, an emerging hallmark of cancer, is also integrally linked to TGF-β-mediated effects on cancer SCs, and that TGF-β-regulated metabolism in particular plays a critical role in the divergent responses to anti-cancer therapies.
RESULTS
An In Vivo Reporter System for Lineage Tracing and Manipulating TGF-β Responding Cells During Malignant Progression
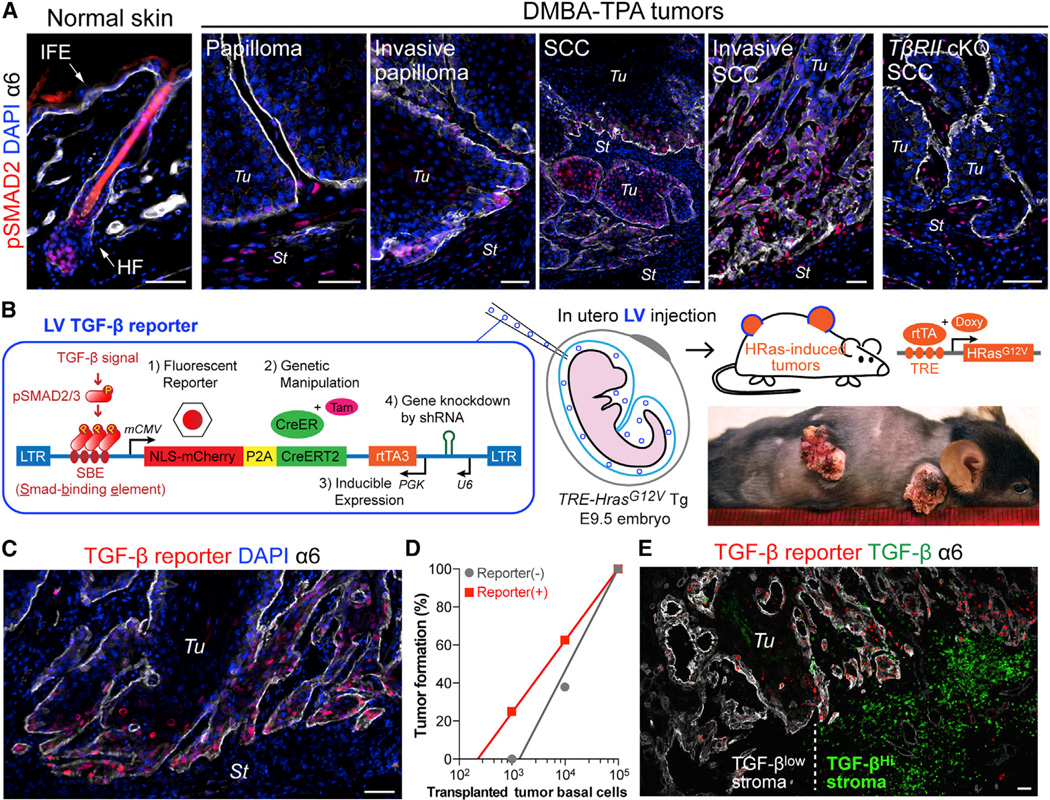
To identify putative TGF-β-responding cells within skin tumors, we first performed anti-phospho (active) SMAD2 immunofluorescence on mouse skin at various stages following classical carcinogenic protocols with tumor-initiator 7,12-dimethyl-benz(a)anthracene (DMBA) and tumor-promoter 12-O-tetradecanoyl-phorbol-13-acetate (TPA) (Figure 1A). As reported previously (Oshimori and Fuchs, 2012b), nuclear pSMAD2 was barely detectable in interfollicular epidermis. In normal skin, it appeared transiently in follicular stem cells (SCs) at the start of a new hair cycle. As benign papillomas formed, pSMAD2 immunolabeling remained low in epithelium but was found in some stromal cells. As papillomas transitioned to malignant SCC, marked nuclear pSMAD2 appeared in epithelial cells at the invasive tumor front. Keratin 14 (K14)-Cre-mediated ablation of Tgfbr2 specifically in skin epithelium resulted in complete loss of pSMAD2 in SCCs, but not surrounding stroma. These findings underscored the efficacy of the antibody and the dependence of SMAD2/3 activation in SCCs on TGF-β/TβRI/II signaling, rather than pathways triggered by Nodal or Activins.
Figure 1. Lentiviral TGF-β Reporter System for Probing Malignant Transformation In Vivo.
(A) pSMAD2 immunolocalization in normal mouse skin and at different stages of DMBA-TPA-induced malignant progression to SCC. Integrin α6 denotes the boundary of tumor epithelia (Tu) and stroma (St). IFE: interfollicular epidermis, HF: hair follicle.
(B) Schematic of LV-mediated in vivo TGF-β reporter and KO/KD system. NLS-mCherry and CreER are under the control of TGF-β signaling. shRNA and rtTA3 transcription factor are under constitutive promoter regulation. LV transduction of surface epithelium of live E9.5 TetO-HrasG12V X Rosa-YFP embryos was achieved by in utero ultrasound-guided microinjection into the amniotic sac. Doxy-induction of HRasG12V initiates tumorigenesis. When desired, CreER is activated by Tam to induce recombination-dependent Rosa-YFP.
(C) Epifluorescence detection of TGF-β–pSMAD2 signaling in HRasG12V SCC.
(D) Limit-dilution orthotopic transplantation of primary tumor basal cells ± TGF-β reporter activity (103 and 104 cells; n=8, 105 cells; n=3).
(E) Epifluorescent TGF-β reporter activity with pan-anti-TGF-β and anti-α6 immunofluorescence shows that basal tumor cells with high TGF-β reporter activity are juxtaposed to stroma with high TGF-β (right). Note heterogeneity demarcated by vertical dotted line.
Scale bars, 50 µm. See also Figure S1.
To monitor TβRI/II–pSMAD2 signaling in vivo, we designed a lentiviral (LV) TGF-β reporter system that used an enhancer composed of multimerized pSMAD2/3 binding elements (SBE) to drive a P2A-based bicistronic transcript encoding nuclear (NLS) mCherry and tamoxifen (Tam)-activatable CreER recombinase (herein called TGFβ-CreER). We inserted a polymerase III-driven promoter in the opposite direction to simultaneously drive an shRNA to achieve knockdown (KD) of a desired transcript (Figure S1A). We first tested this reporter in primary keratinocytes (10MKs) cultured from Rosa26-lox-STOP-lox-EYFP Cre reporter (Rosa-YFP) mice bred on either a Tgfbr2+/+ or Tgfbr2fl/fl background (Figure S1B). Upon TGF-β treatment, reporter-transduced 10MKs expressed NLS-mCherry. Tam then activated CreER in TGF-β-responding MKs, leading to constitutive Rosa-YFP expression. Upon Tgfbr2 ablation, cells lost TGF-β responsiveness, and in turn extinguished NLS-mCherry expression (Figure S1B).
We brought the TGF-β reporter system into an in vivo setting by injecting LV into the amniotic sac of E9.5 mouse embryos. This in utero method allowed titer-dependent (1→>90%) selective transduction of the unspecified surface epithelial progenitors that give rise to skin epithelia (Beronja et al., 2010). By adding a reverse tetracycline transactivator (rtTA) cassette (PGK-rtTA3) to our LV TGF-β reporter construct, we could transduce TRE-Hras1G12V transgenic mice (Chin et al., 1999) and then induce oncogenic HRasG12V with doxycycline (Doxy) (Figure 1B).
In the first experiment, we crossed TRE-Hras1G12V and TRE-H2BGFP mice, and sparsely delivered TGF-β reporter to surface ectoderm of transgenic embryos. Postnatally, doxy-dependent HRasG12V induction occurred exclusively in LV-transduced cells. Within 1–2 months, papillomas formed, which often rapidly progressed to SCC in all or part of the tumor. Tumor epithelia were GFP+, reflecting their derivation from rtTA-expressing, LV-transduced cells (Figure S1C). Additionally, the low levels of virus used (MOI<<1) suggests that these tumors were clonally derived.
In remaining studies, we used TRE-Hras1G12V X Rosa-YFP X Tgfbr2 (+/fl or fl/fl) mice. TGF-β reporter activity (NLS-mCherry) co-localized with pSMAD2 and was particularly intense in a subset of basal tumor cells at invasive fronts (Figure 1C and S1D). Serial transplantation assays previously revealed that the tumor-stromal interface is where cells exist that have long-term, tumor-initiating potential, defined as SCC-SCs (Schober and Fuchs, 2011; Lapouge et al., 2012). To address whether the heterogeneity of TGF-β signaling at this interface might be relevant to SCC-SCs, we used fluorescence-activated cell sorting (FACS) to fractionate CD44+α6hi basal tumor cells according to mCherry expression. mCherry+ cells frequently expressed SC marker CD34 (Figure S1E). In vitro, the colony forming efficiency of TGF-β-responding basal cells was higher than non-responding counterparts, while in direct transplantation assays, FACS-purified TGF-β reporter+ basal cells displayed ~5X higher tumor-initiating frequency than reporterneg counterparts (Figure 1D and S1F). Together, these data indicate that the TGF-β-responding subset of CD44+CD34+α6hi basal tumor cells was enriched for SCC-SCs.
Immunostaining revealed heterogeneity in TGF-β ligand distribution in the stroma that correlated well with basal tumor cell heterogeneity in TGF-β signaling (Figure 1E). Of the various cell types surrounding the tumor, TGF-β immunolabeling best overlapped with CD11b+Ly6C+ monocytic myeloid cells (Figure S1G), in agreement with a prior report that human peripheral blood monocytes secrete TGF-β (Grotendorst et al., 1989). Interestingly, TGF-β+ cells often localized near vasculature, while nuclear pSMAD2 gave a complementary pattern in nearby SCC-SCs (Figure S1G and S1H). The spatial relation between epithelial TGF-β reporter activity and tumor vasculature was exemplified by three-dimensional (3D) microscopic imaging of the tumor-stromal interface (Movies S1 and S2). These results suggest that TGF-β ligand distribution coincides with vasculature and immune cell heterogeneity in tumor microenvironment, and this in turn, generates regional TbRI/II–pSMAD2 signaling within nearby malignant epithelial progenitors at the tumor-stroma interface.
Lineage Tracing Unveils Distinct Behaviors of TGF-β-Responding Versus Non-Responding SCC-SCs
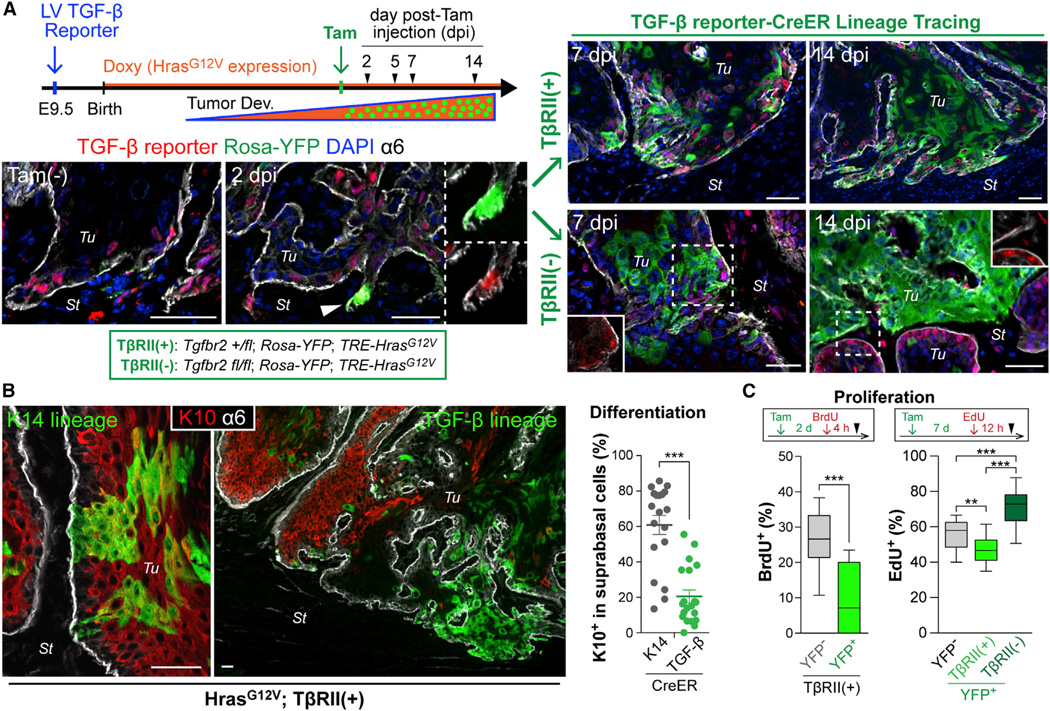
To track the fate of TGF-β-responsive cells during early tumor progression in vivo, we performed TGF-β signaling-dependent lineage tracing by transducing our TGF-β reporter at low MOI as before and then administering Doxy at birth to induce tumorigenesis. Once tumors reached ~7 mm in size, a single low-dose of Tam was then administered systemically to trigger ~24 hr of CreER activity in a small subset of TGF-β-reporter-activated tumor cells (diagram in Figure 2A). Prior to Tam injection, emerging tumors showed mCherry but no YFP, underscoring the dependency of mCherry/CreER bicystronic expression on TGF-β but the reliance of CreER activation on Tam.
Figure 2. TGF-β Signaling-Driven Lineage Tracing during Tumor Development.
(A) Experimental scheme and representative images of TGF-β signaling-driven lineage tracing. Once tumors form (>7 mm), NLS-mCherry is detected in TGF-β-responding transduced cells (Tam-). A single i.p. injection of low-dose Tam elicits Rosa-YFP recombination in a small subset of these cells within 2d post-Tam injection (2dpi). Note YFP/mCherry double-positive cell at invasive front. YFP-marked cells undergo clonal expansion, evident at 7 and 14 dpi, and now independent of TGF-β reporter activity (right). Note difference in clonal expansion rate and morphology based upon whether the initial marked cell is from a tumor initiated on a Tgfbr2 (fl/+) (top) or (fl/fl) (bottom) genetic background.
(B) Immunolabeling (left) and quantifications (right) show that suprabasal differentiation marker K10 preferentially marks YFP+ clones (n=19 analyzed) derived from K14-CreER-induced basal tumor cells. Note that TGFβ-CreER-induced lineage tracing marks TGF-β-responding basal cells that yield YFP+K10neg clones (n=21). Data are mean ± SEM.
(C) S-phase analysis during lineage tracing. BrdU or EdU was administered at 2 or 7 dpi, respectively (see Figures S2H and S2I), and YFPneg and YFP+ basal tumor cells were quantified for nucleotide incorporation (n = 11–15).
Data are box-and-whisker plots. Scale bars, 50 µm. See also Figure S2.
At 2 days post-Tam injection (dpi), single or small clusters of 2–4 YFP+ cells were found at the tumor-stroma interface (Figure 2A). YFP+TβRII+ clones grew markedly over the subsequent 2 weeks and showed enrichment at invasive protrusions. Surprisingly, however, cells within these clones were highly scattered (shown).
On the Tgfbr2fl/fl background, Tam resulted in Tgfbr2 ablation specifically in the few random TGF-β-responding tumor cells that had activated CreER and YFP. YFP+TβRIInegpSMAD2neg cells underwent clonal growth faster, and clones were more tightly packed and less intrusive than TβRII+ counterparts (Figure 2A). Importantly, because we ablated Tgfbr2 in TGF-β-responsive cells, this difference was not attributable to microenvironmental heterogeneity but rather to intrinsic SC differences arising from loss of TGF-β signaling.
The increased scattering and invasiveness of clones derived from TGFβ-responsive tumor cells was accompanied by several features typically associated with epithelial to mesenchymal transition (EMT), including an elongated cell shape, reduced E-cadherin, and enhanced ZEB2 and HMGA2 (Figure S2A–S2D, Movie S3) (Thuault et al., 2006; Hanahan and Weinberg, 2011). By 3D reconstruction of Z-stack images, the clonal nature of the expanding colonies was still discernable, as was their close proximity to tumor vasculatures (Movie S4).
Serendipitously, additional functional lineage tracing unveiled a role for TGF-β signaling in generating aberrant differentiation during malignant progression. When Tam was given to tumors from K14-CreER X Rosa-YFP X TRE-HrasG12V mice transduced with LV-rtTA3, many YFP+ clones from total basal tumor cells (K14hi/K5+) displayed a differentiation keratin K10 suprabasally (Figure 2B). By contrast, in TGFβ-CreER-driven YFP+ clones, K10 was rarely detected, and leading edge cells showed reduced K5 (Figure S2E). Conversely, K13 and K18, ectopically induced in skin SCCs (Nischt et al., 1988; Yamashiro et al., 2010), were readily detected in TGFβ-CreER-driven YFP+ clones, but not in most K14-CreER-driven ones nor in unmarked, TGFβ-non-responsive basal tumor cells from TGFβ-CreER animals (Figure S2F and S2G).
As shown in Figures 2C and S2H, fewer TGF-β-responding (YFP+) basal cells were in S-phase relative to TGF-β non-responding (YFPneg) basal cells. Interestingly, however, as YFP+ basal cells clonally expanded, their SCs remained less proliferative, suggestive of a prolonged slower-cycling state within the TGF-β lineage. By contrast, mosaic YFP+TβRIIneg basal clones showed high cycling rates compared to their YFPnegTβRII+ neighbors (Figures 2C and S2I).
Taken together, our in vivo data provided compelling evidence that TGF-β signaling is directly responsible for generating a pool of slower-cycling SCC-SCs. Moreover, these data further suggest that TGF-β is involved in a non-genetic mechanism that underlies the emergence of tumor heterogeneity at perivascular regions and which leads to simultaneous invasiveness, cell dissemination, and aberrant differentiation, at the expense of SC proliferation and tumor growth.
TGF-β Protects SCC Progenitors From Anti-Cancer Drugs
It has long been suggested that slower-cycling SCs might be refractory to chemotherapeutic anti-proliferative cancer drugs. One of the most widely used anti-cancer drugs, cisplatin [cis-di-amminedichloroplatinum (II)], is the standard chemotherapy for head and neck SCC, and has been used to treat advanced cutaneous SCC. However, tumor recurrence is a major problem.
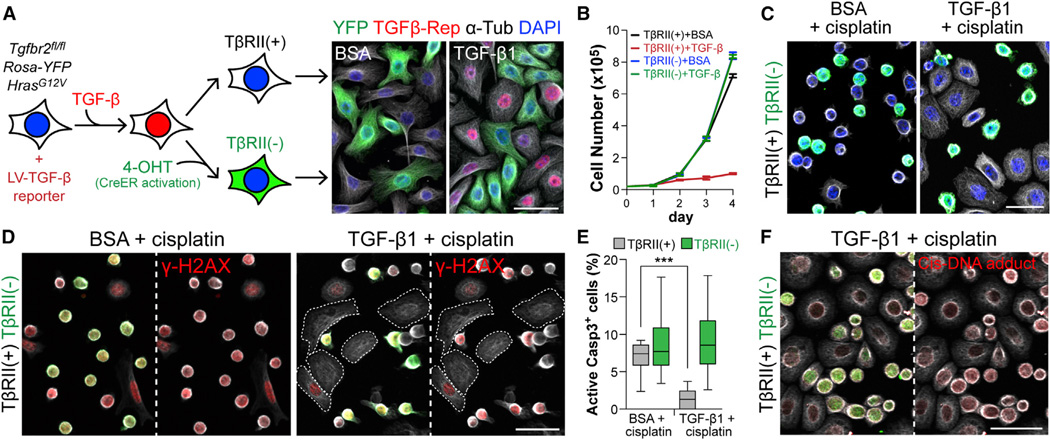
To test whether TGF-β signaling might be involved in drug resistance, we first conducted a series of in vitro experiments. We prepared HRasG12V-expressing 10MKs from Tgfbr2fl/fl X Rosa-YFP mice and transduced them with TGF-β reporter-CreER. After TGF-β1 ± Tam, YFPnegTβRII+ and YFP+TβRIIneg isogenic MKs were co-cultured to assess phenotypes under identical conditions. As expected, TGF-β1 caused reporter activation and growth arrest in YFPnegTβRII+, but not YFP+TβRIIneg MKs (Figure 3A and 3B).
Figure 3. Active TGF-β Signaling in Basal Tumor Cells Increases Their Protection against Cisplatin-Induced Apoptosis.
(A) Experimental scheme to derive isogenic HRasG12V-expressing YFPnegTβRII+ and YFP+TβRIIneg MKs from 10 cultures. (Right) Epifluorescence of cultures ± TGF-β1. Note that only YFPnegTβRII+ cells are TGF-β reporter+ (mCherry+).
(B) Growth curves of TβRII+ and TβRIIneg cells ± 100pM TGF-β1. Data are mean ± SEM.
(C) Immunofluorescence of BSA- or TGF-β1-pretreated (36 hr) YFPnegTβRII+ and YFP+TβRIIneg MKs treated 10 hr with cisplatin. Note that YFP+TβRIIneg MKs frequently displayed signs of apoptotic rounding.
(D) γ-H2AX detection of DNA double-strand breaks induced by cisplatin treatment. Note that both YFPnegTβRII+ and YFP+TβRIIneg MKs show γ-H2AX signal in control, but TGF-β1 selectively spares TβRII+ cells, which remain spread.
(E) Quantifications of AcCasp3+ cells in the same experiment as in (C) and (D) (n=15 microscopic image fields). Data are box-and-whisker plots.
(F) Immunodetection of adduct formed between cisplatin and DNA. Note that YFP+TβRIIneg cells have more cisplatin-modified DNA (red). Anti-tubulin (white). Scale bars, 50 µm.
Cisplatin exerts its cytotoxicity by forming DNA-cisplatin adducts that are recognized by DNA damage recognition complexes which trigger apoptosis (Kelland, 2007). Consistent with these effects, cisplatin caused apoptotic rounding and γ-H2AX (marking DNA double-strand breaks) throughout proliferating cultures (Figure 3C and 3D). Interestingly, after exposure to TGF-β1 for 24–36 hr, most YFPnegTβRII+ cells remained spread, with markedly reduced γ-H2AX compared to YFP+TβRIIneg counterparts. Quantifications with active Caspase-3 (AcCasp3) indicated that TGF-β1 pre-treatment significantly reduced cisplatin-induced apoptotic death (Figure 3E). Moreover, an antibody recognizing DNA-cisplatin adducts showed preferential immunolabeling of YFP+TβRIIneg MKs in cisplatin-treated cultures (Figure 3F). These results suggested that TGF-β signaling enables cultured SCC-SCs to better withstand cisplatin-induced apoptosis.
TGF-β-Responding SCC Progenitors Are Responsible for Drug Resistance and Tumor Recurrence
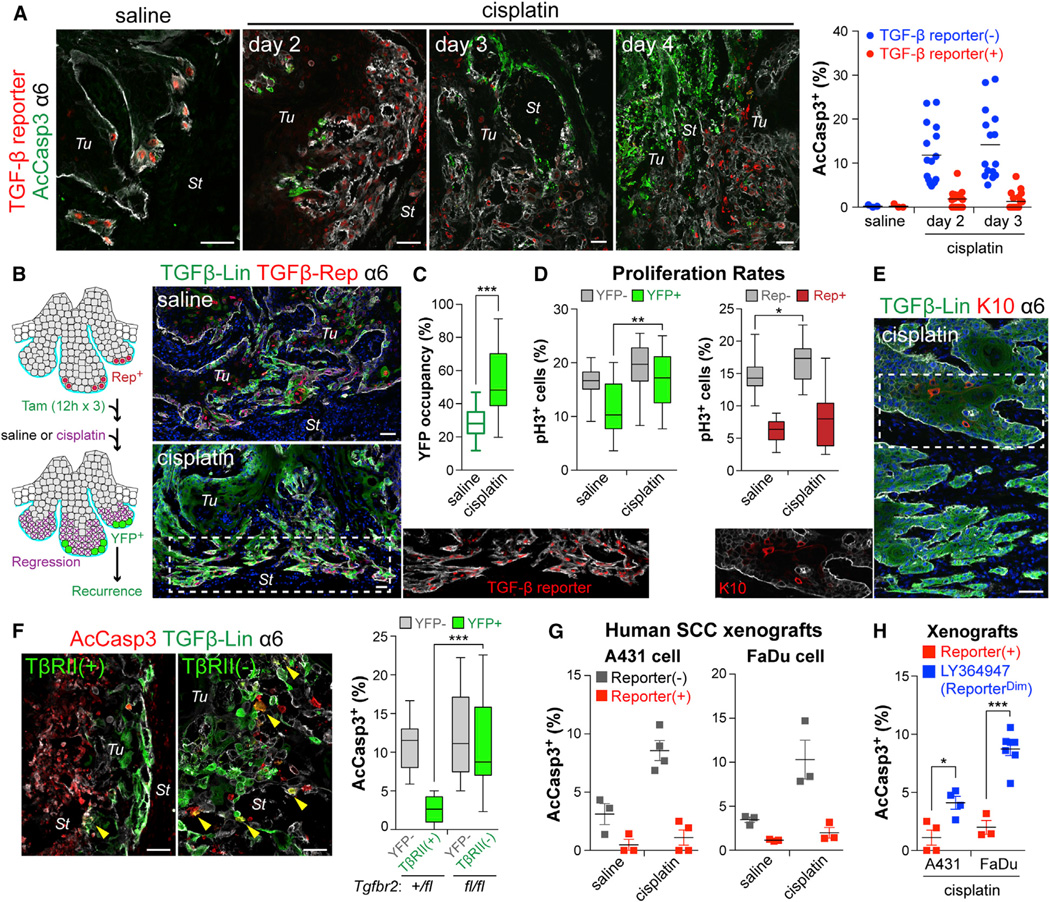
To test TGF-β’s protective qualities in vivo, we challenged tumor-bearing TGF-β reporter mice with systemic cisplatin. While few AcCasp3+ cells were detected in saline-injected control mice, cisplatin significantly increased apoptosis within skin tumor cells. Strikingly, fewer basal tumor cells with active TGF-β signaling were apoptotic (Figure 4A and Movie S5).
Figure 4. TGF-β-Responding SCC-SCs Show Enhanced Drug Resistance In Vivo.
(A) Immunodetection of AcCasp3 (green) and TGF-β reporter (red) in tumors from mice administrated with saline or cisplatin. (Right) Quantifications revealing that TGF-β signaling protected basal tumor cells from cisplatin-induced apoptosis. (3 tumors analyzed; >15 microscopic image fields per tumor).
(B) Experimental scheme and representative examples of lineage tracing to monitor the fate of the TGF-β reporter+ subset of basal tumor cells after cisplatin treatment. (Right) Note that resistant SCCs are largely contributed by TGF-β-responding cells that survived cisplatin.
(C) Quantifications show that the % of YFP+ basal tumor cells increases after cisplatin, suggestive of their preferential survival.
(D) Quantifications of basal tumor cells in G2/M phase (phospho-H3+) that are YFPneg vs YFP+ (left) and reporterneg vs reporter+ (right). Note: although proliferation of basal tumor cells that resist cisplatin is generally elevated, TGF-β-responding SCC-SCs are still slower-cycling.
(E) Recurring tumor clones that resist cisplatin are largely YFP+ and express little K10, consistent with their derivation from TGF-β reporter+ basal cells.
(F) Quantification of apoptotic cells in cisplatin-treated tumors from LV-transduced Tgfbr2+/fl or Tgfbr2fl/fl mice treated as in (B). Note enhanced survival of TβRII+ progenies.
(G and H) Quantification of AcCasp3+ cells after cisplatin treatment of xenografts of (G) TGF-β reporter transduced human SCC cells (reporter+ vs reporterneg), and (H) xenografts pre-treated ± LY364947 (reporter+ vs reporterdim) (n>3).
Data are box-and-whisker plots (C–F) and mean ± SEM (G and H). Scale bars, 50 µm. See also Figure S3.
If TGF-β signaling contributes to drug therapy failure, TGF-β-responding cells should remain during treatment and outgrow their non-resistant peers over time. To test this hypothesis, we conducted TGF-β reporter-driven lineage tracing on TβRII+H-RasG12V tumors during systemic cisplatin treatment. To ensure that we were monitoring TGF-β reporter+ progeny, we carried out Tam treatment (3 doses, 12 hr each) until 1.5 days prior to cisplatin administration. Relative to saline controls, cisplatin injections resulted in a striking reduction in tumor volume within 10–12 days (Figure S3A).
Most notably, the remaining tumors, some of which already showed recurrent growth (Figure S3A), were disproportionately maintained by YFP+ progenies of TGF-β-responding cells (Figure 4B and 4C). This was notable since recurrent tumors still exhibited TGF-β reporter activity, and since even though overall proliferative rates of YFP+ SCC-SCs were elevated in these recurring tumors, the TGF-β reporter+ cohort of basal tumor cells still remained slower-cycling relative to TGF-β-reporterneg counterparts (Figure 4C and 4D). Finally, as in the primary tumor clones derived from TGF-β reporter+ lineages, K10 was broadly absent in YFP+ suprabasal cells, suggesting that recurring tumors were enriched for malignant TGF-β–pSMAD2-signaling basal cells that had survived cisplatin treatment (Figure 4E).
If TGF-β signaling confers increased cisplatin protection to SCC-SCs in vivo, then ablating TGF-β signaling in these cells should confer increased sensitivity to apoptosis. To test this hypothesis, we repeated the experiment shown in Figure 4B, this time on Tgfbr2+/fl (control) and Tgfbr2fl/fl (test) backgrounds. To ensure optimal Tgfbr2 allele targeting, we extended Tam treatments until 3d prior to cisplatin. As shown in Figure 4F, loss of TGF-β signaling resulted in enhanced cisplatin sensitivity.
We obtained similar results when we transduced human SCC lines with TGF-β reporter LV and xenografted them in Nude mice (Figure S3B). Upon cisplatin treatment, TGF-β reporterneg human basal tumor cells showed greater sensitivity than their TGF-β re-porter+ counterparts (Figure 4G). When the experiment was repeated with TGF-β inhibitor LY364947, a dramatic increase in cisplatin sensitivity was observed (Figure 4H and Figure S3C). Together, these findings suggest that both in mouse and human, TGF-β signaling is heterogeneous and increases protection of SCC-SCs against cisplatin.
Transcriptome Analysis Uncovers a Link between TGF-β Signaling and Glutathione Metabolism in SCC Stem Cells
The ability of TGF-β signaling to confer enhanced survival to SCC-SCs of cisplatin-treated tumors was consistent with the cancer stem cell hypothesis for tumor recurrence. A priori, TGF-β’s power could arise from its impact on slow-cycling behavior and/or its effect on transcription. To gain further insights, we used FACS and RNA sequencing (RNA-seq) to purify, transcriptionally profile and compare TGF-β-responding (mCherry+) versus non-responding (mCherryneg) SC populations from >7 mm HRasG12V-induced tumors (Figure S4A–S4C). Our purification scheme was optimized by depleting stromal cell types and dead cells, and then positively selecting for high surface expression of markers of SCC-SCs.
Independent duplicates yielded highly reproducible RNA-seq data (Figure 5A). Dendrogram analysis indicated that despite similarities across SCC-SC profiles, TGF-β reporter+ samples clustered together and separated from the other clustered counterpart. We defined our TGF-β-responsive SCC-SC signature as genes whose transcripts at an FPKM ≥ 1 were differentially expressed by log2 fold change ≥ |1| and which displayed a statistical significance(p< 0.05, q <0.05) across datasets. Our signature consisted of 632 up- and 478 downregulated genes (Figure S4D).
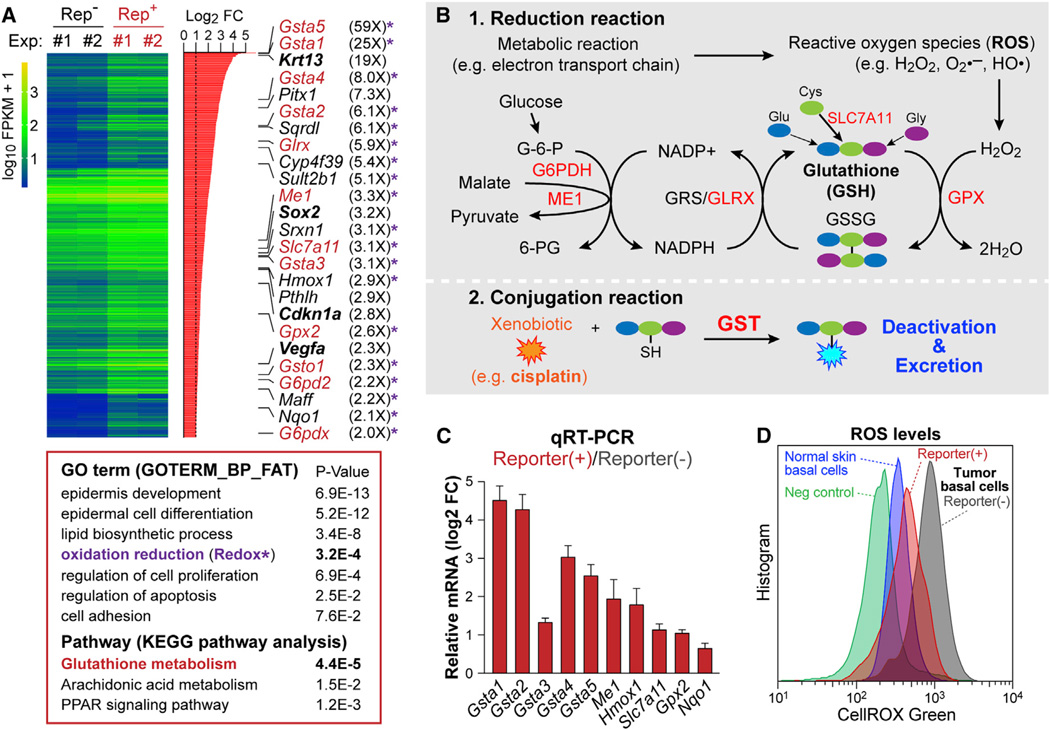
Figure 5. Transcriptional Profiling Reveals a Link between TGF-β Signaling and Glutathione Metabolism.
(A) Summary of transcriptional profiling of TGF-β reporter+ vs reporterneg tumor basal cells by RNA-seq. Significantly upregulated genes are listed on the right side; note genes involved in glutathione metabolism (red), Redox (asterisk). Other genes relevant to text are bolded. (Bottom) Gene ontology (GO: biological function) and KEGG pathway analyses.
(B) Schematic of glutathione metabolic pathway: 1)Reduction reaction; 2)Conjugation reaction. Genes in red are significantly upregulated in TGF-β reporter+ SCs.
(C) RNA-seq validation by qRT-PCR of independently derived in vivo tumor basal cell RNA samples. Data are mean ± SEM.
(D) Flow cytometry analysis of ROS levels in basal cells in normal skin epidermis and tumor epithelia ± TGF-β reporter activity. See also Figure S4.
Several noteworthy alterations were immediately evident in the TGF-β signature. Consistent with our findings thus far, cancer-related differentiation genes, e.g. Krt13, were upregulated. As expected, there was significant overlap with prior signatures from purified SCC-SCs but independent of TGF-β status (Schober and Fuchs, 2011; Lapouge et al., 2012). Notably however, Sox2, Pitx1, Vegfa and other genes typifying these SCC-SC signatures (Boumahdi et al., 2014; Siegle et al., 2014) were more enriched in the TGF-β reporter+ subset than in the total basal SC population. There was also overlap (28%) between our TGF-β reporter+ SCC basal cell RNA-seq and the signature obtained from VEGF-over-expressing papilloma basal cells (Beck et al., 2011).
Gene ontology (GO) term analysis provided insights into biological processes enriched in TGF-β-responding SCC-SCs. In addition to epidermal development; lipid metabolism; and cell proliferation, “reduction and oxidation” (Redox) genes surfaced among top GO-terms. This was especially interesting given that the top upregulated gene pathway in KEGG analysis was “glutathione metabolism” (Figure 5A). Notably, glutathione is the most abundant intracellular antioxidant in animal cells and involves two important metabolic processes (Figure 5B) (Lushchak, 2012). The first is a reduction reaction, which prevents damage from reactive oxygen species (ROS) by exhausting ROS through the conversion of reduced glutathione (GSH) to its oxidized state (glutathione disulfide, GSSG). The second key process is a conjugation reaction regulated by glutathione S-transferase (GST), which is known to metabolize cisplatin (Kelland, 2007).
Our RNA-seq data indicated that genes involved in GSH-conjugation, GSH-mediated reduction and GSH-recycling processes were broadly upregulated in TGF-β reporter+ SCs. This list included GST genes (Gsta1–5, Gsto1) (Figure 5A). qRT-PCR on RNAs from independent tumor samples confirmed their enhanced expression in TGF-β reporter+ basal tumor cells (Figure 5C). Moreover, as judged by CellROX green, a cell-permeant dye that is brightly fluorescent only upon oxidation by ROS, ROS levels were significantly lower in TGF-β reporter+ tumor cells, consistent with the high expression of genes involved in glutathione metabolism (Figure 5D).
TGF-β Induces p21 which In Turn Stabilizes and Activates NRF2-Dependent Transcription in SCC-SCs
The myriad of glutathione metabolism genes upregulated in TGF-β-responding SCC-SCs necessitated further insights before we could abrogate the pathway and assess the consequences to cisplatin resistance. Interestingly, the enhancer/promoters of these genes contained antioxidant response elements (AREs), which are the consensus binding motifs for transcription factor NRF2 (Gorrini et al., 2013) (Figure 6A). Additionally, as judged by immunofluorescence, nuclear NRF2 was prominent in TGF-β reporter+ cells at the tumor-stroma interface (Figure 6B). However, neither RNA-seq nor qRT-PCR data showed TGF-β reporter-dependent differences in NRF2 gene (Nfe2l2) transcription (Figure 6A), indicating that other steps must be involved in upregulating the antioxidant gene response in this SCC-SC subset.
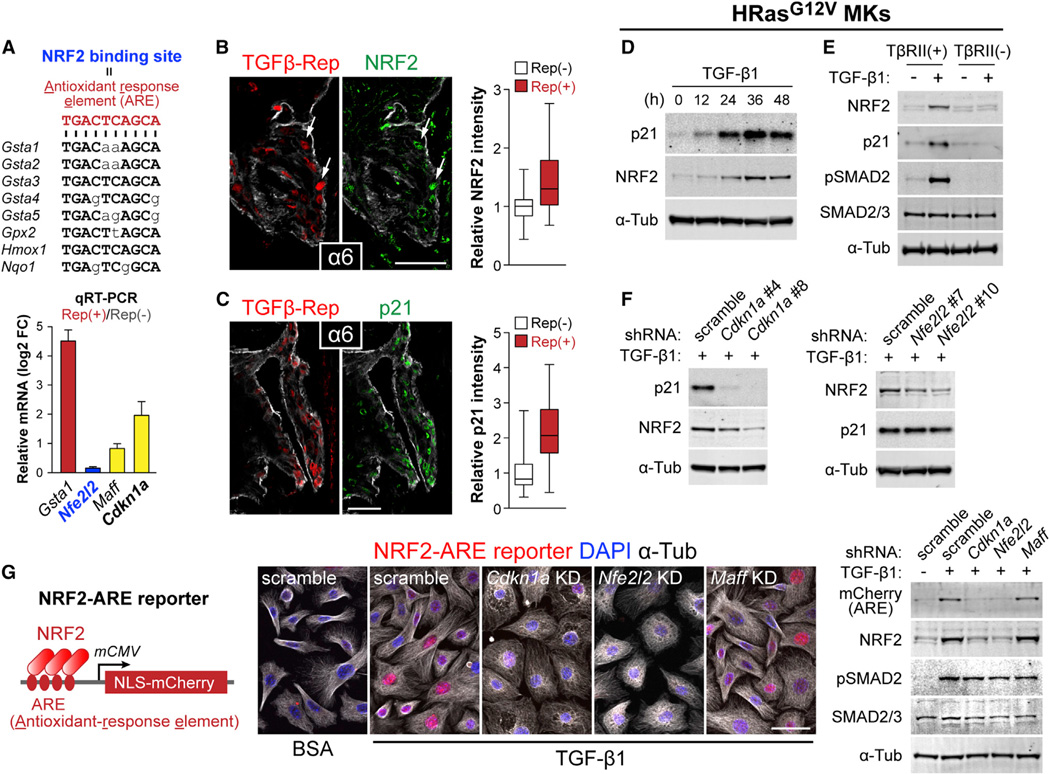
Figure 6. TGF-β Target p21 Is Required for NRF2-Dependent Activation of Antioxidant Genes.
(A) Nucleotide sequence of NRF2 binding motifs within the 5′-upstream region of Gst and other NRF2 target genes. Nucleotides in capital letters are those shared by the antioxidant response element (ARE) consensus sequence. (bottom) qRT-PCR analysis of in vivo tumor basal cell RNA samples. Data are mean ± SEM.
(B and C) Co-expression of NRF2 or p21 (green) and TGF-β reporter (NLS-mCherry) at tumor-stroma interface of TβRII+ tumor sections. Fluorescent intensities of NRF2 and p21 staining in TGF-β reporter+ and reporterneg cells were quantified (NRF2: n=78 and 57 cells, p21: n=101 and 71 cells). Data are box-and-whisker plots.
(D) Immunoblotting of lysates prepared from HRasG12V-overexpressing TβRII+ 10MKs stimulated with TGF-β1 for indicated times.
(E) Immunoblotting of lysates prepared from HRasG12V-overexpressing TβRII+ and TβRIIneg 10MKs stimulated with TGF-β1 for 36 hr.
(F) Immunoblotting of lysates prepared from 36 hr TGF-β1-treated HRasG12V-overexpressing 10MKs transduced with scramble, Cdkn1a or Nfe2l2 shRNAs.
(G) LV NRF2-ARE reporter. (Right) Immunofluorescence and immunoblots of NRF2-reporter transduced HRasG12V-induced MKs expressing scramble (control), Cdkn1a, Nfe2l2, or Maff (control) shRNAs ± TGF-β1 stimulation (36 hr). Note that ARE-reporter activity is abolished upon Cdkn1a or Nfe2l2 but not control KD. Scale bars, 50 µm. See also Figure S5.
Probing mechanism, we first considered KEAP1, which normally binds to and targets NRF2 for proteosome-mediated degradation. TGF-β activity did not affect Keap1 mRNA levels, diminishing the likelihood that KEAP1 absence might underlie NRF2 stabilization. By contrast, cyclin-dependent kinase inhibitor p21 purportedly competes with KEAP1 for NRF2 binding (Chen et al., 2009), and Cdkn1a, encoding p21, is an established target of TGF-β–SMAD signaling (Seoane et al., 2004; Koinuma et al., 2009). Indeed, RNA-seq and qRT-PCR showed Cdkn1a mRNA was upregulated in TGF-β reporter+ SCC-SCs in vivo (Figure 5A and Figure 6A). Immunofluorescence further revealed a strong correlation between TGF-β signaling activity and p21 expression at the tumor-stroma interface (Figure 6C). Moreover, whereas NRF2 and p21 were readily detected in TGF-β-responding tumor cells at the tumor-stromal interface, their expression was not seen in neighboring TβRIIneg tumor cells derived from TGFβ-CreER activation nor in early papillomas, which do not show appreciable TGF-β signaling (Figure S5A–S5C). In vitro studies corroborated these findings (Figure S5D).
Delving deeper, we treated HRasG12V-transformed 10MKs with TGF-β1 and then checked by qRT-PCR for temporal changes in levels of select mRNAs (Figure S5E and S5F). In contrast to Nfe2l2, whose transcripts remained constant during the experiment, Cdkn1a transcripts rose 1–3 hr after TGF-β1 treatment, reaching ~5X normal levels by 48 hr. Moreover by immunoblot, NRF2 and p21 were elevated upon prolonged TGF-β stimulation (Figure 6D). These effects were abrogated in TβRIIneg HRasG12V 10MKs (Figure 6E), supporting the view that p21 induced by TGF-β/pSMAD2 signaling mediates NRF2 protein stabilization.
To rigorously test the hierarchical relation between TGF-β, p21 and NRF2, we used LVs harboring Cdkn1a and Nfe2l2 shRNAs (Figure S5G). Cdkn1a shRNAs not only efficiently depleted p21 but also reduced NRF2 protein in TGF-β stimulated cells (Figure 6F). By contrast, Nfe2l2 depletion did not affect p21 levels induced by TGF-β/pSMAD2 signaling. These data place p21 downstream of TGF-β signaling and upstream of NRF2 stabilization. Indeed, as judged by immunoprecipitation, the interaction between p21 and NRF2 was dependent upon TGF-β1 (Figure S5H).
To test the functional significance of this hierarchical relation, we created a LV NRF2 reporter (Figure 6G). NRF2 reporter activity was potently induced in 10MKs not only by the classical ROS, hydrogen peroxide (H2O2) (Figure S5I) but also by TGF-β stimulation (Figure S5J). Comparative analyses showed that while not as robust as the maximal effect achieved with 500 µM H2O2, the effects of TGF-β1 were comparable to 100–200 µM H2O2 (Figure S5J). Moreover, NRF reporter activation by either H2O2 or TGF-β1 was abolished when these cells were transduced with LVs harboring Cdkn1a or Nfe2l2 shRNAs (Figure 6G and Figure S5K). Together, these results provide compelling evidence that p21 is required for the NRF2-mediated target gene expression that occurs downstream of TGF-β/pSMAD2 signaling in SCC-SCs.
A Role for Glutathione Metabolism in the Cisplatin Resistance of TGF-β-Responding SCC-SCs
To address the physiological relevance of the p21/NRF2 pathway that we unearthed in vitro, we first showed that GSTα, one of the highly upregulated glutathione pathway genes, was indeed upregulated at the protein level in TGF-β reporter+ cells at the tumor-stroma interface of invasive SCCs (Figure 7A). To test whether p21 mediates TGF-β-induced drug resistance, we conducted in vivo KDs by introducing Cdkn1a shRNAs into our LV TGF-β reporter constructs (Figure 7B). In Cdkn1a KD tumors, p21 expression was abrogated in TGF-β reporter+ cells. Importantly, GSTα was also downregulated upon Cdkn1a KD (Figure 7C). Similar results were seen in tumors with Nfe2l2 KD (shown). Together, these findings showed that a key component of glutathione metabolism was dependent upon NRF2 and TGF-β-regulated Cdkn1a.
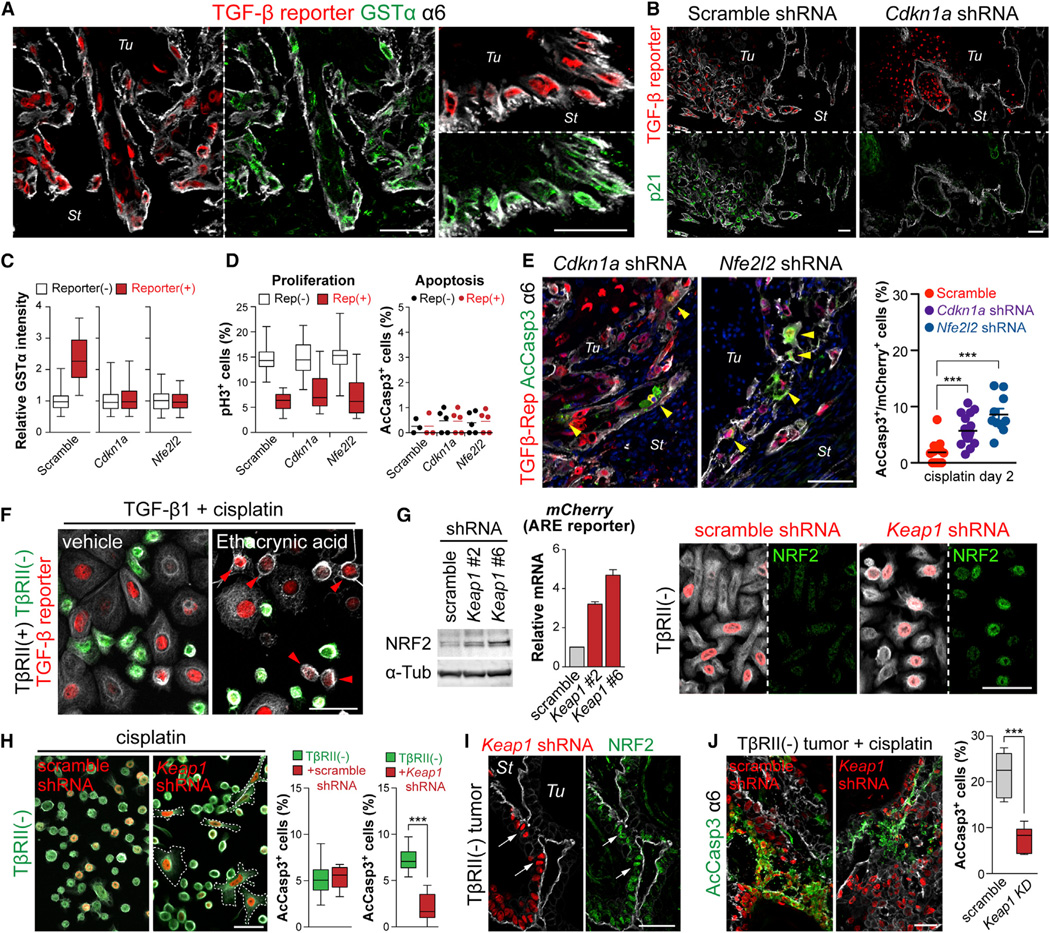
Figure 7. TGF-β-Induced Glutathione Metabolism Confers Enhanced Anti-Cancer Drug Resistance In Vivo.
(A) Coimmunolabeling of TGF-β reporter and GSTα in representative sections of SCCs from LV-transduced mice. Note that nuclear TGF-β reporter signal (red) and cytoplasmic GSTα (green) overlap in invasive cells at the tumor-stroma interface.
(B) Immunofluorescence of TβRII+ tumor sections from transductions of our LV reporter harboring scramble control or Cdkn1a shRNAs. p21 (green) correlates well with TGF-β reporter activity (red) in scramble control and is silenced by Cdkn1a shRNA expression.
(C) Quantifications of GSTα immunofluorescence intensities of TGF-β reporter+ and reporterneg basal tumor cells of mice transduced with scramble, Cdkn1a or Nfe2l2 shRNAs (n=144, 145 or 121 cells).
(D) Quantifications of proliferation (pH3+) and apoptosis (AcCasp3+) in basal cells from HRasG12V-derived SCCs of mice transduced with LVs harboring scramble control, Cdkn1a, or Nfe2l2 shRNAs.
(E) Coimmunolabeling and quantifications (n=3 tumors, >16 microscopic image fields) of AcCasp3 (green, cytoplasmic) and TGF-β reporter activity (red, nuclear) in Cdkn1a or Nfe2l2 KD basal cells of SCCs from cisplatin-treated mice. Yellow arrowheads denote double-labeled cells. Note that without p21 or NRF2, TGF-β reporter+ cells are no longer able to resist apoptosis in response to cisplatin.
(F) TGF-β1-pretreated MKs were exposed to cisplatin ± a potent GST inhibitor, ethacrynic acid. Note that most YFPnegTβRII+ MKs (red arrowheads) still exhibited robust TGF-β reporter activity, but now showed apoptotic rounding like their YFP+TβRIIneg counterparts.
(G and H) KEAP1 stabilizes NRF2 in TβRIIneg SCC-SCs and renders them resistant to cisplatin. (G) Immunoblotting of lysates prepared from HRasG12V-over-expressing 10MKs transduced with scramble or Keap1 shRNAs. (middle) qRT-PCR of NRF2-reporter (mCherry mRNA) expression. (right) NRF2 immunofluorescence (green) in either scramble- or Keap1 shRNA-transduced (H2B–RFP+) TβRIIneg cells. Note that NRF2 is readily detected in TβRIIneg cells only if transduced with Keap1 shRNA. (H) Immunofluorescence of transduced YFP+TβRIIneg HRasG12V-MKs treated with cisplatin for 10 hr. Note that Keap1 KD (H2B–RFP+) suppresses apoptotic rounding. (right) Quantifications of AcCasp3+ cells in the same experiment.
(I) Anti-NRF2 (green) co-immunolabeling of Keap1 KD (H2B–RFP+) cells in TβRIIneg HRasG12V allograft tumors.
(J) AcCasp3+ quantifications show that Keap1 but not scramble shRNA protects TβRIIneg HRasG12V allograft SCC-SCs against cisplatin.
Data are box-and-whisker plots (C, D, H and J) and mean ± SEM (E and G). Scale bars, 50 µm. See also Figure S6.
Notably, the slow-cycling behavior of TGF-β-responding SCC-SCs was not affected appreciably by loss of either p21 or NRF2 (Figure 7D), providing a means of selectively reducing glutathione metabolism without compromising slow-cycling status in TGF-β-responding SCC-SCs. We therefore proceeded to address whether Cdkn1a and Nfe2l2 KD would affect their cisplatin resistance. In the scramble control, TGF-β-responding SCC-SCs showed little apoptosis. In contrast, when mice bearing tumors knocked down for either Cdkn1a or Nfe2l2 were treated with cisplatin, many basal tumor cells were double-positive for AcCasp3 and TGF-β-reporter (Figure 7E). Similarly, drug inhibition of GST increased the sensitivity of TGF-β-responding tumor cells to cisplatin in vitro (Figure 7F). Thus, under circumstances where SCC-SCs were still slow-cycling and responsive to TGF-β, the normalization of glutathione metabolism was sufficient to abolish the survival advantage of TGF-β responding basal cells to cisplatin treatment.
Finally, we addressed the converse, namely whether by enhancing NRF2 stabilization, we could confer enhanced resistance of TβRII-null SCC-SCs to cisplatin. In vitro, both NRF2 protein and NRF2 reporter activity rose in TβRII-null HRasG12V-transformed MKs transduced by either of two different Keap1 shRNAs (Figure 7G). Correspondingly, this resulted in enhanced resistance to cisplatin (Figure 7H). When equal numbers of non-transduced and either scramble- or Keap1 shRNA-transduced TβRII-null HRasG12V-transformed MKs were mixed and then engrafted in vivo, tumors arose which displayed increased NRF2 specifically in Keap1 shRNA-transduced MKs (Figure 7I). Most importantly, fewer of these basal tumor cells apoptosed after cisplatin treatment (Figure 7J). Overall, these data underscore the importance of this pathway in imparting enhanced drug resistance to TGF-β-responding SCC-SCs.
DISCUSSION
Functional and phenotypic heterogeneity among tumor cells have long been recognized, and dynamic changes in genetic, epigenetic, tumor microenvironmental and systemic factors affect subpopulations of tumor cells to acquire advantages for proliferation, survival, spread, and resistance to anti-cancer therapeutics. In studying stem cells of SCCs, we realized that they vary in cycling rates, and that SC numbers increase when TGF-β signaling is abrogated. These findings raised the possibility that TGF-β signaling might be at the root of tumor heterogeneity and that it might impact directly on cancer SCs.
In the present study, we established an in vivo LV delivery system, which allowed us to address roles of TGF-β signaling in the early stages of tumor progression. Our findings provide compelling in vivo evidence of TGF-β’s contributions to the emergence of tumor heterogeneity in the tumor-initiating cells that drive SCC. The heterogeneity in TGF-β-responsiveness is rooted in the congregation of myeloid cells near the tumor vasculature. While TGF-β is secreted in a latent form and deposited in ECM, TGF-β can be activated and released by a variety of mechanisms, which include activated integrins. The paracrine activation of TβRI/II-pSMAD2 in a subset of nearby (integrinhi) SCC-SCs reflected active TGF-β signaling in these cells.
TGF-β represses normal epithelial growth, thereby functioning as an early tumor suppressor. These effects have been extensively studied, as have TGF-β’s late-stage roles in metastasis. However, in the absence of tools to explore intermediate stages of primary tumor progression, the prior speculation as to TGF-β’s dual function in these intermediate steps has been that TGF-β’s cytostatic effects are lost during tumor progression due to activation of oncogenic pathways such as Ras-MAPK, PI3K and c-Myc, which then override TGF-β’s growth inhibitory effects (Chen et al., 2002; Seoane et al., 2004; Gomis et al., 2006; Bruna et al., 2007). At later stages, other TGF-β responses then purportedly prevail that are unrelated to TGF-β’s cytostatic effects but which favor tumor invasion and metastasis.
By designing a functional TGF-β reporter system and coupling Cre activity to TGF-β signaling, we could monitor and compare the behaviors and fates of the subset of basal tumor SCs that activate TβRI/II-pSMAD2 signaling (or which we blocked from doing so) in developing tumors expressing HRasG12V, commonly mutated in human skin cancers. Our lineage tracing analyses of the clonal growth of these cells revealed clearly that TGF-β’s cytostatic and tumor-promoting effects are not mutually exclusive. Rather, TGF-β-responding cells display morphological and biochemical features of invasiveness and malignant conversion during a period when they are in a slow-cycling proliferation state. Moreover, and most importantly, our findings show that these slow-cycling invasive tumor cells gain a marked advantage over hyperproliferative, more tightly clustered TβRII-null SCC-SCs in that they are better protected against DNA damaging agents such as cisplatin.
Cisplatin resistance is a hallmark of head and neck SCCs. Suggested mechanisms for resistance include reduced drug uptake, increased drug efflux, inactivation by GSH conjugation, increased DNA damage repair, and failure to induce apoptosis (Kelland, 2007). Another proposed route is the failure of anti-cancer therapies to target slower-cycling cancer SCs (Meacham and Morrison, 2013). Since our results show that TGF-β reporter+ skin tumor cells are indeed slower-cycling, it seemed plausible that at least in part, the enhanced protection against anti-proliferative cancer drugs might be attributable to the slower-cycling status of TGF-β-responding SCs. However as we learned, this was only the tip of the iceberg in what TGF-β responsiveness is able to achieve, and in fact slower-cycling status appears to be secondary to SCC-SC drug resistance.
Given that the alterations provoked by TGF-β in SCC-SCs extended beyond cycling rates, we asked whether additional changes in transcriptomes might explain the enhanced resistance of TGF-β-responding SCC-SCs to cisplatin. The TGF-β signature encompassed genes already known to play a key role in stemness. However, the signature also show-cased genes involved in glutathione metabolism. Delineating mechanism, TGF-β induced Cdkn1a transcription, leading to p21-mediated NRF2 stabilization and induction of a cohort of glutathione metabolism genes. Most importantly, when Cdkn1a or Nfe2l2 were knocked down, SCC-SCs were still responsive to TGF-β and were still slower-cycling, but their survival in the face of cisplatin was normalized. Further bolstering the importance of this pathway, KD of Keap1 in TβRII-null SCs resulted in not only enhanced NRF2 target gene activity but also enhanced survival in cisplatin treated SCCs.
The role of enhancing antioxidant reactions and glutathione metabolism is still obscure. Although inflammatory cells can have anti-tumorigenic roles, they can also release ROS, which is actively mutagenic, thereby accelerating the genetic evolution of nearby cancer SCs (Grivennikov et al., 2010). In the present study, we showed that even though immune cells concentrate near the vasculature, ROS levels are preferentially reduced in the subset of TGF-β-responding SCC-SCs, and that this is attributable to the TGF-β/p21/NRF2 pathway we delineated here.
In closing, since not all tumor-initiating cells localize to the perivasculature where TGF-β is concentrated, our studies show that this can be advantageous to the tumor. On the one hand, SCC-SCs that do not respond to TGF-β are faster-cycling and can greatly accelerate tumor growth and differentiation. On the other hand, TGF-β-responding SCC-SCs cycle more slowly but show enhanced invasiveness and increased glutathione metabolism, thereby increasing the likelihood not only of metastasis but also long-term survival in the face of ROS and anti-cancer drugs. Given that perivasculature is an emerging niche for many types of SCs, it will be interesting to see in the future if the mechanisms we’ve unearthed here will extend to other cancers. It will also be important to know whether these mechanisms are operative in human SCCs. In this regard, an analysis of the TCGA database revealed poor prognosis for patients with SCCs that upregulate this cohort of glutathione metabolism genes (Figure S6). Whether these tantalizing parallels between mouse and human SCCs will continue to hold must await future cancer databases on tumor-initiating cells as opposed to whole tumor samples. If so, these hitherto underappreciated roles for TGF-β signaling in tumor heterogeneity and anti-cancer resistance could serve as a foundation for designing chemotherapeutics that might overcome drug resistance for this devastating cancer.
EXPERIMENTAL PROCEDURES
In Vivo LV Transduction, Tumor Formation, and Drug Treatment
LV production, concentration, and ultrasound-guided in utero transduction were performed as described (Beronja et al., 2010). Briefly, female mice at day 9.5 of gestation were anesthetized with isoflurane (Hospira). In utero, 0.5 ml LV was microinjected into each embryo’s amniotic sac. To induce tumor formation, rtTA3 was activated by feeding adult mice with Doxy (2 mg/kg) chow. Cre was activated by intraperitoneal (i.p.) injection of Tam (Sigma) in corn oil: TGFβ-CreER, 25 µg/g low-dose, 3×100 µg/g high-dose; K14-CreER, 17.5 µg/g. Cisplatin (Sigma) was dissolved in saline (1 mg/ml) and administrated by i.p. injection (10 mg/kg). TβRI kinase inhibitor (LY364947, Tocris) was i.p. injected (1 mg/kg), 3X/wk. For limit-dilution transplantation and xenotransplantation, 1.0 3 103–105 mouse primary tumor cells and 1.0 × 105 human SCC cells were subcutaneously injected with Matrigel (BD) in Nude mice. Tumor size was calculated using the formula 4/3π × L/2 × W/2 × D/2. For cell proliferation analysis in vivo, BrdU (50 mg/g) or EdU (25 mg/g) was injected i.p. 4 or 12 hr before lethal administration of CO2. All procedures were performed with IACUC-approved protocols.
In Vitro Cell Culture Experiments
Cells were cultured in E medium with 15% fetal bovine serum (FBS) and 50 µM CaCl2 (10 MKs) or 1.5 mM CaCl2 (human SCC lines) at 37°C, 7.5% CO2. For stimulation experiments, media were supplemented with either recombinant mouse TGF-β1 (100 pM = 2.5 ng/ml, R&D Systems), 4-hydroxytamoxifen (1 µM, Sigma), cisplatin (20 µM, Sigma), H2O2 (1–1,000 µM, Fisher), or ethacrynic acid (50 µM, Abcam). For colony formation assay, FACS-isolated 1.03 104 α6hiCD44+mCherryneg and α6hiCD44+mCherry+ primary tumor basal cells were plated onto mitomycin-treated mouse 3T3 fibroblasts in 6-well dishes in E medium with 15% FBS and 300 µM CaCl2. Colony number was counted after 14 day culture.
Statistics
Data were analyzed and statistics performed (unpaired two-tailed Student’s t test) in Prism5 (GraphPad). Significant differences between two groups were noted by asterisks or actual p values (*p < 0.05; **p < 0.01; ***p < 0.001). Quantification data were presented in mean value ± SEM or in box and whisker plots with the dimensions of the box encompassing the 25th–75th percentile, the horizontal bar representing the median, and the error bars representing minimum and maximum values.
Supplementary Material
Highlights.
We devise a system to monitor, manipulate, and track TGF-β signaling in SCCs in vivo
Perivascular TGF-β causes signaling-based heterogeneity among SCC stem cells
TGF-β slows proliferation but aids in malignancy and anti-oxidant metabolism
TGF-β-responding cells resist anti-cancer therapeutics, leading to tumor recurrence
ACKNOWLEDGMENTS
We thank B. Reva for helpful consultation regarding TCGA data analyses; S. Karlsson for floxed-Tgfbr2 mice; F. Costantini for Rosa-YFP mice; L. Chin for TetO-Hras mice; and L. Polak, J. Levorse, and others for assistance in the mouse facility. We are grateful to extensive interactions and helpful discussions with former and present Fuchs lab members. We appreciate the assistance of RU’s Comparative Bioscience Center (an AAALAC facility) for expert care and housing of our mice, the RU Flow Cytometry Resource Center (S. Mazel, Director) and Weill Cornell Medical College Genomics Resources Core Facility for Illumina sequencing (J. Xiang, Director). N.O was supported by Human Frontier Science Program and the Japan Society for the Promotion of Science and is now supported by NIH K99-R00 pathway to independence award. E.F. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the NIH and from the New York State Department of Health (NYSTEM-C029559) to E.F. and from the National Cancer Institute (K99-CA178197) to N.O.
Footnotes
ACCESSION NUMBERS
RNA-seq data have been submitted to NCBI-GEO under the accession number GSE64867.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.01.043.
REFERENCES
- Beck B, Driessens G, Goossens S, Youssef KK, Kuchnio A, Caauwe A, Sotiropoulou PA, Loges S, Lapouge G, Candi A, et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- Berns A. Stem cells for lung cancer? Cell. 2005;121:811–813. doi: 10.1016/j.cell.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen W, Sun Z, Wang X-J, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- Derynck R, Miyazono K. The TGF-β family. Woodbury, New York: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcón C, Nadal C, Van Poznak C, Massagué J. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in meta-static breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotendorst GR, Smale G, Pencev D. Production of transforming growth factor β by human peripheral blood monocytes and neutrophils. J. Cell. Physiol. 1989;140:396–402. doi: 10.1002/jcp.1041400226. [DOI] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFβ signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CVE, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-β signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol. Cell. Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Lapouge G, Beck B, Nassar D, Dubois C, Dekoninck S, Blanpain C. Skin squamous cell carcinoma propagating cells increase with tumour progression and invasiveness. EMBO J. 2012;31:4563–4575. doi: 10.1038/emboj.2012.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S-L, Herrington H, Reh D, Weber S, Bornstein S, Wang D, Li AG, Tang C-F, Siddiqui Y, Nord J, et al. Loss of transforming growth factor-β type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006;20:1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak VI. Glutathione homeostasis and functions: potential targets for medical interventions. J. Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, Huelsken J. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, et al. Transforming growth factor β receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- Nischt R, Roop DR, Mehrel T, Yuspa SH, Rentrop M, Winter H, Schweizer J. Aberrant expression during two-stage mouse skin carcinogenesis of a type I 47-kDa keratin, K13, normally associated with terminal differentiation of internal stratified epithelia. Mol. Carcinog. 1988;1:96–108. doi: 10.1002/mc.2940010205. [DOI] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. The harmonies played by TGF-β in stem cell biology. Cell Stem Cell. 2012a;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012b;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-β and integrin/focal adhesion kinase (FAK) signaling. Proc. Natl. Acad. Sci. USA. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le H-V, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Siegle JM, Basin A, Sastre-Perona A, Yonekubo Y, Brown J, Sennett R, Rendl M, Tsirigos A, Carucci JA, Schober M. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat. Commun. 2014;5:4511. doi: 10.1038/ncomms5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin C-H, Moustakas A. Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro Y, Takei K, Umikawa M, Asato T, Oshiro M, Uechi Y, Ishikawa T, Taira K, Uezato H, Kariya K. Ectopic coexpression of keratin 8 and 18 promotes invasion of transformed keratinocytes and is induced in patients with cutaneous squamous cell carcinoma. Biochem. Bio-phys. Res. Commun. 2010;399:365–372. doi: 10.1016/j.bbrc.2010.07.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.