Abstract
Collision-induced dissociation (CID) remains the predominant mass spectrometry based method for identifying phosphorylation sites in complex mixtures. Unfortunately, the gas-phase reactivity of phosphoester bonds results in MS/MS spectra dominated by phosphoric acid (H3PO4) neutral loss events, suppressing informative peptide backbone cleavages. To understand the major drivers of H3PO4 neutral loss, we performed robust non-parametric statistical analysis of local and distal sequence effects on the magnitude and variability of neutral loss, using a collection of over 35,000 unique phosphopeptide MS/MS spectra. In contrast to peptide amide dissociation pathways, which are strongly influenced by adjacent amino acid side chains, we find that neutral loss of H3PO4 is affected by both proximal and distal sites, most notably basic residues and the peptide N-terminal primary amine. Previous studies have suggested that protonated basic residues catalyze neutral loss through direct interactions with the phosphate. In contrast, we find that nearby basic groups decrease neutral loss regardless of mobility class, an effect only seen by stratifying spectra by charge-mobility. The most inhibitory bases are those immediately N-terminal to the phosphate, presumably due to steric hindrances in catalyzing neutral loss. Further evidence of steric effects is shown by the presence of proline which can dramatically reduce the presence of neutral loss when between the phosphate and a possible charge donor. In mobile proton spectra the N-terminus is the strongest predictor of high neutral loss, with proximity to the N-terminus essential for peptides to exhibit the highest levels of neutral loss.
Introduction
Reversible protein phosphorylation is involved in the regulation of virtually all aspects of cellular function, with more than 10,000 human proteins known to be phosphorylated [1]. Phosphorylation networks, comprised of complex pathways of kinases, phosphatases and interacting regulatory proteins enable the cell to adapt rapidly to diverse environmental stimuli by transduction of extracellular signals to the nucleus. Mass spectrometry has become the primary technology for large-scale phosphoproteomics, allowing thousands of phosphorylation sites to be monitored simultaneously and providing a global snapshot of complex signaling network responses to diverse biological stimuli [2]. Unfortunately, the unusual chemistry of the phosphoester bond ensures that only a small percentage of these phosphopeptides can be identified by the predominant ion trap dissociation method, collision-induced dissociation (CID)[3]. Furthermore, the exact amino acid position of the phosphate can be localized in only a minority of identified peptides[4]. Phosphoester bonds in phosphoserine (pSer) and phosphothreonine (pThr) are highly labile relative to other bonds in peptides. As a result, phosphopeptide CID spectra are frequently dominated by a single peak corresponding to the neutral loss of phosphoric acid, H3PO4. Neutral losses from fragment ions further complicate the spectra by adding additional peaks to the spectrum, which confound commonly used search engines for phosphopeptide identification.
CID MS3 sequencing of the dominant H3PO4 precursor neutral loss ion yields a higher proportion of sequence-specific fragments. Several groups have found, however, that performing MS3 may lower identification rates [5], likely due to reduced sampling rate. Consequently, most identifications come from MS2 scans or multistage activation scans that combine information from both levels of fragmentation [6]. Electron transfer dissociation (ETD) is a complementary fragmentation method that tends to preserve the phosphoester bond and results in greater fragmentation across peptide backbone sites, but works well only for a subset of cationic phosphopeptides with high charge density [7]. Incidentally, CID performs well for localizing these same types of peptides, limiting the complementarity of ETD.
Loss of 98 Da in an MS2 spectrum is a positive indicator of the presence of phosphorylation, barring a few caveats[8]. In practice, however, the degree of neutral loss is highly variable and in some cases not present at all, depending on the peptide sequence[9]. The inability to predict neutral loss levels in CID MS2 from sequence alone reduces the effectiveness of phosphopeptide identification by common search engines by at least 40%[10]. Accurate models to predict the fragmentation of phosphopeptides would enable more sophisticated and discriminative methods for phosphopeptide identification. However, the gas phase chemistry of phosphopeptides is poorly understood. Peptide fragmentation during CID is generally explained according the mobile proton model[11], which assumes that fragmentation occurs either by charge directed mechanisms that require a proton at the site of cleavage, or charge remote mechanisms that do not require protonation but may involve participation of neighboring side chains. Thus, the pattern of fragmentation is strongly influenced by whether the peptide contains “mobile” protons that are not sequestered by basic residues and, consequently, free to migrate around the peptide.
The inverse correlation between neutral loss of H3PO4 and charge mobility in phosphopeptide MS2 has been well known for over a decade [9]. This led Tholey, et al. to propose a charge remote beta-elimination reaction of phosphate from pSer or pThr to form dehydrobutyric acid or dehydroalanine, respectively [12]. More recent work has shown strong experimental and computational evidence that neutral loss proceeds by a charge directed mechanism involving nucleophilic attack by a backbone carbonyl oxygen to form an oxazoline ring [13–16], which would require protonation of the phosphate. Intuitively, this would predict higher neutral loss in mobile proton phosphopeptides, contrary to previously observed trends. To address this discrepancy, formation of a stable hydrogen bonded structure between the phosphate and a protonated basic residue, usually arginine, in place of a mobile proton, has been proposed for conferring electropositive character to the phosphate when mobile protons are not present. These groups have also found that, despite the uncommonness of sole loss of HPO3, there is a significant competing pathway in which HPO3 is lost from the phosphosite with concurrent or sequential loss of water from elsewhere in the peptide [15, 17, 18].
Further evidence for charge directed, ring-forming mechanisms was found by several groups that independently identified neutral-loss-dependent cleavage of the backbone [19–21]. This diagnostic cleavage occurs between the α-carbon and amide carbon, one bond N-terminal to the ring that was formed by neutral loss. The resulting ion has been variously described as an “x-type ion” [19], a “y+10 ion” [20] and a neutral loss [21]. The diagnostic ion is seen both in neutral loss from pSer and pThr [19] and during neutral loss of water from the non-phosphorylated cognates [20]. Harrison also observed an additional ion attributed to a neutral loss mechanism analogous to oxazoline formation, but leading to a larger cyclized product incorporating one or more residues [21]. Although the influence of surrounding amino acid side chains on dissociation events was not systematically explored, these studies point to previously unappreciated mechanisms underlying neutral loss in phosphopeptides that are currently not accounted for by current peptide identification algorithms.
Previous studies of phosphopeptide fragmentation mechanisms are largely based on small sets of phosphopeptide MS2, limiting their applicability for developing a general model of phosphopeptide neutral loss. In this study, we comprehensively examine neighboring residue effects that determine rates of neutral loss in phosphopeptide CID MS2, using 34,057 spectra from public spectral databases supplemented by spectra obtained in our laboratory from biological and synthetic sources. Using a robust non-parametric statistic based on changes in the quantile distribution of neutral loss levels conditioned on local sequence features, we show that immediately adjacent residues and those up to seven residues distal to the phosphosite influence the total amount of observed H3PO4 neutral loss. Distal basic sites, most notably the N-terminus, show strong effects on neutral loss and suggest mechanisms contrary to the mobile proton model, in which immobile protons participate in charge directed mechanisms by forming secondary structures.
Experimental Procedure
Peptide libraries
Phosphopeptide libraries were commercially synthesized by solid phase synthesis on Wang resin (Genscript for libraries 1–10 or Anaspec for libraries 11–13) (supplementary Figure 1). Degenerate sites in library peptides were generated by adding a mixture of amino acids in certain coupling steps (Genscript) or by splitting the pool of peptides, performing parallel couplings and recombining (Anaspec). The libraries contain either 60 or 720 expected unique sequences with between one and four phosphorylation sites. The library sequences were designed with the following principles: no peptides within the library should be isobaric, sequence correlation should be minimized, and peptides should bear similar characteristics to those observed in biological datasets. Libraries were received as lyophilized powders. Peptides were solubilized with 5% formic acid, 95% water solution, agitated for approximately 2 minutes, then diluted to 0.1% formic acid prior to LC/MS/MS analysis.
WM239A dataset: Samples from cellular extract
Phosphopeptide samples were obtained from the human melanoma cell line, WM239A. Cells were lysed in boiling SDT buffer (100 mM Tris pH7.6, 4% SDS, 100 mM DTT) and lysate was sonicated for 15 s. Buffer exchange, iodoacetamide cysteine alkylation and trypsin digestion was performed by the FASP method [22]. Peptides were desalted on Oasis HLB columns (Waters) and dried by vacuum centrifugation. Peptides were fractionated prior to phosphopeptide enrichment, using ERLIC chromatography [23]. Briefly, peptides were solubilized in 70% acetonitrile, 20 mM ammonium formate (pH 2.2) and loaded onto a 4.6 mm × 150 mm PolyWAX LP column (PolyLC) at 1 mL/min using an Agilent 1100 HPLC (Agilent Technologies). Peptides were eluted as follows, collecting 1mL fractions: 0–5 min with buffer A (70% acetonitrile (MeCN), 20 mM ammonium formate pH 2.2), 5–15 min with a linear gradient to 100% buffer B ( 10% MeCN, 20 mM ammonium formate pH 2.2), 15–20 min linear gradient to 100% buffer C (10% MeCN, 1 M ammonium formate, pH 2.2), 15–20 min linear gradient to 100% buffer D (10% MeCN, 1% TFA), 20–24 min wash with buffer D, followed by re-equilibration of the column with buffer A. Fractions were concentrated by vacuum centrifugation to 12.5 μL.
Phosphopeptides were enriched using a batch method with titanium dioxide beads (Titansphere, GC Sicences) [24]. ERLIC fractions were diluted to a final volume of 400 μL with loading buffer (65% acetonitrile, 2% TFA, 140 mM glutamic acid). Titanium dioxide beads were washed in 65% acetonitrile, 2% TFA, followed by a wash with loading buffer, and added at a peptide-to-bead ratio (w/w) of 1:20 and rotated 15 min at room temperature. Beads were washed once with loading buffer, once with 65% acetonitrile, 0.5% TFA, and twice with 65% acetonitrile 0.1% TFA. Beads were resuspended in 0.1 mL of 65% acetonitrile 0.1% TFA and packed onto the top of a 200 μL C8 Stagetip (ThermoFisher). Phosphopeptides were eluted with 100 μL of 20% acetonitrile, 1% NH4OH into a receiving tube with 20 μL of 25% acetonitrile, 1% TFA to neutralize the pH. Remaining phosphopeptides bound to the C8 resin were eluted with two 100 μL volumes of 65% acetonitrile, 1% NH4OH into the same tube. Samples were dried by vacuum centrifugation.
Mass spectrometry
Dried phosphopeptides were solubilized in 0.1% formic acid and directly injected onto a BEH C18 column (25 cm × 75 μm i.d., 1.7 μm bead, 100 Å pore size, Waters, part #: 186003545) on a 2D nanoAcquity system (Waters) in direct injection mode. Peptides were eluted with a linear gradient from 95% buffer A (0.1% formic acid) to 30% buffer B (0.1% formic acid in acetonitrile) in 120 min at a flow rate of 300 nL/min. Mass spectrometry analysis was performed on a LTQ-Orbitrap (ThermoFisher). Survey scans were collected in the Orbitrap at 60,000 resolution (at m/z=300), and MS/MS sequencing was performed by CID in the LTQ in data-dependent mode, using monoisotopic precursor selection and rejecting singly charged and unassigned precursors for sequencing. The 10 most intense ions were targeted. After two observations of a peptide, dynamic exclusion of ±10 ppm mass lasting 180s was applied. The maximum injection time for MS survey scans was 500 ms with 1 microscan and AGC= 1×106. For LTQ MS/MS scans, maximum injection time for survey scans was 250 ms with 1 microscan and AGC= 1×104. Peptides were fragmented by CID for 30 ms in 1 mTorr of N2 with a normalized collision energy of 35% and activation q=0.25.
Phosphopeptide identification
MS/MS spectra were extracted with readw (version 4.3.1) and searched with Mascot (v 2.2, Matrix Science) against a human IPI 3.27 protein database showing up to two missed tryptic cleavages, with fixed modification of carbamidomethyl-cystein and methionine oxidation, N-terminal pyroglutamic acid (Gln), N-terminal acetylation, and phosphorylation on Ser, Thr and Tyr as variable modifications. Precursor m/z error was 20 ppm and fragment m/z error was 0.4 Da. A custom software pipeline was used to extract identifications from Mascot DAT files. Phosphopeptide identifications were accepted at 1% FDR at the peptide level determined by separate search of a database with reversed protein sequences. To ensure correct identifications, we further filtered for spectra that had a Mascot delta score of greater than 5, including to other phosphate localizations. This approximately equated to filtering on an Ascore of 20 with 94% of spectra having an Ascore of greater than 22. From these samples, we derived a dataset of 5,749 unique precursor ions containing a single pSer or pThr. All accepted ions were doubly or triply protonated.
PhosphoPep data set: Spectra from public spectral database
The PhosphoPep project at the Institute of Systems Biology (ISB) [25] provides a public database currently containing more than 30,000 phosphopeptide spectra obtained from several experiments. The database currently contains data derived from yeast, C. elegans, fly and human samples. The exact experimental details vary; however, in general, tryptic phosphopeptides were enriched from cellular extract and then analyzed by LC/MS/MS on either an LTQ-Orbitrap or LTQ-FTICR. Spectra are denoised. Replicates matched to the same peptide sequence and charge state are combined to create consensus spectra. Fragment ions are annotated using the SpectraST toolset [26]. In this study, all of the included spectra are assumed to be correct identifications. We obtained a database of 34,057 unique doubly or triply protonated peptide ions containing a single pSer or pThr. Similar results are obtained when using only the most confident 50% of spectra (xCorr score greater than 3.0, data not shown); however, the consequent reduction in the number of spectra reduces the statistical significance of the results, though all discussed trends remain significant. We found that 84% of the included spectra were well localized (Ascore greater or equal to 22), and the major results remained the same after filtering. We determined, however, that since identification and localization correlated with the amount of neutral loss, it was better to use the unfiltered data as misidentifications are expected to be unbiased.
Two important caveats for the use of consensus spectra became apparent in the analysis of the PhosphoPep data. Consensus spectra in this case were obtained through the averaging of multiple observed spectra of the same peptide. Firstly, the spectra varied widely in the amount of noise and the quality of annotation. When analyzing spectra that deviated greatly from observed trends in neutral loss, many proved to be the result of questionable annotation or especially noisy spectra. Removal of the lowest signal-to-noise spectra from the dataset, however, significantly lowered the number of spectra available. Second, the consensus spectra in the PhosphoPep data showed 12% lower neutral loss than our unprocessed spectra collected in our lab, an effect that we speculate is the result of peak voting procedures [27] that emphasize low-abundance ions and dampen high-abundance ions. This effect appears to be general to consensus spectra and is observed in other libraries that do not contain phosphopeptides (Supplementary Figure 5). Creating data mining metrics that were robust to these artifacts is essential to the effective use of these rich repositories of data.
Data management
Peptide spectra were stored in a custom-built ion-centric relational database (Supplemental Figure 2). For analysis, the sequence of identified peptides was encoded in a phosphosite-centric manner with phospho-residue at position 0. The distances of a phosphosite to each of the termini, the identity of the phosphorylated residue and the identity of each residue from one to 13 residues N-terminal to the phosphosite (positions −1 to −13) and one to thirteen residues C-terminal to the phosphosite (positions 1 to 13) were recorded. The termini were annotated one residue position distal to the terminal residues. Since peptides are of variable length, many peptides do not extend to all 27 residue positions in this encoding. For instance, the peptide XXpSXX has the N-terminus at the −3 position and amino acids at the −2, −1, 1 and 2 positions, while all other positions are outside of the sequence. Any position that is not within the peptide sequence are annotated as missing (‘−‘), thus placing all peptide sequences regardless of length on the same scale. In addition to the sequence information, the charge of the precursor ion was recorded.
Neutral loss analysis
Peaks were annotated with fragment ion assignments using a priority system similar to that used in Sun, et al [28]. Peaks were initially identified with a stringent tolerance of ±0.125 Thompson (Th) plus 200 parts per million (ppm). This mass window was determined from analysis of a few dozen problematic spectra to sensitively match most expected peaks; however, it often missed peaks that had been merged by the centroiding algorithms, most notably ammonia and water losses from multiply charged ions. Identifications matching within this window were then assigned to the ion with the following priority: neutral loss from the precursor, singly charged b- or y-ions, singly charged a-ions provided the corresponding b-ion was present, singly charged b- or y-ions with a single neutral loss provided the intact b- or y-ion was present, singly charged a-ions with a single neutral loss provided the corresponding a-ion was present, singly-charged b- or y-ions with multiple neutral losses provided the intermediate neutral loss ions are present. This list was then repeated for doubly- and triply-charged ions. If no matches were found, the mass tolerance was increased to 0.250 Th plus 400 ppm to account for poorly centroided peaks, and the process was repeated. If multiple identifications of the same priority were possible, multiple identifications were assigned. The total neutral loss in a spectrum is the total intensity annotated as having lost phosphoric acid divided by the total identified intensity. Peaks that have ambiguous neutral loss state had their intensities divided between the identifications.
Visualizing neutral loss patterns
To facilitate analysis robust to the presence of outliers and noise, we developed a nonparametric test to identify sequence-based factors that influence neutral loss. In this method, the overall frequency distribution of neutral loss intensity among all spectra is divided into three equal classes: the third of the spectra with the highest neutral loss is classified as high neutral loss, the middle third of the data is excluded, and the bottom third of the spectra is classified as low neutral loss. The metric is an odds ratio of high neutral loss to low neutral loss conditioned on putatively explanatory factors. Formally, this ratio is expressed as
| (eq. 1) |
where p(NL = high|AAi = X) is the probability that a spectrum is among the tercile of spectra with the highest neutral loss, given that it was chosen from the subset of spectra in which the amino acid at position i is X; p(NL = low|AAi = X) is the conditional probability for a spectrum being in the lowest tercile, and p(NL = high/p(NL = low) represents the prior odds of high versus low neutral loss. Since the data were divided into equal terciles, the prior odds are by definition 1, simplifying the equation to
| (eq. 2) |
Since we are using phosphosite-centric positional indexing, there is no guarantee that any given position is actually within the extent of the peptide. For example, if the phosphate is at the second position, there is no amino acid at the −2 to −7 positions. Rather than allowing sparse data, we explicitly encode these positions as “−“. Because the position of the phosphate within the peptide is a confounding factor to discovering sequence-specific effects, we corrected the prior odds at each position to exclude peptides that did not have an amino acid at the position. The resulting odds-ratio, which we use for the remainder of this work to measure the importance of an effect, is
| (eq. 3) |
A p-value for the significance of the observations can be obtained from a two-tailed binomial distribution. This p-value corresponds to the likelihood that a result at least as extreme as the observed would be obtained by chance, if the criterion had no effect on neutral loss. Since we are simultaneously testing 22 amino acids at each of 14 positions around the phosphate, the p-value was adjusted for 308 multiple hypotheses using the Sidak method [29].
The spectra were further divided by proton mobility. Mobile proton spectra are defined as having a charge greater than the sum of the number of arginines, lysines and histidines in the sequence. Immobile proton spectra were defined as having a charge less than or equal to the number of arginines in the sequence. All other spectra were classified as partially mobile proton. Once these subsets of spectra are generated, they are binned by tercile (likely with different cutoffs from before) and tested for odds-ratio, as above.
Results
Evaluation of the types and amounts of neutral loss observed
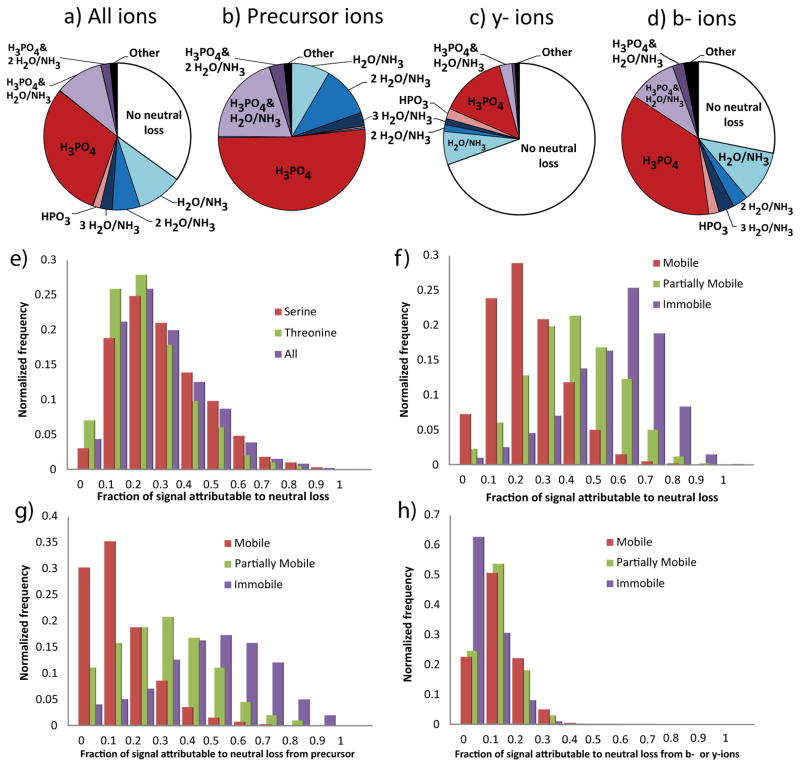
Heteroatom-containing side chains frequently exhibit neutral losses upon CID, providing competitive fragmentation pathways to H3PO4 loss in phosphopeptides. The resulting complexity in fragment ion patterns complicates de novo sequencing and identification by database search algorithms. Thus, improved understanding of the relative rates of major neutral loss events is essential for improving phosphopeptide MS2 interpretation. H2O and NH3 are the most commonly observed neutral losses during CID of protonated peptides. We first examined the prevalence of H2O and NH3 neutral loss events in combination with H3PO4 loss in a large set of 5749 validated phosphopeptide MS2 collected on an LTQ-Orbitrap instrument (WM239A dataset), comparing the ion intensity attributable to each neutral loss event normalized to the total ion signal (Figure 1a). Only 32% of ion intensity represented peaks showing no neutral loss. As expected, neutral loss of H3PO4 was the most common neutral loss event, followed by water and ammonia neutral losses, which differ by one Da and are often indistinguishable in low mass resolution CID, and thus are considered together in this analysis. Furthermore, all combinations of two distinct neutral losses accounted for significant signal, while ions displaying three or more neutral losses were uncommon.
Figure 1.
Neutral loss characteristics of the fragmentation spectra of 5749 unique tryptic peptide ions containing a single pThr or pSer derived from samples of the human cell line WM239A. (ad) Distribution of identifiable ion intensity by type of neutral loss observed. (a) represents the total signal within the spectra, including b- and y-ions that do not contain the site of phosphorylation. (b) Precursor ions include only those ions that do not have a cleavage at the backbone. (c) b-ions and (d) y-ions include only those ions of the given series that contain the site of phosphorylation. (e–h) The distribution of the fraction of identifiable signal annotated to result from the neutral loss of phosphoric acid, binned by increments of 10% of all signal. Separated by identity of the phosphorylated residue (e) or the charge mobility of the precursor ion (f). (g) Signal annotated as precursor ion neutral loss. (h) Same as (f) except that only signal annotated to have at least one cleavage of the peptide backbone counted.
When neutral loss from precursor ions and b-ion and y-ion series were treated separately, trends strongly deviated from the overall distribution of neutral loss (Figure 1b–d). Remaining intact peptide was not observed (Figure 1b), as would be expected due to resonant activation of the precursor ion. Interestingly, neutral loss of H3PO4 was more than 3-fold higher for b-ions compared with y-ions. Only 22% of y-ion intensity shows neutral loss, compared to 72% for b-ions, confirming trends suggested by a previous statistical learning study that examined over 3,000 spectra [30]. This indicates that ions that have undergone neutral loss form a product that is more susceptible to b-ion formation, or that b-ions are more likely to undergo neutral loss. One potential explanation is the difference in composition of b- and y-ions in tryptic peptides, which is absence of a C-terminal basic residue in b-ions. This difference in basicity would lower mobility of the remaining proton, or result in gas-phase interaction with the resulting H3PO4. However, the dramatic difference in b- and y-ion neutral loss persisted when ions were partitioned by the presence of basic residues. Importantly, this effect is not a result of the bias to the presence of basic residues on the C-terminus as it persists in peptides with non-tryptic C-termini and those constrained to have N-terminal arginine or lysine (Supplemental Figure 3). One implication of the greater observed stability of phosphate on y-ions in CID spectra is that precise localization of phosphorylation sites relies more heavily on y-ion series, since observation of intact phosphoresidues is required for unambiguous localization.
Variation in neutral loss between peptides
Models that estimate neutral loss based on global averages have been useful for increasing the number of phosphopeptides identifications when incorporated into search algorithms [30]. However, if neutral loss rates vary widely between peptide MS2 in a sequence dependent manner, such methods are likely to introduce biases against outliers to the main trend. To assess global variability of H3PO4 loss in phosphopeptide CID, we examined the distribution of neutral loss over all spectra in the WM239A dataset (Figure 1e). For peptides containing either pSer or pThr, the observed neutral loss of H3PO4 varies from undetectable to accounting for the entire signal. pSer shows slightly more neutral loss than pThr, as noted previously [31]. Interestingly, this trend is opposite of the analogous neutral loss of water from unmodified serine and threonine [31], suggesting that the loss of water and loss of H3PO4 have different rate limiting factors. That pSer, while less basic than pThr, shows higher neutral loss, suggests that proton availability is not rate limiting in the neutral loss of H3PO4 as would be predicted for charge directed mechanisms involving direct protonation of the phosphate. While the difference between neutral losses from pSer and pThr is useful for inferring mechanism, the magnitude of the difference is very small compared to the overall variation in neutral loss. Thus, other factors must contribute to the variability of H3PO4 loss.
Previous studies have noted an inverse correlation between proton mobility and the amount of H3PO4 neutral loss of in small sets of phosphopeptide MS2, with lower charge mobility correlating with higher neutral loss [9]. To quantify this effect over a significantly larger data set, we examined the distribution of neutral loss in spectra stratified by proton mobility (Figure 1f). The differences between mobility groups accounted for 24% of the observed variance in neutral loss, a much greater effect than was observed between pSer and pThr. However, the variance within each group is still large, suggesting other factors are important in loss of H3PO4.
The vast majority of the difference in neutral loss between charge mobility classes is the result of differences in neutral losses from the parent, the ‘M-98’ peak and its derivatives (Figure 1g). All classes of charge mobility demonstrate similar levels of sequence ions displaying neutral loss (Figure 1h). Sequence ions displaying neutral loss are especially problematic for peptide identification, since the presence of multiple correlated ion series poses a significant risk of spurious matching for identification algorithms. Anecdotally, the spectra that suffered the worst depression of identification scores were those which displayed intermediate levels of neutral loss, meaning no one series was dominant.
Effects of local sequence on neutral loss
We next examined whether local sequence influences neutral loss. Although the WM239A dataset was nearly twice as large as the largest previous study on phosphopeptide neutral loss [30], we found that statistically meaningful estimates of sequence effects required much larger sets of phosphopeptide MS2. The Institute for Systems Biology maintains the PhosphoPep project as a repository of MS2 spectra obtained from large-scale phosphoproteomics studies. By extracting spectra containing a single pSer or pThr from this data, we developed a dataset containing 34,057 unique phosphopeptide spectra, more than 10 times larger than in previous studies.
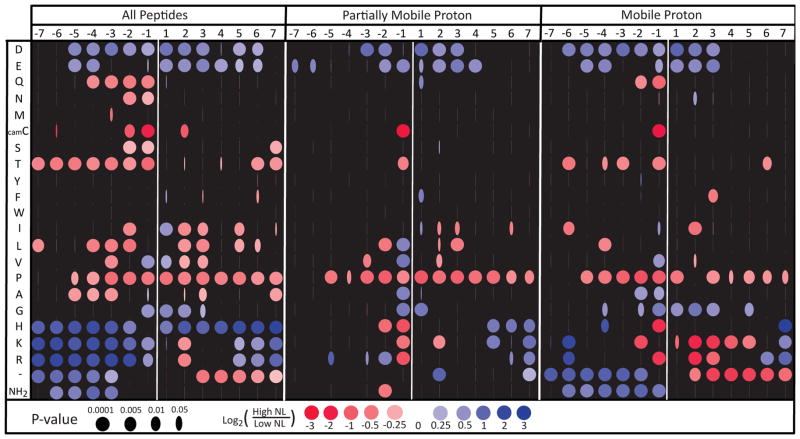
To assess the effect of localized residues in the presence of intensity biases, we developed a metric that is robust to wide variability in the underlying data, poor spectral quality, mis-annotation, and distorted intensity distributions. We found that problems associated with variability and noise were solved by considering only large changes in neutral loss using a non-parametric quantile-based statistic. Spectra were binned by tercile: high neutral loss contained the third of the spectra with the highest neutral loss, while low neutral loss contained the third with the lowest. Within a subset of the spectra (for instance those spectra that contain alanine immediately N-terminal to the phosphosite), the ratio of the number of spectra in the upper versus the lower terciles defines the odds ratio on neutral loss. This metric, combined with the size of the PhosphoPep dataset, allowed us to assess the positional effects on neutral loss of each amino acid within seven residues of the phosphosite while retaining statistical significance (Figure 2, left panel). Multiple hypothesis correction was essential for controlling false discovery rate; an uncorrected version of Figure 2 is included as Supplementary Figure 6.
Figure 2.
The effects of peptide sequence adjacent to the site of phosphorylation on 34,057 unique spectra from the ISB PhosphoPep database. The horizontal axis represents the position of residues from 7 sites N-terminal to 7 sites C-terminal relative to the phosphorylated residue. The vertical axis denotes the presence of a particular amino acid at the position. ‘−‘ indicates that the peptide is not long enough to contain the site, thus ‘−‘ serves as a cumulative marker for the peptide terminus. NH2 represents the N-terminus. A blue dot indicates that the peptides with the specified amino acid at the specified position are more likely to have high neutral loss than low neutral loss. Red dots indicate that low neutral loss is more likely. Low neutral loss is defined as having neutral loss less than at least two-thirds of the peptides in the dataset (bottom tercile). High neutral loss is defined as greater than at least two-thirds dataset (top tercile). The width of the iris represents the multiple-hypothesis-corrected probability that this ratio could occur by chance if the indicated residue had no effect on neutral loss.
Residues immediately adjacent to the phosphosite show the strongest effect on neutral loss of H3PO4. We examined all mobility classes in aggregate, observing that when proximal to pSer or pThr, glycine, basic amino acids and acidic amino acids appear to increase neutral loss (Fig. 2, left panel). Proline reduces neutral loss, but other aliphatic side chains increase neutral loss when immediately adjacent. Threonine, serine, carbamidomethyl-cysteine, glutamine, and asparagine all reduce neutral loss when N-terminal to the phosphosite. The effects of distal residues were surprisingly important. Twelve of the twenty amino acids significantly affect neutral loss even when only considering positions more than five residues away from the phosphosite.
Simpson’s paradox: controlling for charge mobility, proximal basic residues suppress H3PO4 neutral loss
The number of basic residues in a protonated peptide negatively correlates with charge mobility, and as shown previously in Figure 1, neutral loss increases with decreasing charge mobility. Thus, the positive effect of nearby basic residues on neutral loss could simply reflect a correlation with previously shown effects of charge mobility. To assess this possibility, we repeated the flanking residue analysis with the data stratified by charge mobility (Figure 2, center and right panels, Supplementary Figure 4). The lack of available data from immobile proton spectra reduces the number of results that attain significance after multiple testing corrections (Supplementary Figure 4). The only significant results for immobile proton cases show that arginine at the −1 position strongly inhibits neutral loss and that proximity to the C-terminus enhances neutral loss. Mobile and partially mobile cases are observed at high enough frequency that many significant effects can be observed.
Surprisingly, in contrast to the positive effect of proximal basic residues on neutral loss in the global analysis, mobile and partially mobile MS2 show decreased neutral loss when basic residues are adjacent to the phosphoresidue. Separating the effects by charge mobility reveals that the basic residue effect observed in the global analysis is reversed within each mobility subgroup, driven by an underlying correlation between the presence of basic residues and the charge mobility of the peptides. This counterintuitive trend is an example of Simpson’s paradox, which arises when a correlation present in the aggregate population disappears or reverses in each and every subgroup upon stratification [32]. We propose that basic residues directly slow neutral rates when proximal to the phosphoresidue. However, in non-mobile proton MS2, the lack of available protons slows the aggregate of other backbone fragmentation pathways to a greater degree, increasing apparent neutral loss of H3PO4 due to reduced fragmentation pathway competition. Thus, the previously published claims that basic residues increase the probability of H3PO4 neutral loss may be based on a statistical artifact of aggregating data across mobility classes.
By controlling for the shifts in overall backbone fragmentation caused by changes in proton mobility, we can more closely examine the direct variation in neutral loss caused by basic residues. The only observed trend with basic residues that was universal across all charge mobility classes is that basic residues at the −1 position strongly inhibit neutral loss. We hypothesize that this effect occurs because the protonated basic residue will form a very stable hydrogen-bonded structure with the phosphate, producing steric hindrance that inhibits formation of an oxazoline and subsequent neutral loss. In mobile proton spectra, basic residues from the −2 to the +5 position inhibit neutral loss, although some of these are not significant after multiple-testing correction. We suspect that proximity of basic residues to the phosphoester facilitates hydrogen bond formation, leading to stable hydrogen-bonded phosphates. These bonded complexes prevent the phosphate from interacting with mobile protons or less stable charge donors, such as the N-terminus, that could better catalyze the neutral loss reaction. As proton mobility decreases, the presence of nearby basic residues is less detrimental to neutral loss, suggesting that, absent other reactive acidic sites, the hydrogen bonding of protonated-bases to the phosphate is less inhibitory. At the lowest levels of proton availability, the base-catalyzed mechanisms may be the prevalent method of neutral loss.
The N-Terminus is the primary driver of neutral loss in mobile proton peptides
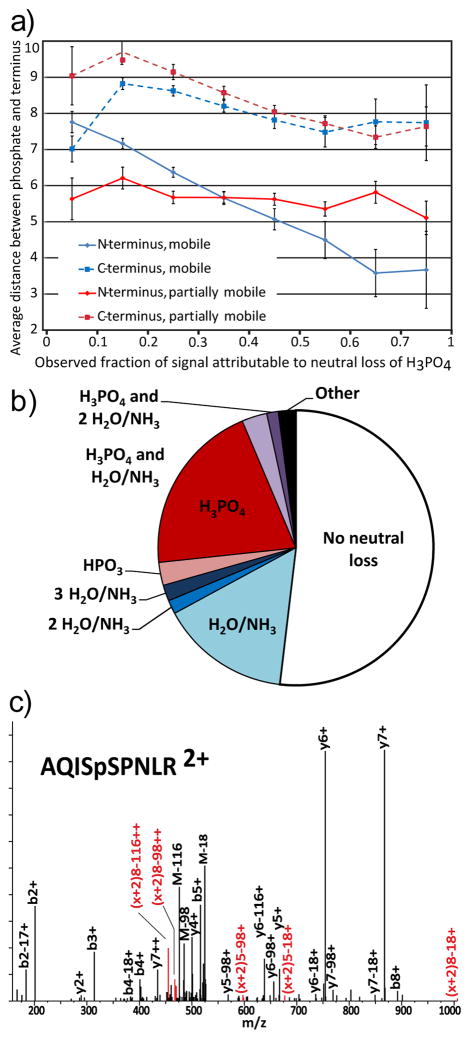
The effect of the N-terminus on neutral loss of H3PO4 varies depending on proton mobility states. In spectra with immobile protons, proximity to the N-terminus inhibits neutral loss, with partially mobile protons the terminus has minimal effect, and with mobile protons the N-terminus strongly enhances neutral loss when 1 to 6 residues away. To test the extent of this effect, we stratified the spectra by level of neutral loss and examined the average distance of the phosphosite to the termini (Figure 3a). For peptides with mobile protons, those with low neutral loss tended to be more than seven residues from the N-terminus, while being within four residues was required for the highest levels of neutral loss. While proximity of the phosphoresidue to the N-terminus strongly enhances neutral loss, it is not sufficient; many such peptides still exhibit low neutral loss, indicating that the N-terminus alone is not sufficient for the elevation of neutral loss. Despite this lack of sufficiency, proximity to the N-terminus explains 4% of the variance in neutral loss from phosphopeptides with mobile protons, a larger effect than was observed for any other explanatory factor (Supplementary Table 1).
Figure 3.
a)The observed changes in proximity to the N-terminus (solid lines) and the C-terminus (dashed lines) in mobile (blue) and partially mobile (red) based on the observed amount of neutral loss displayed by a peptide. b) Distribution of neutral loss in b-ions derived from 37 peptides with acetylated N-terminii. c) Spectrum of doubly protonated AQISpSPNLR. Red markers with the ion series denoted (x+2)indicate ions annotated as the x-type ions that have been reported as markers of oxazoline or macrocycle formation.
Enhancement of neutral loss by N-terminal proximity suggests involvement of the N-terminal amine group in the mechanism of H3PO4 loss. However, peptide length, proximity to the C-terminus, or other correlated factors could also explain the observed effects. To assess the importance of the N-terminal amine, we examined 37 mobile proton phosphopeptides from the WM239A dataset with acetylated N-termini. Acetylation of the N-terminus reduces the basicity of the N-terminus. Consistent with the hypothesis of direct action, the acetylated peptides displayed on average 42% less neutral loss than the rest of the data. Interestingly, the distribution of neutral losses from the acetylated b-ions is more similar to that of amino-N-terminated y-ions than amino-N-terminated b-ions (Figure 3b). Thus, the bias of high neutral loss from b-ions is partially explained by the effects of proximity to the N-terminus. The effects of N-terminal acetylation imply that direct action by the N-terminal amine is responsible for the activation of neutral loss.
The N-terminus may participate in neutral loss of H3PO4 by nucleophilic attack or charge stabilization. Mechanisms involving nucleophilic attack by the N-terminus, such as the formation of diketopiperazine b-ions, also show maximal effect at the 2nd peptide bond [33]. However, ab initio modeling studies indicate that nucleophilic attack by the N-terminus on the α-carbon of pSer and pThr to be higher energy than oxazoline pathways for neutral loss, even under optimal conditions [34]. Furthermore, nucleophilic attack requires the N-terminus to be uncharged. Since the N-terminus is predominantly protonated in mobile but not in partially mobile cases, such mechanisms would be expected to be at least as prevalent in partially mobile cases as they are in the mobile cases. Consequently, the observation that proximity to the N-terminus enhances neutral loss in mobile proton spectra but not in partially mobile implies that the enhancement is dependent upon action by a protonated N-terminus. As has been suggested by several previous studies[14–16], this indicates that the neutral loss of phosphoric acid by the charge directed oxazoline mechanism is more efficient when the charge is stabilized on a more basic site. However, the increased effects of the N-terminus interacting with the phosphate (Scheme 1) rather than other charge donors are novel. Similarly to the inhibition by proximal basic residues, this indicates that, especially as charge mobility rises, small changes is the stability of base-phosphate pairs can be critical in determining whether the interactions stabilize the phosphate or catalyze its loss.
Interaction of a charged N-terminus and the phosphoester group might bring the neighboring N-terminal amide carbonyl in proximity to the α-carbon of the phosphorylated residue, facilitating nucleophilic attack and formation of large cyclic structures after elimination, similar to those that Harrison [21] proposed in neutral loss of water from unmodified serine and threonine. To assess this possibility, we looked for the x-type ion, here denoted (x+2), derived from cleavage at the first alpha carbon-carbonyl carbon bond, reported previously as a marker for this macrocyclic-cleavage [21]. While less common than the x-type ion immediately N-terminal to the site of phosphorylation [19], peaks consistent with macrocycle x-type ions were found in MS/MS of several peptides. The spectrum of doubly protonated AQISpSPNLR shows an especially expressive example (Figure 3c).
The (x+2)8 ions suggest the formation of a macrocycle, while the (x+2)5 ions indicate the formation of an oxazoline. Unexpectedly, in cases where an x-ion was present, we also observed a charge-reduced form, presumably due to loss of H3O+. The presence of x-ions distal to the site of phosphorylation suggests that charge stabilization is leading to the formation of macrocycles in the neutral loss of phosphoric acid (Scheme 2).
Proline provides a competitive pathway and reduces backbone flexibility required for distal basic interactions
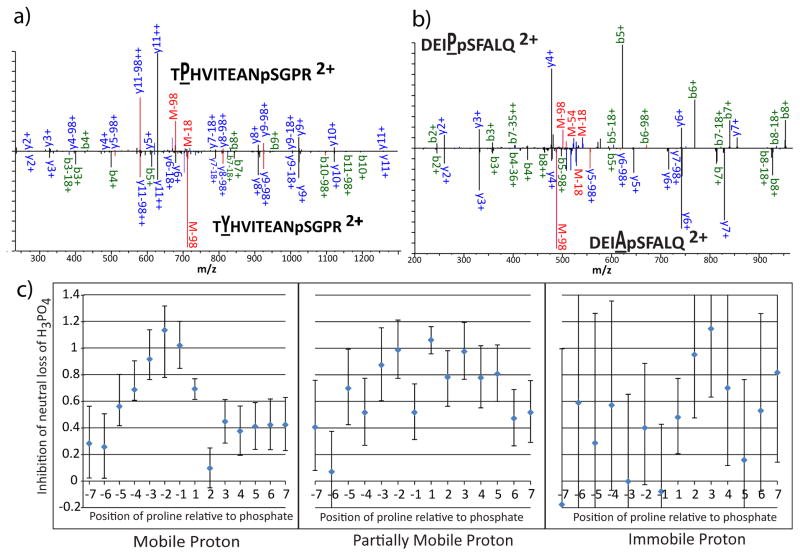
The analysis of the effects of flanking residues (Figure 2) reveals that proline strongly inhibits neutral loss regardless of position. We initially ascribed this effect to competition from the well-known enhancement of backbone cleavage N-terminal to proline [35]. We examined cases from the synthetic phosphopeptide library dataset where the effect of single proline substitutions could be evaluated, and found evidence for competition in some spectra. An extreme example of this effect can be seen in Figure 4a, which compares the spectra of doubly protonated TPHVITEANpSGPR and TYHVITEANpSGPR, differing only by the substitution of tyrosine for proline at the 2nd position. Proline at the second position strongly activates formation of the y11++ ion relative to a tyrosine at that position. The ratio of the M-98 peak to other sequence ions is similar between the two spectra. Spectra such as this indicate that competition is an important factor in the inhibition by neutral loss; however, it does not fully explain the effect. A second group of proline-containing spectra shows a decrease in neutral loss peaks relative to all other ions. For instance, Figure 4b shows a spectrum in which the change of an alanine to a proline one residue N-terminal to the phosphosite significantly reduces the amount of neutral loss relative to the b- and y-ion series and neutral losses of water and ammonia. Competition does not explain the change in ion ratios, indicating that these prolines may interfere with the mechanisms for neutral loss.
Figure 4.
The effects of proline on neutral loss. a) Comparison of the spectra of doubly-protonated TXHVITEANpSGPR where X=P in the top panel and X=Y in the bottom panel. For lettering: blue denotes y-ions, green denotes b-ions, and red denotes losses from the precursor. For ion markers: black denotes no neutral loss, red denotes loss of H3PO4, blue denotes loss of water or ammonia, purple denotes loss of H3PO4 and water or ammonia. b) Comparison of the spectra of doubly protonated DEIXpSFALQ where X=P in the top panel and X=A in the bottom panel. c) Representation of the increased chances of peptides from the ISB dataset being in the lowest tercile of neutral loss, given that there is a proline at the indicated position relative to the phosphosite. The y-axis is in units of log2 of the enrichment. Error bars represent the 95% confidence interval for the mean.
Proline’s 5-membered ring reduces the flexibility of peptide backbones, creating steric hindrance and preventing the formation of cyclic conformations, or “loops”, that facilitate intramolecular charge solvation. Given the evidence that neutral loss is enhanced by structures that bring the phosphate into proximity with immobilized protons, we hypothesized that proline’s direct reduction of neutral loss was caused by inhibiting loop formation. To examine this possibility, we plotted the magnitude of inhibition as a function of position relative to the phosphosite (Figure 4c). In peptides with mobile protons, proline shows strong inhibition when N-terminal to the phosphosite, consistent with interference with formation of loops required for interaction between the phosphate and the N-terminus. Proline slightly inhibits neutral loss when C-terminal to the phosphosite or when it is more than five residues N-terminal. In peptides with partially mobile protons, the presence of proline reduces neutral loss regardless of whether the proline is-terminal or N-terminal to the phosphate. This suggests that, in addition to the enhancement by the N-terminus that was observed with mobile protons remains, loops that allow the phosphate to interact with the basic residue at the C-terminus encourage neutral loss in partially mobile cases. In immobile cases, proline shows most significant inhibition of neutral loss when C-terminal to the phosphosite. This can be rationalized under the loop-forming hypothesis since the N-terminus is expected to be uncharged in these peptides, leaving tryptic C-terminus as the only site that is generally protonated in these peptides.
Aspartic and glutamic acid
Aspartic and glutamic acid both increase the amount of neutral loss from position −6 to +5. It is possible for a carboxylic acid to initiate neutral loss through nucleophilic attack, yielding an acid anhydride, which would rapidly eliminate phosphoric acid under CID conditions. This has been proposed as the mechanism for neutral loss of HPO3 in pTyr [36, 37]. Reactions of this kind would be compatible with previous experiments which indicate that a competing neutral loss reaction pathway exists in which HPO3 is lost from the phosphoresidue concomitantly with the loss of water from elsewhere in the peptide [17]. This reaction would be isobaric and indistinguishable from neutral loss of phosphoric acid. Acid-mediated direct attack would elegantly explain the experimental prevalence of the combined pathway with the rarity of observing the loss of only HPO3. Alternatively, a carboxylate group could directly donate a proton to catalyze the reaction or simply compete with the phosphate for hydrogen bonding with basic sites, thus disrupting conformations that stabilize the phosphate rather than catalyze neutral loss.
To test the plausibility of direct action by acidic residues, we examined the phosphopeptide libraries for cases where a single acidic residue replaced another residue. Contrary to the strong enhancement suggested by the analysis of the PhosphoPep database, substitution incorporating single acidic residues has minimal effect on the observed amount of neutral loss. Indeed, single replacements tend to slightly lower the observed neutral loss rather than increase it. This is in agreement with the observations of Cui, et al who found that mutating acidic residues did not significantly affect neutral loss [17]. It should be noted the presence of one acidic residue greatly increases the chances of finding more acidic residues due to the presence of acidophilic kinase motifs in biological data sets. Thus, the increased neutral loss observed for nearby acidic residues in the global analysis shown in Fig 2 may be an aggregate effect of multiple acidic residues, rather than an effect particular to that position. We attribute the rise in the rate of neutral loss to the creation of an extensive hydrogen bond network within the peptide that is able to modulate or stabilize the transfer of protons to the phosphate.
Neutral loss of water and ammonia from threonine and other amino acids interferes with the loss of H3PO4
While aspartic and glutamic acid showed a tendency to increase neutral loss, other amino acids with propensity for loss of water or ammonia from their side chains reduced the observed neutral loss of H3PO4, especially when these amino acids are N-terminal to the phosphate and in peptides with mobile protons (Figure 2). Because this N-terminal bias is similar to the N-terminal enhancement of neutral loss of H3PO4, we hypothesized that a similar mechanism leads to analogous loss of water from modified and unmodified Ser and Thr, along with the loss of water and ammonia from asparagine, glutamine, and carbamidomethyl-cysteine.
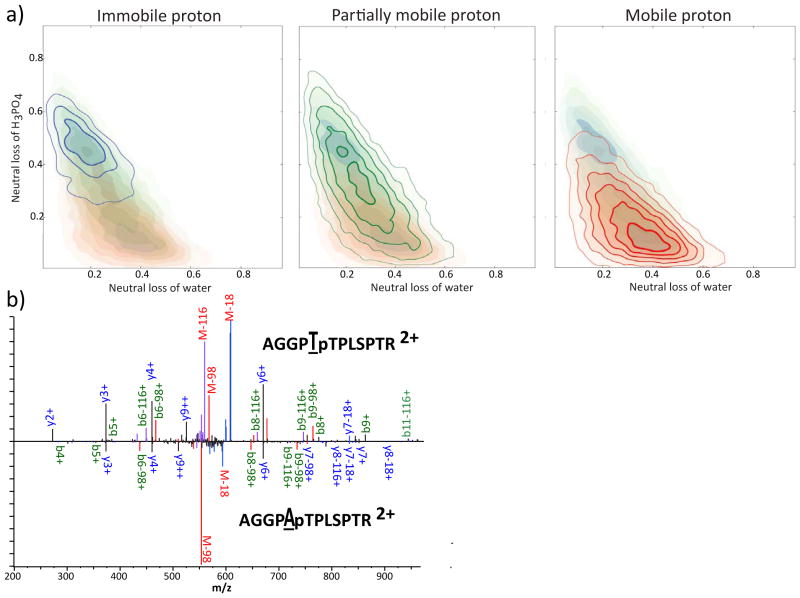
If neutral loss of water and ammonia competes with the loss of H3PO4, there should be negative correlation between the two pathways. To assess the relationship between the pathways, we examined the distribution of the neutral losses of water and H3PO4 among spectra (Figure 5a), revealing a negative correlation between the intensity of the two neutral loss events. Immobile proton cases favor the neutral loss of H3PO4, while mobile cases show higher loss of water. The reduction in neutral loss of water in peptide ions with fewer mobile protons implies that proton availability is a rate-limiting factor. While the difference in neutral loss between proton mobility groups is striking, within groups there remains a strong negative correlation between water and H3PO4 loss pathways. The immobile and partially mobile cases exist over a broad range of neutral loss propensity for both H2O and H3PO4. In spectra with mobile protons, those peptides with high neutral loss of H3PO4 show negative correlations between the neutral loss of H2O and H3PO4 similar to the correlations seen in immobile and partially mobile peptides. However, for low neutral loss of H3PO4, neutral loss of water becomes uncorrelated with loss of H3PO4. This suggests two regimes of neutral loss in mobile proton cases: one in which there is high water loss that has effectively shut off loss of H3PO4 and another where the two neutral loss pathways compete.
Figure 5.
a) Contour plots showing the density of spectra when plotted by neutral loss of water or ammonia (x-axis) and neutral loss of H3PO4 as a fraction of overall ion current. Ions that showed loss of both were ignored. Shaded areas duplicate the data in the other graphs for ease of comparison. Contours with blue shading indicate cases with immobile protons, green indicate partially mobile, and red indicate mobile. The contour lines increase with the square of density of spectra; the innermost contour has 25-fold more density than the outer in mobile proton and partially mobile proton cases, and 9-fold more density in immobile proton cases. b) Comparison of the spectra of doubly-protonated AGGPXpTPLSPTR where X=T in the top panel and X=A in the bottom panel.
To determine what characteristics might be unique to these ions showing very low neutral loss of H3PO4, we examined the sequences of these peptides. When there is a mobile proton, peptides that exhibit high water neutral loss (fraction of signal showing neutral loss of water greater than 0.4) and low phosphoric acid neutral loss (less than 0.2) contain 17% more serines and 85% more threonines than the average peptide in the dataset. Threonines that are N-terminal to the site of the phosphate are especially enriched, being more than twice as prevalent as in other peptides. The number of threonines N-terminal to the phosphate explains 3.6% of the variance in the neutral loss of H3PO4 from mobile proton phosphopeptides. This competition explains the general decrease of neutral loss of phosphoric acid when there is a threonine N-terminal to the phosphate (Figure 2). As an example of this effect the spectra of AGGPTpTPLSPTR and AGGPApTPLSPTR, which differ only by the substitution of threonine for alanine at position 5, are shown in Figure 5b. The addition of the threonine adds a highly active pathway for the neutral loss of water, as evidenced by the appearance of very strong M-18 and M-116 ions. This shows that the mechanism for the reduction of neutral loss of H3PO4 is competition with the loss of water.
While it is possible for the N-terminus to perform nucleophilic substitution directly [38], it must be unprotonated to do so. Consequently, we would expect that this mechanism would predominate in partially mobile and immobile proton cases, not mobile proton cases. Therefore, it is likely that the N-terminus is stabilizing the charge required for neutral loss of water from unphosphorylated threonine, in the same way proposed for phosphorylated residues (Scheme 3). It is possible that coordination of the N-terminus and the phosphate brings the terminus into closer contact with the unmodified alcohol side chain, enhancing the loss of water. The mobility-dependent reduction of neutral loss of water implies that the availability of protons is a rate-limiting factor for neutral loss of water. Thus, we hypothesize that threonine is more effective than serine because the gas-phase basicity of its side chain is about 14 kJ/mol higher than serine [39], allowing the proton to be more readily transferred to threonine. Competition for a charge-stabilizing partner could explain the strong negative correlations that are observed between the neutral loss pathways. The inhibition of neutral loss of phosphoric acid by N-terminal alcohols, implies that N-terminally catalyzed neutral loss proceeds through the direct loss of H3PO4, rather than combined loss of water and HPO3, in accordance with the observation that the combined loss becomes less prevalent as charge mobility increases [17].
Discussion
In this work we present a large statistical study of the effects of peptide sequence on the neutral loss of phosphoric acid from phosphopeptides under CID. While stereotypical images of phosphopepdide spectra involve dominant peaks representing loss of phosphoric acid from the precursor, it exceedingly rare for more than two-thirds of ion current to be sequestered in that peak. In practice the degree of neutral loss is highly variable, with spectra presenting as many as six competing neutral loss series. Here we show that much of the variability can be explained by the peptide sequence near the phosphate. The data suggests that, as has been shown previously, neutral loss is primarily catalyzed by charge pairing between the phosphate and nearby bases. However, small fluctuations in the nature of this pairing can have dramatic effects on the resulting neutral loss. Consequently, nearby bases may counterintuitively reduce neutral loss through the formation of stable charge solvation on the phosphate. Neutral loss is preferential if less stable pairings, most notably with the N-terminus are possible. These effects may be further modulated by reduced peptide flexibility due to proline and direct competition with the neutral loss of other small molecules.
Previous studies have struggled to reconcile the charge-directed model of H3PO4 neutral loss with the observation that neutral loss pathways decrease when protons become more available [9, 12, 13]. The paradox has been explained by direct catalysis by protonated basic residues via non-covalent interactions between the phosphate and nearby arginines [14]. At first glance, this view appears to be supported by our evidence when examined over all spectra that the presence of basic residues increases the amount of neutral loss. However, in an example of Simpson’s paradox, when we stratify the spectra by charge-mobility, we find evidence for the converse, that nearby bases inhibit neutral loss regardless of mobility class. Furthermore these effects were not limited to adjacent residues but extended across the entire peptide. Additionally, the N-terminus exerts a strong positive effect on H3PO4 neutral loss when both proximal and distal to the phosphorylation site, making it the primary predictor of neutral loss in mobile proton peptides.
These observations suggest a model in which the phosphate coordinates with protons that are immobilized at basic sites. Because these complexes are constitutively present, the basicity of the phosphate itself becomes unimportant in determining the rate of neutral loss. By this model, the rate is dominated by the basicity of the hydrogen bonded basic group. This gas-phase basicity then determines the partial charge that resides on the phosphate and consequently the susceptibility of the phosphorylated side chain to nucleophilic attack. The N-terminus, therefore, becomes the ideal driver of neutral loss because it is the least basic site that is protonated with high occupancy, making it the ideal catalyst. This equilibrium may be further modified by the participation of other side chains, such as by proton donation from the carboxylic acids on Glu and Asp. Because these base-mediated complexes would be stable in the presence of mobile protons, the reaction rate of H3PO4 neutral loss increases more slowly with charge availability than the rate of backbone fragmentation.
The neutral loss of water from Ser and Thr is inversely correlated with the loss of H3PO4 from pSer and pThr, suggesting that the formation of stable complexes between protonated bases and the alcohol side chains is less favorable. Consequently, the formation of the complex becomes rate-limiting, while even small partial charges on the Ser or Thr side chain are enough for the neutral loss of water. Interaction with the N-terminus still appears to be an important determinant for these neutral losses and the competition for charge pairing with the terminus defines the neutral loss pattern. It further appears that other amino acids with side chains capable of neutral loss show effects when near the N-terminus, suggesting that interaction of side chains with immobilized protons may be the general mechanism of neutral loss in peptides
It is worth noting that neutral loss reactions facilitated by intramolecular interactions with charge donors cannot be usefully described by the mobile proton model. The mobile proton model posits three reaction mechanisms: charge remote mechanisms, charge directed mechanisms involving a mobile proton, and charge directed mechanisms involving a proton immobilized at the site of the reaction. We suggest that neutral loss reactions are catalyzed by a proton that is immobilized distal to the reaction site; however, by the adoption of secondary structures, it interacts with the reaction site. Under this model, the peptide acts more like an irregular solvation shell around the proton, than a series of discrete protonation sites. While the conjugation of charge between multiple bases is important in all CID reactions, the evidence here suggests that it may be especially important for neutral losses. This may explain some of the difficulties in the prediction of neutral loss using kinetic models based on the mobile proton model [40–42].
This study demonstrates the importance of using large datasets to achieve the statistical significance necessary to detect sequence based factors that influence neutral loss propensity. Large datasets permit testing of multiple hypotheses at the scale necessary to guard against inflation of type I errors without excessively lowering sensitivity. While obtaining large libraries of spectra locally is prohibitively difficult, we exploited the recent availability of large curated MS2 phosphopeptide libraries, which enabled large-scale analysis of local sequence effects on H3PO4 neutral loss. Robust, non-parametric statistics were required to overcome the reduction in control of both data acquisition and data processing inherent in publically available curated data.
Peptide identification algorithms can be improved with more accurate models of MS/MS intensities predicted with empirical kinetic models of peptide fragmentation [28, 43]. Accurate prediction of neutral loss levels of H3PO4 in phosphopeptide CID is problematic, even with sophisticated kinetic models [42], likely due to distal sequence effects documented here. The additional ions and poorly understood effects on product ion abundances due to these neutral loss pathways lowers the accuracy of conventional peptide identification algorithms, since the neutral loss provides information that is at best uninformative and usually confounding. This proves especially true with algorithms that attempt to use the intensity patterns in fragment ions to infer sequence information. The effects of neighboring residues could be incorporated easily into existing statistical or kinetic models to improve the prediction of the amount neutral loss. Furthermore, knowledge of site specific variations in expected neutral loss could be incorporated into phosphorylation site localization methods to increase confidence levels. By these methods the variability of neutral loss becomes informative in phosphopeptide identification rather than a detriment.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 CA155453 (W.M.O.). We would like to acknowledge the assistance of Veronica Bierbaum in preparing and editing this manuscript.
Literature Cited
- 1.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268:5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 2.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5:4052–61. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 3.Old WM, et al. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34:115–31. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 5.Villen J, Beausoleil SA, Gygi SP. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics. 2008;8:4444–52. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulintz PJ, Yocum AK, Bodenmiller B, Aebersold R, Andrews PC, Nesvizhskii AI. Comparison of MS(2)-only, MSA, and MS(2)/MS(3) methodologies for phosphopeptide identification. J Proteome Res. 2009;8:887–99. doi: 10.1021/pr800535h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8:4466–83. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann WD, Kruger R, Salek M, Hung CW, Wolschin F, Weckwerth W. Neutral loss-based phosphopeptide recognition: a collection of caveats. J Proteome Res. 2007;6:2866–73. doi: 10.1021/pr060573w. [DOI] [PubMed] [Google Scholar]
- 9.DeGnore JP, Qin J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:1175–88. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 10.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 11.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. J Mass Spectrom. 2000;35:1399–406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Tholey A, Reed J, Lehmann WD. Electrospray tandem mass spectrometric studies of phosphopeptides and phosphopeptide analogues. J Mass Spectrom. 1999;34:117–23. doi: 10.1002/(SICI)1096-9888(199902)34:2<117::AID-JMS769>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Reid GE, Simpson RJ, O’Hair RA. Leaving group and gas phase neighboring group effects in the side chain losses from protonated serine and its derivatives. J Am Soc Mass Spectrom. 2000;11:1047–60. doi: 10.1016/S1044-0305(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 14.Palumbo AM, Tepe JJ, Reid GE. Mechanistic insights into the multistage gas-phase fragmentation behavior of phosphoserine- and phosphothreonine-containing peptides. J Proteome Res. 2008;7:771–9. doi: 10.1021/pr0705136. [DOI] [PubMed] [Google Scholar]
- 15.Palumbo AM, Reid GE. Evaluation of gas-phase rearrangement and competing fragmentation reactions on protein phosphorylation site assignment using collision induced dissociation-MS/MS and MS3. Anal Chem. 2008;80:9735–47. doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 16.Rozman M. Modelling of the gas-phase phosphate group loss and rearrangement in phosphorylated peptides. J Mass Spectrom. 2011;46:949–55. doi: 10.1002/jms.1974. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Yapici I, Borhan B, Reid GE. Quantification of competing H3PO4 versus HPO3 + H2O neutral losses from regioselective 18O-labeled phosphopeptides. J Am Soc Mass Spectrom. 2014;25:141–8. doi: 10.1007/s13361-013-0744-4. [DOI] [PubMed] [Google Scholar]
- 18.Cui L, Reid GE. Examining factors that influence erroneous phosphorylation site localization via competing fragmentation and rearrangement reactions during ion trap CID-MS/MS and -MS(3.) Proteomics. 2013;13:964–73. doi: 10.1002/pmic.201200384. [DOI] [PubMed] [Google Scholar]
- 19.Kelstrup CD, Hekmat O, Francavilla C, Olsen JV. Pinpointing phosphorylation sites: Quantitative filtering and a novel site-specific x-ion fragment. J Proteome Res. 2011;10:2937–48. doi: 10.1021/pr200154t. [DOI] [PubMed] [Google Scholar]
- 20.Kilpatrick LE, Neta P, Yang X, Simon-Manso Y, Liang Y, Stein SE. Formation of y + 10 and y + 11 ions in the collision-induced dissociation of peptide ions. J Am Soc Mass Spectrom. 2012;23:655–63. doi: 10.1007/s13361-011-0277-7. [DOI] [PubMed] [Google Scholar]
- 21.Harrison AG. Pathways for water loss from doubly protonated peptides containing serine or threonine. J Am Soc Mass Spectrom. 2012;23:116–23. doi: 10.1007/s13361-011-0282-x. [DOI] [PubMed] [Google Scholar]
- 22.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 23.Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80:62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- 24.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–35. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 25.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep--a database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–40. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–67. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 27.Lam H, Deutsch EW, Eddes JS, Eng JK, Stein SE, Aebersold R. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–5. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Meyer-Arendt K, Eichelberger B, Brown R, Yen CY, Old WM, Pierce K, Cios KJ, Ahn NG, Resing KA. Improved validation of peptide MS/MS assignments using spectral intensity prediction. Mol Cell Proteomics. 2007;6:1–17. doi: 10.1074/mcp.M600320-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Sidak Z. Rectangular Confidence Regions for Means of Multivariate Normal Distributions. Journal of the American Statistical Association. 1967;62:626. [Google Scholar]
- 30.Payne SH, Yau M, Smolka MB, Tanner S, Zhou H, Bafna V. Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J Proteome Res. 2008;7:3373–81. doi: 10.1021/pr800129m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, Yu C, Qiao Y, Lin Y, Dong G, Liu C, Zhang J, Zhang Z, Cai J, Zhang H, Bu D. Deriving the probabilities of water loss and ammonia loss for amino acids from tandem mass spectra. J Proteome Res. 2008;7:202–8. doi: 10.1021/pr070479v. [DOI] [PubMed] [Google Scholar]
- 32.Hernan MA, Clayton D, Keiding N. The Simpson’s paradox unraveled. Int J Epidemiol. 2011;40:780–5. doi: 10.1093/ije/dyr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savitski MM, Falth M, Fung YM, Adams CM, Zubarev RA. Bifurcating fragmentation behavior of gas-phase tryptic peptide dications in collisional activation. J Am Soc Mass Spectrom. 2008;19:1755–63. doi: 10.1016/j.jasms.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Gronert S, Li KH, Horiuchi M. Manipulating the fragmentation patterns of phosphopeptides via gas-phase boron derivatization: determining phosphorylation sites in peptides with multiple serines. J Am Soc Mass Spectrom. 2005;16:1905–14. doi: 10.1016/j.jasms.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Breci LA, Tabb DL, Yates JR, 3rd, Wysocki VH. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal Chem. 2003;75:1963–71. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- 36.Metzger S, Hoffmann R. Studies on the dephosphorylation of phosphotyrosine-containing peptides during post-source decay in matrix-assisted laser desorption/ionization. J Mass Spectrom. 2000;35:1165–77. doi: 10.1002/1096-9888(200010)35:10<1165::AID-JMS44>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 37.Edelson-Averbukh M, Shevchenko A, Pipkorn R, Lehmann WD. Gas-phase intramolecular phosphate shift in phosphotyrosine-containing peptide monoanions. Anal Chem. 2009;81:4369–81. doi: 10.1021/ac900244e. [DOI] [PubMed] [Google Scholar]
- 38.Salek M, Lehmann WD. Neutral loss of amino acid residues from protonated peptides in collision-induced dissociation generates N- or C-terminal sequence ladders. J Mass Spectrom. 2003;38:1143–9. doi: 10.1002/jms.531. [DOI] [PubMed] [Google Scholar]
- 39.Hunter EPL, Lias SG. Evaluated gas phase basicities and proton affinities of molecules: An update. Journal of Physical and Chemical Reference Data. 1998;27:413–656. [Google Scholar]
- 40.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal Chem. 2004;76:3908–22. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides with three or more charges. Anal Chem. 2005;77:6364–73. doi: 10.1021/ac050857k. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z. Prediction of collision-induced-dissociation spectra of peptides with post-translational or process-induced modifications. Anal Chem. 2011;83:8642–51. doi: 10.1021/ac2020917. [DOI] [PubMed] [Google Scholar]
- 43.Yen CY, Meyer-Arendt K, Eichelberger B, Sun S, Houel S, Old WM, Knight R, Ahn NG, Hunter LE, Resing KA. A simulated MS/MS library for spectrum-to-spectrum searching in large scale identification of proteins. Mol Cell Proteomics. 2009;8:857–69. doi: 10.1074/mcp.M800384-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.