Abstract
Introduction
Peri-operative window trials provide an opportunity to obtain intact tumor samples at 2 different time-points for evaluation of potential surrogate biomarkers. We report results of a pilot trial designed to determine if treatment-mediated changes in gene expression can be detected in formalin-fixed paraffin-embedded (FFPE) samples after 10-day exposure to anastrozole in estrogen receptor (ER)-positive breast cancer compared to untreated controls.
Methods
Paired tumor samples (biopsy, surgical) were obtained from 26 postmenopausal women with ER-positive breast cancer. Patients were assigned anastrozole (1mg/dy) for 10 days immediately prior to surgery (13 cases) or no treatment(13 controls). 502 cancer-related genes were examined by the Illumina cDNA-mediated annealing, selection, extension, and ligation, FFPE cDNA array(moderated t-test,p≤.005). Surrogate biomarkers reflecting changes in gene expression were examined by immunohistochemistry(Wilcoxon rank-based test,p<.05).
Results
Sufficient RNA was available from 19 paired samples (8 controls,11 cases). Frozen tissue and FFPE showed good correlation (r=0.82). Within each group, 18 genes, reflecting roles in proliferation, angiogenesis, and apoptosis, showed differential expression from biopsy to surgery (p<0.005). Estrogen-related genes were dysregulated in the treated group only. A reduction in Ki-67 was observed in 7 (54%) treated cases and in 1 (7.7%) control patient.
Conclusions
10-day exposure to anastrozole resulted in dysregulation of 18/502 cancer-related genes, and Ki-67 was reduced in 54% of cases. FFPE samples demonstrated good correlation with frozen samples. However, changes in gene expression and increased Ki-67 in the control group suggest local effects of wound healing may represent a confounding factor in the interpretation of peri-operative window trials.
Keywords: Aromatase inhibition, preoperative window trials, breast cancer, gene expression, proliferation
Introduction
Estrogen receptor-α is the most well-established predictive marker in breast cancer treatment and the key factor driving decisions regarding hormonal treatment for patients with invasive disease. However, despite optimal therapy, a significant proportion of estrogen receptor (ER)-positive patients will exhibit either de novo anti-estrogen resistance or present with recurrent disease after a prolonged disease-free interval (1).
Advances in molecular biology have facilitated genomic characterization of tumors, which has proven to be a powerful predictive biomarker for patients with invasive breast cancer. For example, a 21-gene panel, which aids identification of patients with node-negative, ER-positive disease with a high risk of recurrence, has been validated for use in the clinical setting to select patients for chemotherapy (2, 3). However, identification of reliable molecular predictors of response to endocrine therapy alone remains challenged by the extended natural history of ER-positive breast cancer and the long duration of treatment.
Peri-operative window trials, using molecular biomarkers as surrogate endpoints for treatment response, offer investigators a unique opportunity to obtain intact tumor samples at 2 different time points and thus evaluate potential biomarkers over a short period of time (4, 5). Challenges to this approach in the present clinical setting include defining the optimal length of the treatment window in order to identify measurable changes in the surrogate biomarker without delaying standard clinical management, the natural variability and intrinsic heterogeneity of tumors that may be underappreciated in a limited tissue sample, and the reliance on more readily available paraffin-embedded material as opposed to frozen tumor samples. Moreover, some of the observed changes can be related to the biopsy procedure itself, and the omission of such measurements may compromise interpretation of the perceived treatment response and represents a notable limitation of many studies.
To address these challenges, we designed a peri-operative window trial that included an untreated control group to determine if treatment-mediated changes in gene expression could be detected after a 10-day exposure to anastrozole in ER-positive invasive breast cancer using Illumina cDNA-mediated annealing, selection, extension, and ligation (DASL), formalin-fixed paraffin-embedded (FFPE)─based cDNA array.
Materials and Methods
Our institutional review board approved this pilot study. Eligible patients were enrolled during their preoperative surgical consultation and gave informed, written consent. Eligibility criteria included postmenopausal women with intact, histologically confirmed ER-positive (≥ 10% nuclear staining) invasive breast cancer, ≥ 1 cm in size. Ineligibility criteria included hormone-replacement therapy within 3 months of diagnosis, prior tamoxifen for chemoprevention, a prior history of breast cancer, and medical contraindication(s) to anastrozole (6).
Patients were non-randomly assigned to the treatment (anastrozole) or control arm by their physicians. Patients assigned to the treatment arm were instructed to begin taking anastrozole (1mg/day) 11 days before the scheduled date of surgery and to take the last dose on the day before surgery. Patients assigned to the control arm proceeded to surgery as planned. Paired tumor samples from the core needle biopsy (CNB) and the surgical procedure were obtained from FFPE tissue blocks. Fresh-frozen surgical samples were obtained whenever possible by the study pathologist during gross macroscopic evaluation of the specimen. Time to freeze for all samples was less than 30 minutes.
Standard clinical and tumor characteristics were recorded. If the diagnostic core biopsy was performed elsewhere, ER status was confirmed at our institution. ER and progesterone receptor (PR) positivity was reported as the percentage of tumor cells with positive staining nuclei (CloneSP1 and CloneIE2 Rb-monoclonal antibody, Ventana, respectively). HER2 expression was assessed by the Hercept-Test Kit (Dako Corporation), and samples with a score of ≥ 2+ were confirmed by fluorescence in situ hybridization (FISH).
Gene Expression
The commercially available Illumina DASL expression microarray platform (Cancer Panel TM v1, Illumina Inc, CA), which is optimized for gene expression analysis from FFPE tissue, was used for this study. This platform contains 502 cancer-related genes with unique probe groups for 3 different sites per gene and utilizes DASL technology. Total RNA is converted to cDNA, which is annealed to the probe groups, extended, and then ligated to locus-specific oligonucleotides. In this study, all samples were distributed on one 96-well plate for DASL analysis.
Briefly, 5 freshly cut 8µm sections of FFPE tissue from the CNB and surgical tissue blocks were de-paraffinized, and areas homogeneous for tumor were manually micro-dissected (Optical micro-dissection microscope, Zeiss, Germany); mRNA was extracted using the High-Pure RNA Paraffin Kit Roche (Roche, IN). Care was taken to select surgical sections away from areas of biopsy site changes for all analyses. To test the ability of the Illumina platform to reliably assess gene expression patterns of FFPE samples as compared to intact RNA, matched fresh-frozen surgical tumor samples were also collected, manually microdissected, and RNA extracted (Absolutely RNA Miniprep Kit, Stratagene, CA). The integrity of RNA extracted from the FFPE and fresh-frozen samples was assessed on the Aligent 2100 BioAnalyzer.
The Illumina array data were processed by BeadStudio software. The moderated t-statistic implemented in the limma package of R was used to compare gene expression between sample classes (http://www.bioconductor.org) (7). For gene expression between paired FFPE tumor samples (CNB versus surgery), the moderated t-test for paired samples was performed. As this analysis included 502 genes, a p-value of < 0.005 was considered statistically significant. We acknowledge that at p < 0.005, 2 to 3 of the 502 genes studied could be identified by chance. A subset analysis including only the 25 estrogen-related genes present on the Illumina DASL array was carried out, with a p-value of < 0.01. The Pearson product-moment correlation coefficient was used to assess the degree of correlation between matched surgical FFPE and fresh-frozen surgical samples.
Immunohistochemistry
Immunohistochemistry (IHC) for proliferation index, angiogenesis, and apoptosis was independently performed using Ki-67 (Clone QBEnd/10 mouse monoclonal antibody, Lab Vision), CD34 (Clone SP6, rabbit monoclonal antibody, Lab Vision), and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), respectively. Consecutive 5µm sections from same core biopsy and surgical FFPE blocks used for gene expression analysis were prepared and stained according to manufacturers’ instructions.
Areas homogenous for tumor were reviewed and scored by 2 observers (TK, AP) blinded to study arm. Ki-67 was scored as the percentage of tumor cells with strong nuclear staining. Apoptosis was scored as the percentage of tumor cells exhibiting nuclear fragmentation and staining. Angiogenesis was scored as the number of endothelial cells exhibiting positive staining for CD34. To account for variability among core biopsy specimens and ensure adequate cellular representation, all scoring was assessed in a minimum background of 100 tumor cells and/or one high power field (x20). The individual scores for each biomarker were compared for each pair of tumor samples (CNB versus surgical) to assess for changes in individual tumors. The median change across all samples in the 2 groups (anastrozole-treated versus control) was compared using the Wilcoxon signed rank test.
Results and Discussion
A total of 31 patients were enrolled (14 controls, 17 cases). Paired tumor samples (core biopsy and surgery) were available from 26 patients (13 controls, 13 cases). The remaining samples had insufficient tissue for analysis. Median patient age was 65 years (range, 53–84 years), and 46% of patients had stage I disease. There were no significant differences in age at diagnosis, baseline tumor size, tumor grade, or nodal status between groups (Table 1). The median time interval from CNB to surgery in the control group was 18 days (range, 9–63), compared to 31 days (range, 20–61) in the treated group (p = 0.05). Although the time interval between biopsy and surgery was somewhat longer in the anastrozole-treated group, all patients in this group took anastrozole for the 10 days immediately prior to surgery.
Table 1.
Clinical and pathologic characteristics of the study population.
| Variable | Controls (n = 13) | Anastrozole-treated (n = 13) |
|---|---|---|
| Mean age, years (range) | 64 (53–77) | 68 (53–84) |
| Mean tumor size, cm (range) | 1.7 (1–6) | 1.9 (1–3.5) |
| Grade | ||
| 1 | 0 | 0 |
| 2 | 5 | 3 |
| 3 | 8 | 9 |
| Nodal status | ||
| N0 | 9 | 8 |
| N1 | 4 | 5 |
| Median time interval biopsy → Surgery, days (range) | 18 (9–63) | 31 (20–61) |
Gene expression analysis
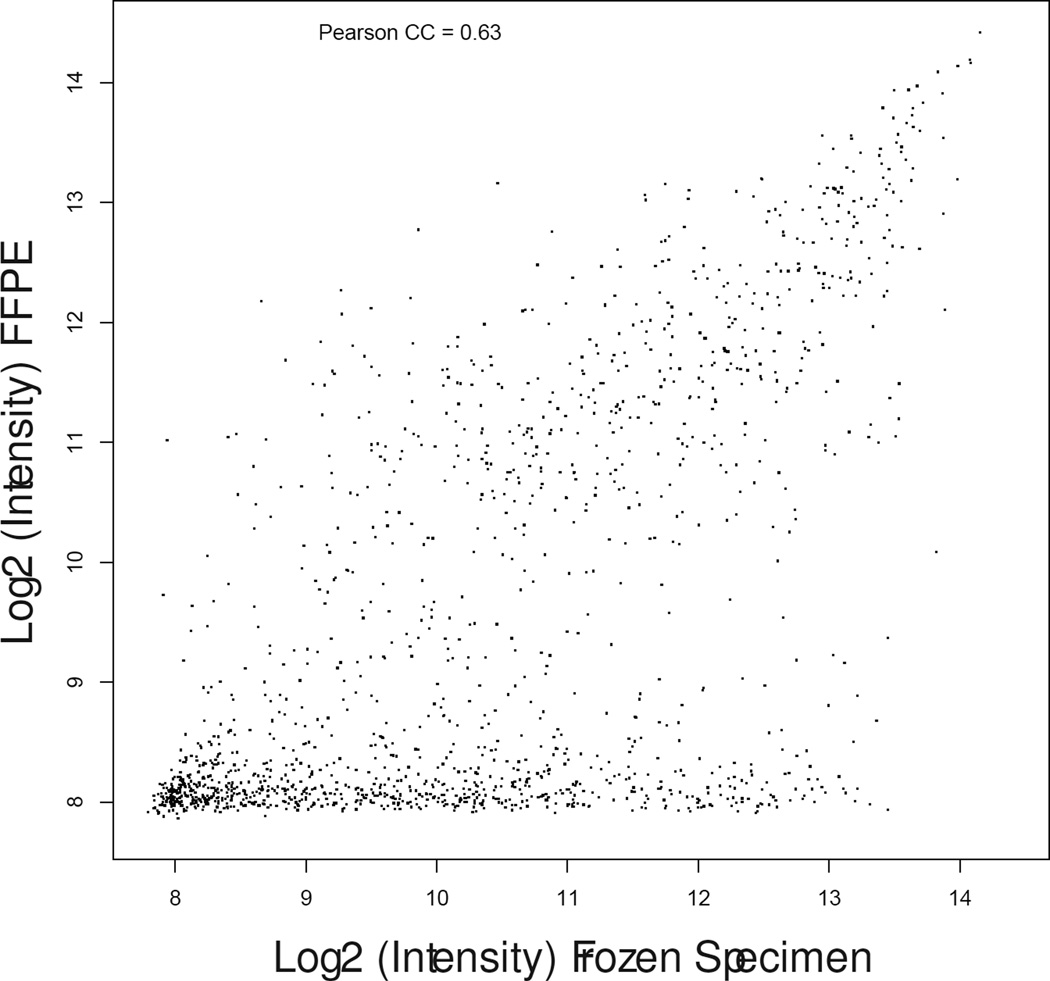
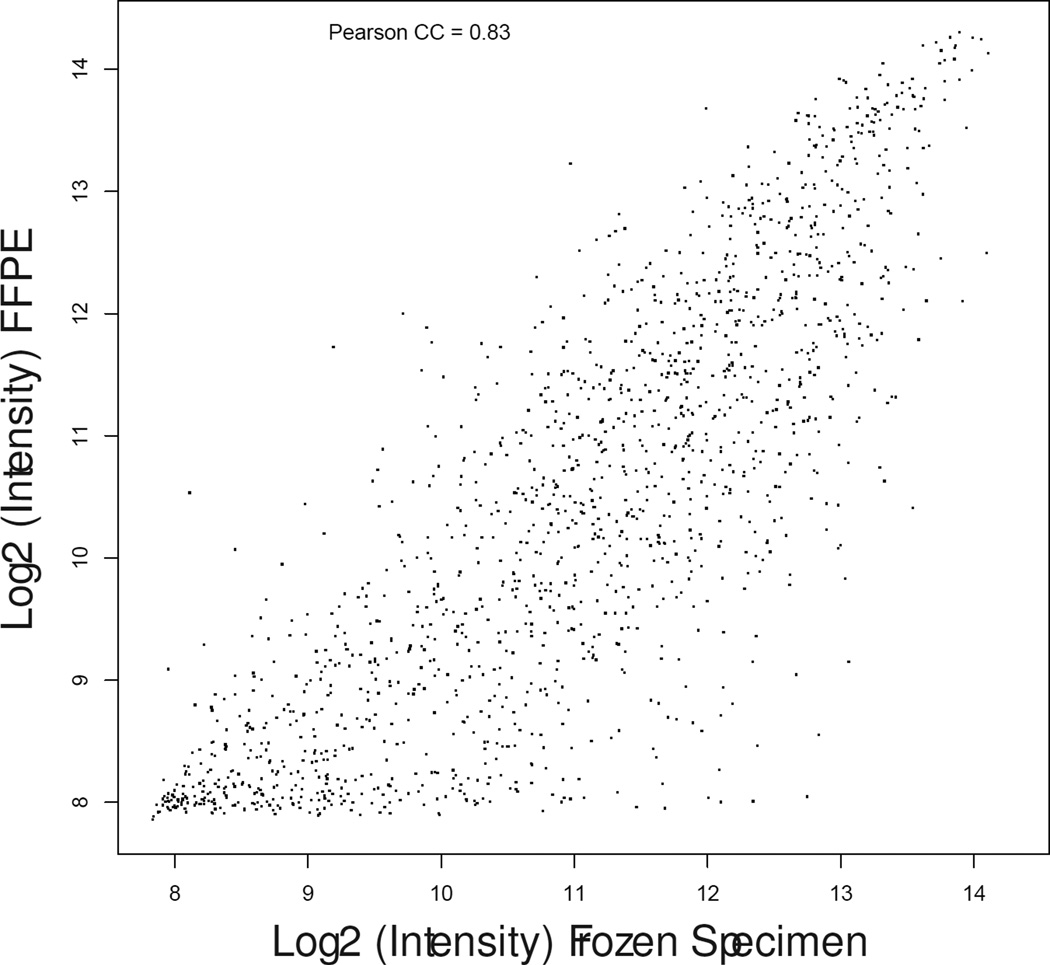
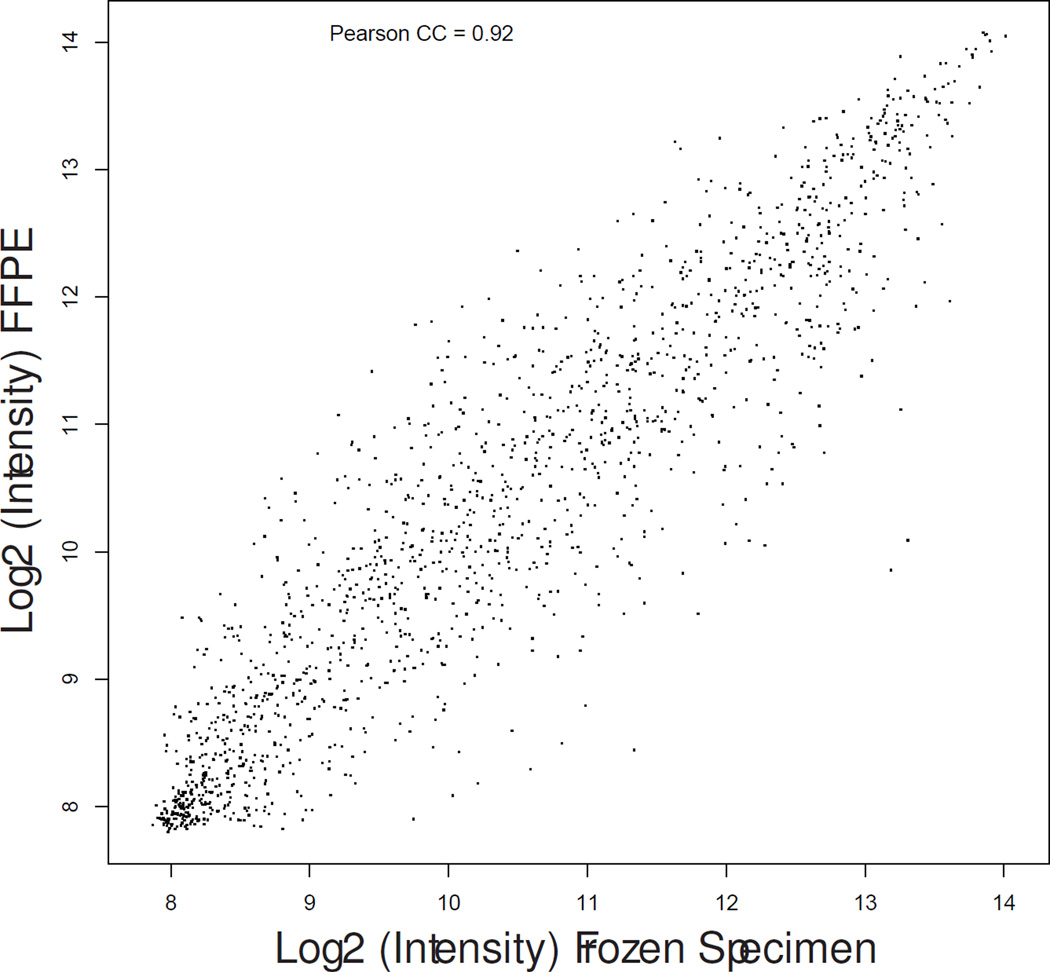
Matched fresh-frozen surgical samples were available for 16 patients (8 controls, 8 cases). Gene expression profiles as measured by DASL from FFPE RNA samples demonstrated consistent biological differences with those measured from fresh-frozen tissue (median correlation coefficient, 0.82; range, 0.63–0.92) as shown in Figure 1.
Figure 1.
Scatter plot graphs showing gene expression correlation between fresh-frozen (X-axis) and formalin fixed paraffin-embedded (Y-axis) specimens in patient matched samples #13-control (a), #17-treated (b) and #19-treated (c) representative of the lowest, median, and highest correlation coefficients.
Sufficient RNA for gene expression analysis was obtained from 19/26 (73%) FFPE paired (biopsy and surgery) samples (8 controls, 11 anastrozole-treated). Selecting a level of significance of p < 0.005, 18 genes were differentially expressed between the biopsy and surgical samples in each patient group. Both groups included genes with known functional roles in cell proliferation, DNA repair, angiogenesis, and apoptosis (Table 2a and 2b). Estrogen-regulated genes (specifically, FOS, PRG, and connective tissue growth factor [CTGF]) were only differentially expressed in the anastrozole-treated group. There were only 2 dysregulated genes in common when comparing the 18 genes identified in the 2 groups MLL and FOSB, both of which play a role in wound healing, among other important functions.
Table 2.
| a. Untreated controls (n = 8): dysregulated genes over time (core needle biopsy to surgery). Probes sorted by ascending fold change. Probe sets sorted by Log2Fold Change. | |||||
|---|---|---|---|---|---|
| Target ID | Symbol | Definition | Function | Log 2 Fold Change |
p-value |
| GI_19924137-S | RAD23A | RAD23 homolog A (S. cerevisiae) (RAD23A); mRNA. |
DNA repair | −1.95 | 0.0020 |
| GI_4885214-S | ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ERBB4); mRNA. |
Cell proliferation; pro-mitotic |
−1.91 | 0.0043 |
| GI_7549801-S | BAG1 | BCL2-associated athanogene (BAG1); mRNA. |
Anti-Apoptosis | −1.85 | 0.0012 |
| GI_19482173-S | CUL2 | cullin 2 (CUL2); mRNA. | Pro-Apoptosis & Cell cycle negative regulator |
−1.78 | 0.0036 |
| GI_19482173-S | CUL2 | cullin 2 (CUL2); mRNA. | Pro-Apoptosis & Cell cycle negative regulator |
−1.75 | 0.0029 |
| GI_34147567-S | RAP1GDS 1 | RAP1; GTP-GDP dissociation stimulator 1 (RAP1GDS1); mRNA. |
Unknown | −1.64 | 0.0031 |
| GI_6466449-A | COMT | catechol-O-methyltransferase (COMT); transcript variant MB-COMT; mRNA. |
Cell metabolism | −1.53 | 0.0037 |
| GI_4505054-S | LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (LYN); mRNA. |
Cell growth/ Maintenance |
−1.47 | 0.0035 |
| GI_38201662-A | ING1 | inhibitor of growth family; member 1 (ING1); transcript variant 4; mRNA. |
Cell cycle - anti-mitotic |
−1.43 | 0.0010 |
| GI_5174568-S | MLL | myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog; Drosophila) (MLL); mRNA. |
Transcription factor |
−1.36 | 0.0030 |
| GI_4758605-S | ILK | integrin-linked kinase (ILK); mRNA. |
Cell-adhesion | −1.24 | 0.0008 |
| GI_19924130-A | RAD50 | RAD50 homolog (S. cerevisiae) (RAD50); transcript variant 1; mRNA. |
DNA repair | −1.10 | 0.0044 |
| GI_19924134-A | RAD51 | RAD51 homolog (RecA homolog; E. coli) (S. cerevisiae) (RAD51); transcript variant 1; mRNA. |
DNA repair | −1.03 | 0.0041 |
| GI_12669916-S | E2F5 | E2F transcription factor 5; p130-binding (E2F5); mRNA. |
Cell-Cycle; negative regulator |
−1.01 | 0.0048 |
| GI_18765747-A | COL18A1 | collagen; type XVIII; alpha 1 (COL18A1); transcript variant 1 ;mRNA. |
Cell adhesion; Cell Cycle- negative regulator |
−1.00 | 0.0046 |
| GI_33239450-A | PCNA | proliferating cell nuclear antigen (PCNA); transcript variant 2; mRNA. |
Cell cycle; pro- mitotic |
−0.96 | 0.0010 |
| GI_16936532-A | CDK4 | cyclin-dependent kinase 4 (CDK4); transcript variant 2; mRNA. |
Cell Cycle; pro- mitotic |
−0.87 | 0.0020 |
| GI_16933566-S | MEL | mel transforming oncogene (derived from cell line NK14)- RAB8 homolog (MEL); mRNA. |
Cell growth/ maintenance |
−0.79 | 0.0050 |
| GI_4504808-S | JUNB | jun B proto-oncogene (JUNB); mRNA. |
Transcription factor |
+1.09 | 0.0008 |
| GI_5803016-S | FOSB | FBJ murine osteosarcoma viral oncogene homolog B (FOSB); mRNA. |
Transcription factor |
+1.17 | 0.0049 |
| b. Anastrozole-treated cases (n = 11): dysregulated genes over time (core needle biopsy to surgery). Probes sorted by ascending fold change. Probe sets sorted by Log2Fold Change. | |||||
|---|---|---|---|---|---|
| Target ID | Symbol | Definition | Function | Log 2Fold Change |
p-value |
| GI_31981491-S | PR | Progesterone receptor (PR); mRNA. |
Transcription factor |
−1.70 | 0.0024 |
| GI_27894367-S | NOTCH1 | Notch homolog 1; translocation-associated (Drosophila) (NOTCH1); mRNA. |
Signal transduction; development |
−1.49 | 0.0007 |
| GI_17017983-S | CDK9 | Cyclin-dependent kinase 9 (CDC2-related kinase) (CDK9); mRNA. |
Cell cycle | −1.35 | 0.0015 |
| GI_4504610-S | IGF2R | Insulin-like growth factor 2 receptor (IGF2R); mRNA. |
Signal transduction |
−1.23 | 0.0045 |
| GI_5174568-S | MLL | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog; Drosophila) (MLL); mRNA. |
Transcription activating factor |
−1.17 | 0.0035 |
| GI_4885360-S | GRPR | Gastrin-releasing peptide receptor (GRPR); mRNA. |
Cell cycle | −1.16 | 0.0039 |
| GI_4557792-S | NF1 | Neurofibromin 1 (neurofibromatosis; von Recklinghausen disease; Watson disease) (NF1); mRNA. |
Cell cycle negative regulator & Cell growth |
−1.08 | 0.0049 |
| GI_4759335-S | FANCG | Fanconi anemia; complementation group G (FANCG); mRNA. |
DNA repair | −1.08 | 0.0016 |
| GI_4504938-S | LAF4 | Lymphoid nuclear protein related to AF4 (LAF4); mRNA. |
Transcription activating factor |
−0.94 | 0.0006 |
| GI_6633806-S | LMO2 | LIM domain only 2 (rhombotin-like 1) (LMO2); mRNA. |
Cell growth and maintenance |
−0.88 | 0.0039 |
| GI_6382057-A | ABL1 | v-abl Abelson murine leukemia viral oncogene homolog 1 (ABL1); transcript variant b; mRNA. |
Pro-apotosis, Cell cycle negative regulator, DNA repair |
−0.84 | 0.0016 |
| GI_4504938-S | LAF4 | Lymphoid nuclear protein related to AF4 (LAF4); mRNA. |
Transcription activating factor |
−0.69 | 0.0047 |
| GI_20127489-S | MADH2 | MAD; mothers against decapentaplegic homolog 2 (Drosophila) (MADH2); mRNA. |
Negative regulation of DNA transcription |
−0.66 | 0.0041 |
| GI_8400737-S | TP53 | Tumor protein p53 (Li- Fraumeni syndrome) (TP53); mRNA. |
Pro-apotosis, Cell cycle negative regulator, DNA repair |
−0.64 | 0.0012 |
| GI_20127459-S | XPC | Xeroderma pigmentosum; complementation group C (XPC); mRNA. |
DNA repair | −0.54 | 0.0022 |
| GI_4503122-S | CTGF | Connective tissue growth factor (CTGF); mRNA. |
Cell adhesion, cell differentiation, regulation cell growth |
+0.76 | 0.0034 |
| GI_4506264-S | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (PTGS2); mRNA. |
Response to oxidative stress |
+1.26 | 0.0012 |
| GI_6552332-S | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS); mRNA. |
Cell growth & Inflammatory response |
+1.32 | 0.0009 |
| GI_5803016-S | FOSB | FBJ murine osteosarcoma viral oncogene homolog B (FOSB); mRNA. |
Cell cycle negative regulator |
+1.54 | 0.0009 |
| GI_6552332-S | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS); mRNA. |
Cell growth & Inflammatory response |
+1.67 | 0.0001 |
| GI_6552332-S | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS); mRNA. |
Cell growth & Inflammatory response |
+1.91 | 0.0002 |
| GI_10834983-S | IL6 | Interleukin 6 (interferon; beta 2) (IL6); mRNA. |
Acute phase response |
+2.15 | 0.0003 |
| GI_5803016-S | FOSB | FBJ murine osteosarcoma viral oncogene homolog B (FOSB); mRNA. |
Cell cycle negative regulator |
+2.37 | 0.0023 |
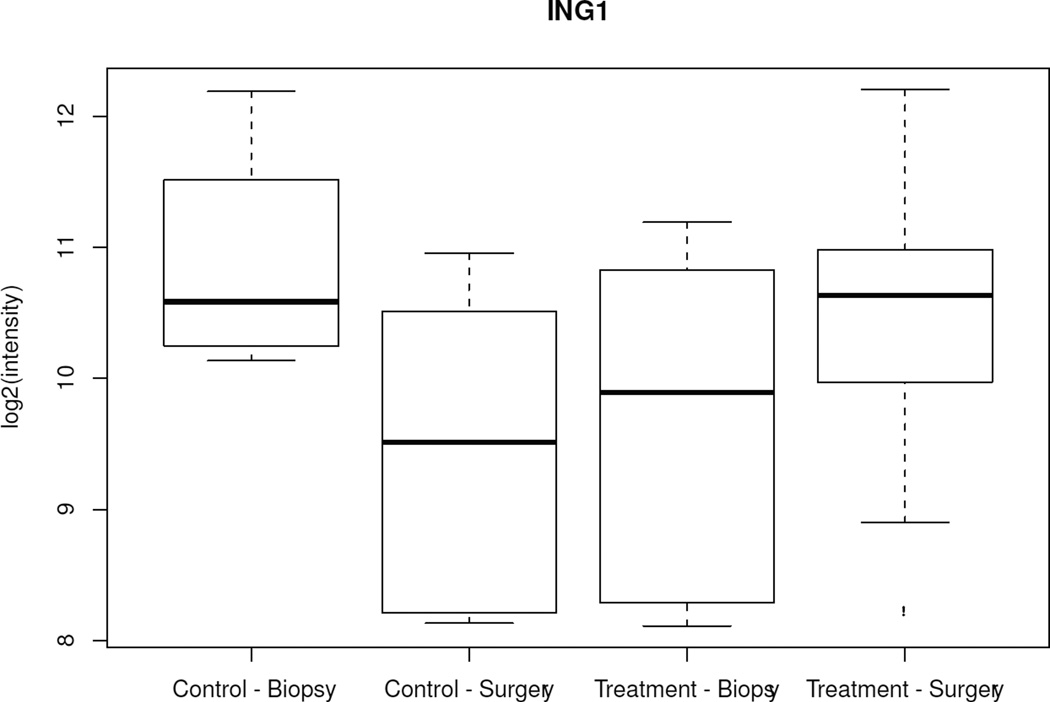
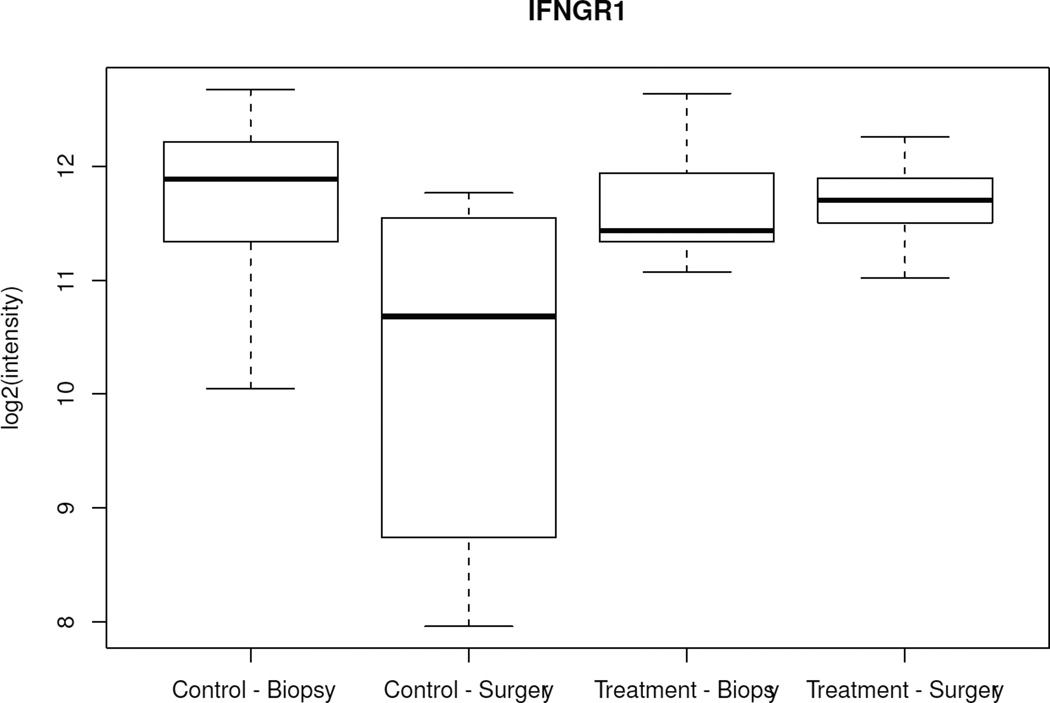
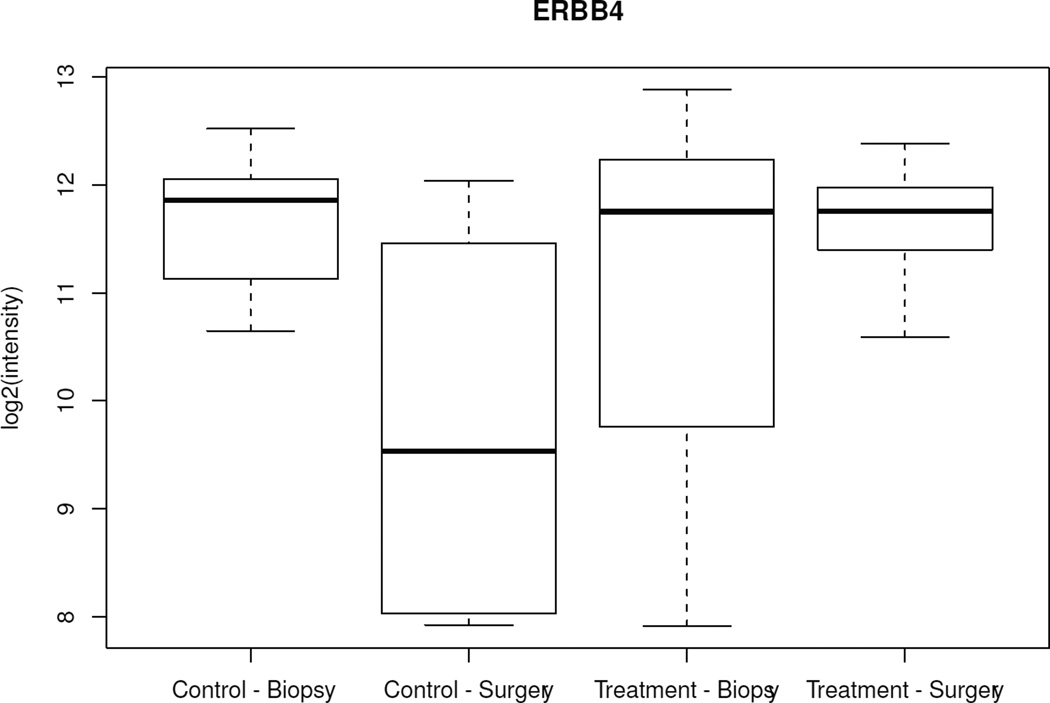
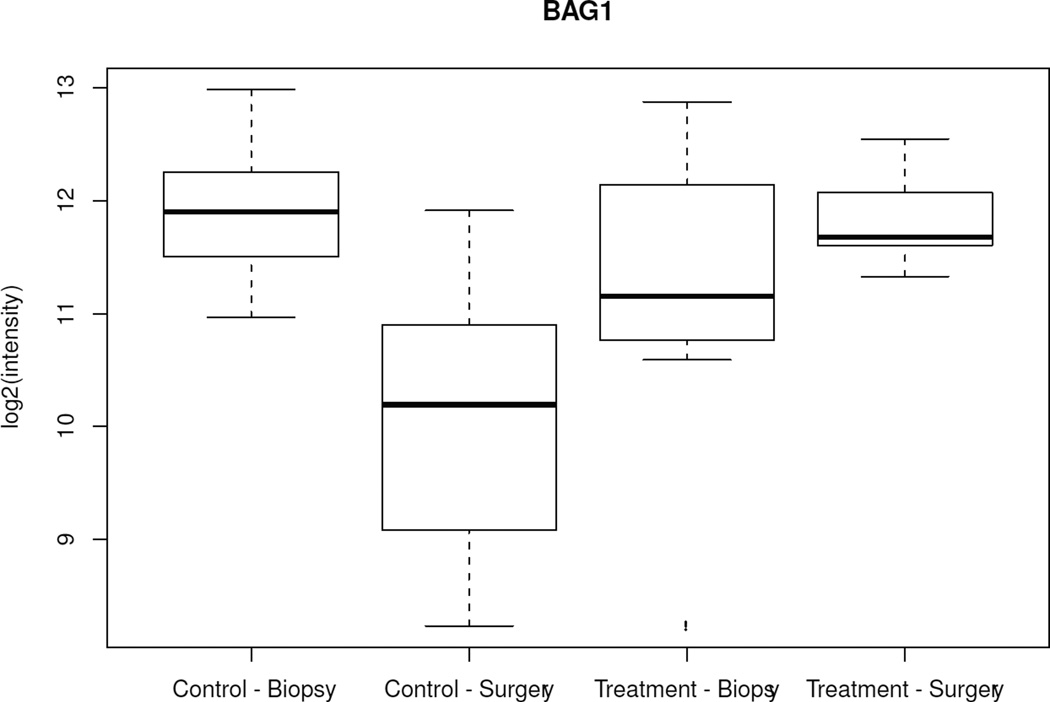
When changes over time (CNB to surgery) among all 502 genes were analyzed between the treated and control groups, only 5 genes showed significantly different variations (ERBB4, ING1, BAG1, IFNGR1, TFDP1) (p < 0.005). Specifically, from CNB to surgery, all 5 genes showed lower mRNA levels in surgical specimens from the control group and either unchanged or higher levels in surgical specimens from anastrozole-treated patients (Figure 2, Table 3). There was no difference in baseline mean expression of these genes between the 2 groups. When the same type of analysis was performed focusing on the 25 estrogen-related genes, only PR showed a significant variation between the 2 groups (p = 0.01); specifically, PR was 1.07-fold (Log 2 scale) downregulated in the anastrozole-treated group compared to controls.
Figure 2.
Box-plot of the 5 genes with significant different variations over time (biopsy to surgery) between the control and treated groups. Boxes represent inferior quartile, median, and superior quartile values for each group.
Table 3.
Genes differentially expressed over time (core needle biopsy to surgery) between anastrozole-treated and control groups (p < 0.005).
| Target ID | Symbol | Definition | Controls Log 2 FC |
Anastrozole treated Log 2 FC |
Treated controls Log 2 FC |
p-value |
|---|---|---|---|---|---|---|
| GI_7549801-S | BAG1 | Homo sapiens BCL2- associated athanogene (BAG1); mRNA. |
−1.85 | 0.77 | 2.615 | 0.0007 |
| GI_4885214-S | ERBB4 | Homo sapiens v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (ERBB4); mRNA. |
−1.91 | 0.69 | 2.597 | 0.0014 |
| GI_4557879-S | IFNGR1 | Homo sapiens interferon gamma receptor 1 (IFNGR1); mRNA. |
−1.49 | 0.01 | 1.498 | 0.0016 |
| GI_4557879-S | IFNGR1 | Homo sapiens interferon gamma receptor 1 (IFNGR1); mRNA. |
−1.41 | 0.11 | 1.516 | 0.0043 |
| GI_38201662-A | ING1 | Homo sapiens inhibitor of growth family; member 1 (ING1); transcript variant 4; mRNA. |
−1.43 | 0.74 | 2.17 | 0.0010 |
| GI_34147667-S | TFDP1 | Homo sapiens transcription factor Dp-1 (TFDP1); mRNA. |
−1.13 | 0.73 | 1.858 | 0.0036 |
FC: Fold change
Correlation between genetic and cellular changes
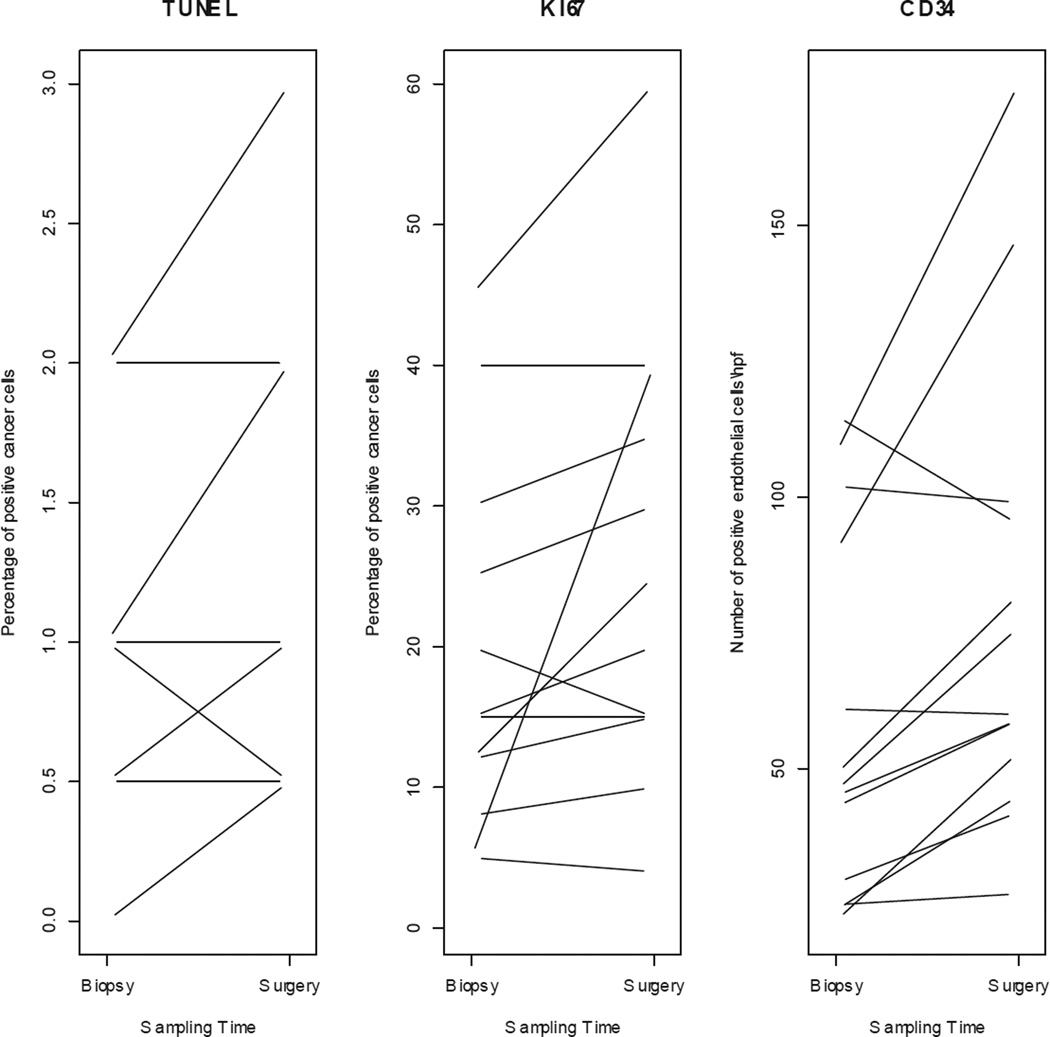
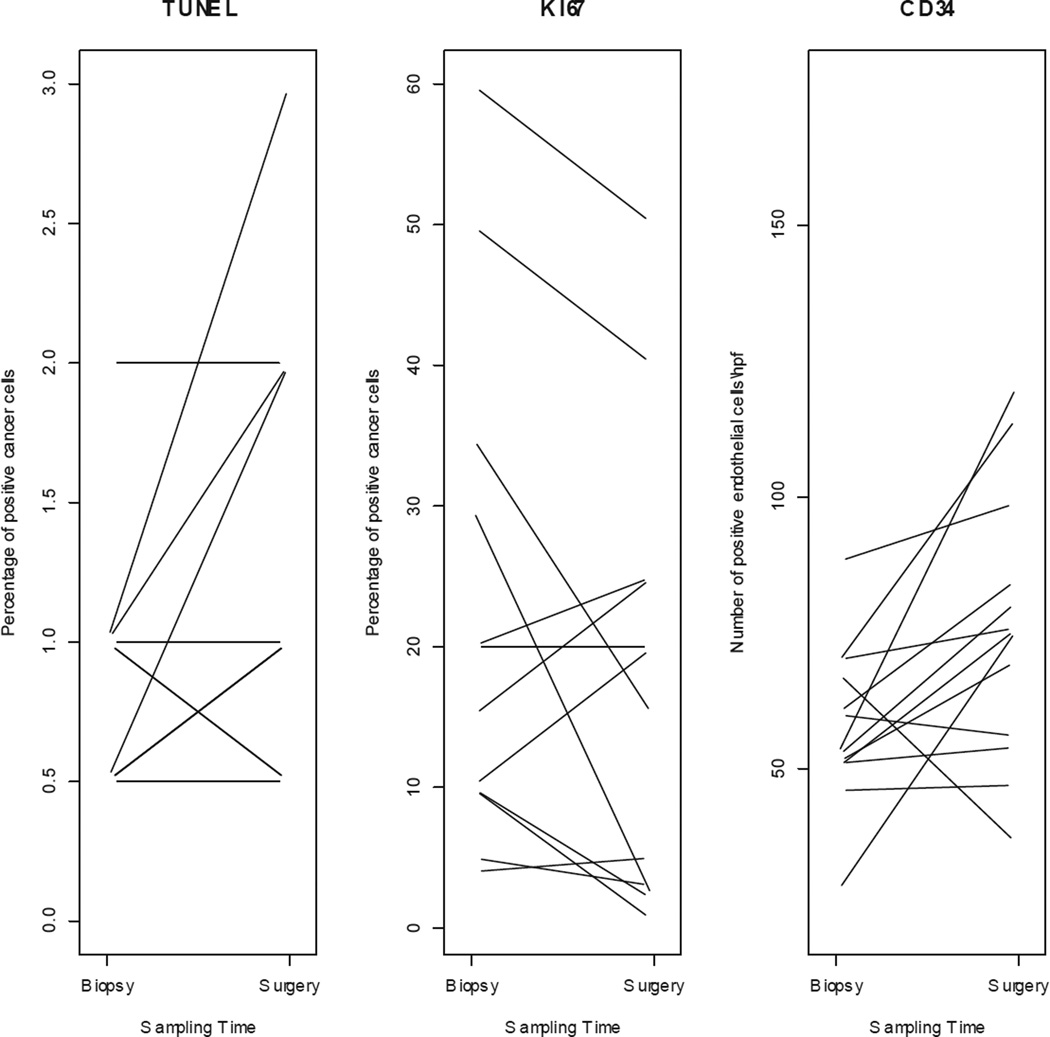
IHC was performed on all paired samples (n = 26). There was no significant difference in mean baseline biomarker expression between controls and anastrozole-treated (Table 4a). Individual variations on TUNEL, Ki-67, and CD34 expressions are shown in Figure 3a and 3b.
Table 4.
Median baseline biomarker immunohistochemistry expression (a) and changes in median biomarker expression from biopsy to surgery (b).
| (a) | ||
|---|---|---|
| Controls (n = 13) |
Anastrozole-treated (n = 13) |
|
| Ki-67, % (range) | 15 (5–45) | 20 (5–60) |
| CD34, No. of cells/hpf (range) | 46 (22–115) | 52 (27–88) |
| TUNEL, % (range) | 1 (0–2) | 1 (0–2) |
| (b) | |||
|---|---|---|---|
| Controls (n = 13) |
Anastrozole-treated (n = 13) |
ΔMedian expression cases versus controls p-value |
|
| Ki-67, % | + 5% (p = .04) | 0% (NS) | 0.03 |
| CD34, No. of cells / hpf | + 14% (p = .01) | + 24% (p = .02) | 0.92 |
| TUNEL, % | 0% (NS) | 0% (NS) | 0.62 |
TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; NS, not significant
Figure 3.
TUNEL, Ki-67 and CD34 expression changes from biopsy to surgery for individual (a) control and (b) treated patients.
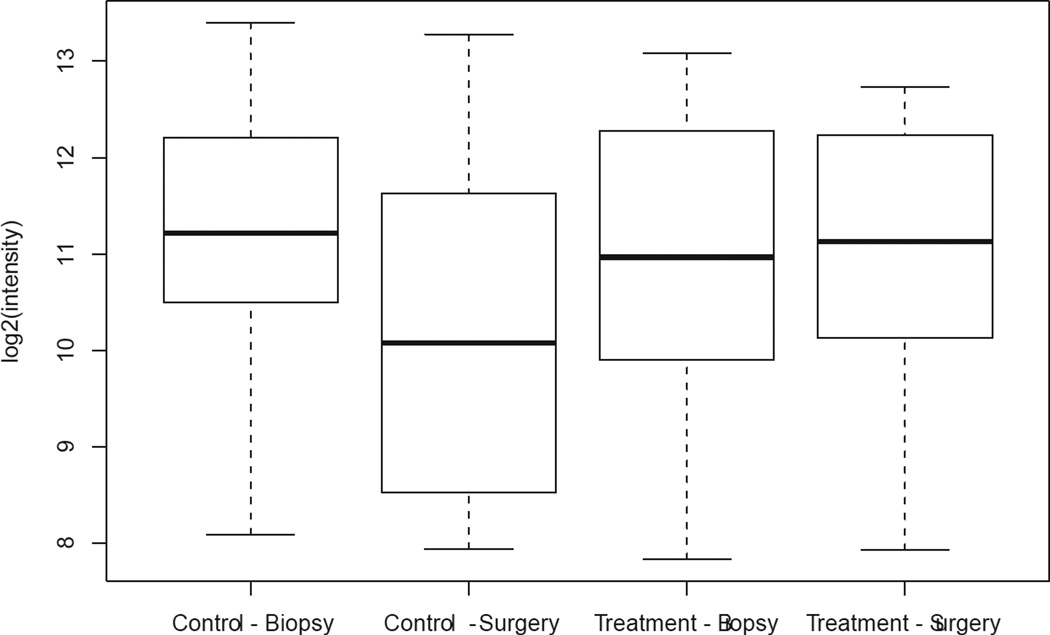
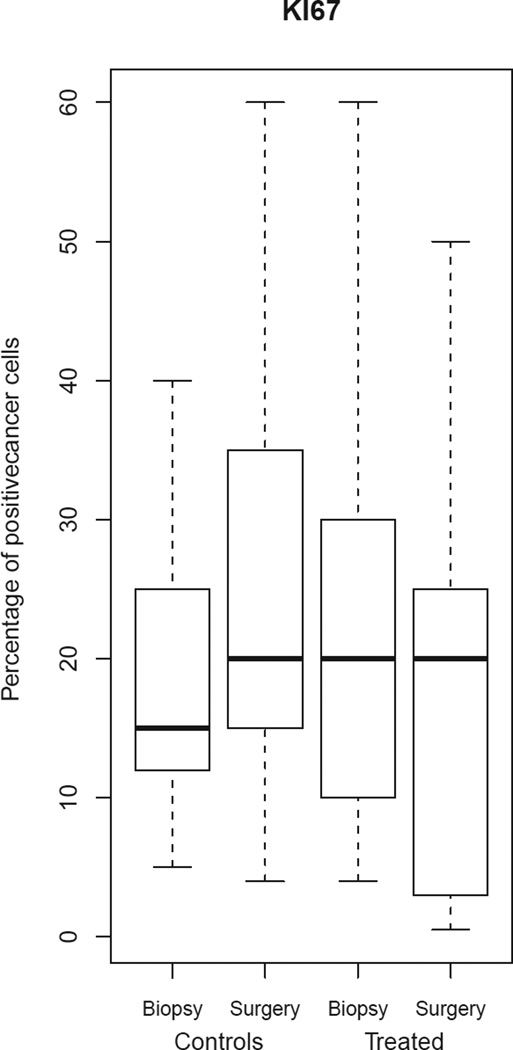
We observed a 2% to 28% decrease in Ki-67 index in 7 out of 13 (54%) patients in the treated group; 2 patients (15%) showed stable Ki-67 index over time, and 4 (31%) patients showed a 1% to 10% increase in the Ki-67 index. Among all anastrozole-treated patients, a change in median Ki-67 expression between the 2 time points was not evident (Figure 4, Table 4b). Among controls, there was a significant 5% increase in the median Ki-67 index from biopsy to surgery (p = 0.04), with 8/13 (62%) control patients exhibiting from a 2% to 35% increase in Ki-67 index, 4 showing stable Ki-67 index, and 1 showing 1% reduction in Ki-67 index (Figure 4, Table 4b). When the change in median Ki-67 expression from biopsy to surgery was compared between groups (controls versus anastrozole-treated), the difference was significant (p = 0.03) and independent of time interval from biopsy to surgery (Figure 4).
Figure 4.
Comparison of Ki-67 expression between biopsy and surgery for the control and treated groups. Boxes represent inferior quartile, median, and superior quartile values for each group.
Among both controls and anastrozole-treated cases there was a significant increase in median CD34 expression from biopsy to surgery; however, there was no significant difference in median CD34 expression between groups (Table 4b). Finally, the baseline level of apoptosis was notably low in both groups, and while minimal variations (1%) were detected in a minority of individual samples (3/13 controls, 3/13 treated patients), there was no appreciable change in median apoptosis index in either group.
Discussion
Peri-operative window trials represent a practical opportunity to rapidly evaluate in vivo treatment response without delaying surgery or requiring repeat tissue sampling. Gene expression profiling allows for an in-depth assessment of early treatment response, and while traditional platforms have been limited by a reliance on fresh-frozen tissue as sources of molecular recovery, newer platforms are optimized for use with FFPE tissue. Here we report the results of a peri-operative window trial that identified significant changes in gene expression profiles over time, indicating that as little as 10-days exposure to anastrozole is sufficient to induce detectable and biologically meaningful changes in ER-positive invasive breast cancer. Specifically, using an FFPE-based gene expression array, we identified distinct gene expression changes that distinguish anastrozole-treated cases versus controls, and we observed a median correlation coefficient of 0.82 for the paired comparison of gene expression in fresh-frozen and FFPE samples, supporting this platform use in this clinical setting.
Among controls there was a change in gene expression over a (median) 18-day time interval from biopsy to surgery, whereby 18/502 cancer-related genes were differentially expressed. Gene ontology of these genes revealed that the dysregulated biologic processes included cell proliferation, angiogenesis, and apoptosis, suggestive of changes induced by the ongoing wound-healing process. Subsequent IHC analysis demonstrated parallel cellular changes in both angiogenesis and proliferation, with an overall upregulation of both processes seen over time, supporting gene array findings.
After a 10-day exposure to anastrozole, the treated group also showed significant change in 18/502 cancer-related genes, including estrogen-related genes that were not identified in the control group. A 10-day exposure to anastrozole was insufficient to detect a change in angiogenesis as measured by IHC for CD34 (p = 0.92) when compared to controls, but we were able to identify a small suppressive effect (5%) on cell proliferation as measured by the Ki-67 index (p = 0.03). Finally, we were unable to demonstrate a change in apoptosis as measured by TUNEL; however, this may be a factor of time as earlier studies of aromatase inhibitors have shown that changes in apoptosis are small and occurred only after 2 weeks of treatment. Several of the genes identified among treated patients in our study have been previously reported as differentially expressed in anastrozole-treated patients. Using Agilent oligonucleotide arrays on fresh specimens, Mello-Grand et al reported similar changes in gene expression of FOS, PR, and CTGF after a 3-month window of treatment (8). Mackay et al also identified upregulation of CTGF after 14 days of treatment with aromatase inhibitor (9).
Comparing the 2 lists of genes differentially expressed over time among controls and anastrozole-treated patients identifies only 2 common genes: FOSB and MLL. These 2 genes may also reflect the effect of wound healing on gene expression. FOSB shows higher expression in early phases of wound healing, with important functions on fibroblast and epithelial cell proliferation and migration (6, 10). The MLL gene encodes a DNA-binding protein that methylates histone H3 and positively regulates expression of target genes, including multiple HOX genes, major players in morphogenesis and vascular remodeling in wound healing (11).
The higher frequency of CD34 positive cells in the surgical specimens of both the control and anastrozole-treated groups compared to the paired core biopsies also supports an increase in vascular proliferation in surgical specimens secondary to biopsy site repair. Notably, these changes were evident despite the fact that we selected areas as far as possible from the biopsy site, implicating that the effects of wound healing may not be restricted to the wound bed. Further evidence of wound healing affecting our gene lists is represented by CTGF, IL-6, and PTGS2 (a.k.a. COX2) upregulation in surgical specimens from the anastrozole-treated group and integrin-linked kinase (ILK) upregulation in the control group. CTGF has been described as playing a central role in wound healing, scarring, and persistent fibrosis (12). Mackay et al (9) found an upregulation of genes related to extracellular matrix remodeling, including CTGF, after 14 days of treatment with aromatase inhibitors. IL-6 is a well-known chemotaxin involved in wound healing. ILK encodes a serine/threonine protein kinase, which associates with the cytoplasmic domain of beta integrins and acts as a proximal receptor kinase regulating integrin-mediated signal transduction. ILK has been associated with fibrosis in benign and malignant diseases (13–15). COX-2 is involved in angiogenesis, cell proliferation, and invasiveness in solid tumors (16), yet has been associated with poor prognostic factors in breast cancer (17–19). Its overexpression may be masking some short-term effects of aromatase inhibitors on cell proliferation in our samples.
The comparison of the cellular changes over time between groups highlighted that the significant increase in proliferation seen in the control group was not evident after short-term exposure to anastrozole, and this trend was independent of the difference in the median time interval between sampling in the 2 groups. While the increase in proliferation over time seen in the control group may be explained by the natural history of the tumor, given the short median time interval between biopsy and surgery (18 days), we hypothesize that the increase in proliferation is a direct consequence of the biopsy. Although minimally invasive for patients, a core-needle biopsy is a traumatic event resulting in intratumoral and peritumoral bleeding. Miller et al (20) also observed an up-regulation of pro-inflammatory genes after anastrozole treatment for 2 weeks, but the absence of a control group in that study does not allow one to measure the effect of the biopsy itself. A wound-healing dynamic is evident within the tumor bed within 48 hours and may last up to 8 weeks (21–23). Our data suggest that a 10-day exposure to anastrozole is sufficient to counteract this increase in proliferation as demonstrated by the lack of change in median Ki-67 from biopsy to surgery when measured among the entire group of treated patients. Although we did observe a reduction in Ki-67 index in 7/13 (54%) patients, analysis by tumor size (< 2 cm versus > 2 cm) or time to surgery also failed to demonstrate a significant difference in median Ki-67 across the treated group (data not shown).
An alternate explanation may be that scoring of proliferation by IHC for Ki-67 on core biopsy specimens may show intrinsic differences due to shorter time of exposure to fixatives and restricted area available for analysis; however, these biases should have impacted both patient groups in the same manner, whereas we saw a different pattern between cases and controls. Although time to surgery was different in the 2 groups, both groups seem to be in the same phase of the wound-healing process, and all treated patients underwent surgery on the 11th day after treatment initiation. In the absence of data from a control group, a lack of change in median Ki-67 among anastrozole-treated cases may have been interpreted differently. Whether the increase in Ki-67 observed in the controls represents intrinsic sample discrepancy, tumor progression, or the biologic effects of wound healing, it is important to note that some patients may need a prolonged exposure to aromatase inhibitors to show a reduction in Ki-67 index, whereas others may truly represent the group of patients that will not respond to aromatase inhibitors as expected in the clinical setting.
Many pro-mitotic genes represented on the Illumina DASL array displayed downregulation variance in treated patients compared to controls. BIRC-5 (surviving) and MYBL2, 2 genes represented in the Oncotype DX test proliferative profile, were downregulated in anastrozole-treated cases as compared to controls 2.08-fold and 1.18-fold (Log2 scale), respectively. These numbers did not reach statistical significance, however, due to sample size. Differences among genes regulated by the ER were also observed between treated cases and controls. Specifically, PR genes showed the greatest negative-fold change in the treated group—1.7-fold (Log2 scale) downregulated after treatment—while no significant change was observed in the control group (p = 0.01). Strong PR downregulation was also noted by Miller et al (24). Other estrogen-regulated genes, BCL-2 and EGFR, were also downregulated after anastrozole exposure 1.76-fold and 1.25-fold (Log2 scale), respectively, as compared to controls, but these levels did not reach statistical significance. Expression levels of PR and BCL2 are both included in the estrogen-signaling profile of the Oncotype DX assay. Colony stimulating factor-2 (CSF2) is a proinflammatory molecule that displayed upregulation after anastrozole administration but downregulation in controls. As beta-estradiol is known to have an anti-inflammatory role through downregulation of CSF2, its upregulation after anastrozole is not unexpected (25). In total, these findings support our hypothesis that short-term (10-day) exposure to anastrozole is sufficient to induce detectable and biologically meaningful changes in ER-positive invasive breast cancer.
Direct comparison of the changes in gene expression over time between controls and cases identified 5 genes (ERBB4, ING1, BAG1, IFNGR1, and TFDP1) that were differentially dysregulated between the groups. There was no difference in baseline expression of these 5 genes between treated cases and controls, but 3 of them (ERBB4, ING1, BAG1) were also seen in the list of differentially expressed genes over time in the control group. This finding highlights that the wound-healing process may contribute significantly to dysregulated genes identified in window treatment trials, and it is important to be aware of this phenomenon to avoid misinterpretations.
Despite our small sample size, the strengths of this study are: 1) the availability of matched frozen tissue in 16 of 19 cases demonstrating good correlation between the gene expression profiles of FFPE versus fresh-frozen tissue; and 2) the inclusion of a control group to account for natural variability over time with respect to changes at both the level of gene expression and IHC. The latter point warrants further discussion. In peri-operative window trials, interpretation of the chosen endpoint must reliably capture the underlying biologic activity with respect to both, natural variability (or occurring as a result of biopsy) and changes related to the intervention being studied. In our IHC analysis, we demonstrated that over a short time interval between sampling, there was a statistically significant increase in Ki-67 observed in the untreated controls, but no significant change appreciated among cases.
Ki-67 is a biomarker that has been studied extensively as a marker of response to therapy in many solid tumors. As anti-estrogen therapy classically results in decreased cell proliferation, changes in Ki-67 after short-term (2 weeks) endocrine therapy have been used as a surrogate endpoint for treatment response (26–29) and, in some studies, as a predictor of clinical outcome (30, 31). For example, both the IMPACT and P024 studies reported that post-treatment Ki-67 levels correlated with 5-year recurrence-free survival (30), relapse-free survival (31), and breast cancer-specific survival (31). As such, Ki-67 data has been included in a multiplex prognostic model, the Preoperative Endocrine Prognostic Index (PEPI), which incorporates tumor size, nodal status, ER, and Ki-67 expression following 3–4 months of neoadjuvant endocrine therapy to predict the long-term outcome in ER-positive breast cancer patients (29). However, what has not been addressed in studies to date is the significance of an unchanged Ki-67 level after treatment compared to an increase or decrease in expression. In the context of our data, we report a significant increase in the mean of Ki-67 index among untreated controls compared to no change from baseline in the treated group, suggesting that stable Ki-67 expression over time may still reflect a treatment response and thus warrants further consideration.
Of note, in this study, we chose a 10-day exposure to aromatase inhibition to avoid any unnecessary delay in surgical treatment beyond what is normally encountered at our institution (average institutional interval between diagnosis and surgery, 10.4 days; range, 1–46 days). Since previous studies have shown good correlation between observed physiologic and molecular changes after short (2 weeks) and intermediate (12–16 weeks) duration estrogen deprivation, we hypothesized that a 10-day exposure to aromatase inhibition would be adequate to induce an observable treatment response (9, 29). Furthermore, since the half-life of anastrozole is approximately 48 hours, the effect of pharmacokinetics in the setting of a 10-day exposure time is minimal. While we were unable to do so in this pilot study, incorporating serum estradiol levels into future preoperative window trials of anastrozole would be a valuable means to measure aromatase inhibition and confirm this effect. Similarly, although there were no significant clinical differences between anastrozole-treated cases and controls in this pilot study, we acknowledge that randomization to treatment or control arms would have been the preferred study design.
Because our gene expression platform was limited to 502 cancer-related genes (at the time of this study the whole-genome DASL assay was not available), there may be additional effects on other biological systems or other known estrogen-responsive genes that our study did not capture. Finally, due to limited material remaining in the core biopsy specimens, we were unable to perform reverse transcription-polymerase chain reaction to validate the changes in gene expression over time; however, we were able to demonstrate concordant changes in proliferation by IHC. The significance of changes in genes related to angiogenesis and apoptosis remains unclear.
Conclusions
In summary, short-term aromatase inhibition with anastrozole led to early detectable genetic changes in FFPE samples supporting the rationale for peri-operative window trials to evaluate molecular mechanisms of treatment response. However, in the absence of treatment, changes in gene expression were noted simply when sampling was performed at 2 different time points, suggesting that the wound-healing response may be a potential confounder in interpreting data from window trials. When using Ki-67 as a biomarker of treatment response, 54% of our patients showed a reduction on Ki-67, 15% remained stable, and 30% showed an increased Ki-67 index varying from 1% to 10%. Our gene expression data support the hypothesis that the wound-healing response has a significant effect on early gene expression and that short-term exposure to anastrozole can suppress this effect in ER-positive breast cancer.
Acknowledgments
Acknowledgements and Funding
This work was supported by AstraZeneca, Investigator-Sponsored Study IRUSANAS0036.
List of Abbreviations
- ER
estrogen receptor
- DASL
cDNA-mediated annealing, selection, extension, and ligation
- FFPE
formalin-fixed, paraffin-embedded
- CNB
core needle biopsy
- PR
progesterone receptor
- FISH
fluorescence in situ hybridization
- IHC
immunohistochemistry
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- CTGF
connective tissue growth factor
- ILK
integrin-linked kinase
- CSF2
colony stimulating factor-2
- PEPI
pre-operative endocrine prognostic index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest
The author(s) declare that they have no competing interests.
Author Contributions
Morrogh M contributed to the conception and design, collection and assembly of data, data analysis and interpretation of statistics, and manuscript writing.
Andrade V contributed to the collection and assembly of data, data analysis and interpretation of statistics, manuscript writing, and final approval of the manuscript.
Patil A contributed to the collection and assembly of data.
Qin L contributed to the data analysis and interpretation of statistics and manuscript writing.
Mo Q contributed to the data analysis and interpretation of statistics.
Sakr R contributed to the conception and design.
Arroyo C contributed to the collection and assembly of data.
Brogi E contributed administrative support and helped provide study material or patients.
Morrow M contributed administrative support.
King TA contributed to the conception and design, provision of study material or patients, collection and assembly of data, manuscript writing and final approval of the manuscript.
References
- 1.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Chang JC, Makris A, Gutierrez MC, Hilsenbeck SG, Hackett JR, Jeong J, Liu ML, Baker J, Clark-Langone K, Baehner FL, Sexton K, Mohsin S, Gray T, Alvarez L, Chamness GC, Osborne CK, Shak S. Gene expression patterns in formalin-fixed, paraffin-embedded core biopsies predict docetaxel chemosensitivity in breast cancer patients. Breast Cancer Res Treat. 2008;108:233–240. doi: 10.1007/s10549-007-9590-z. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Dixon M. A study looking at the changes in the breast cancer of postmenopausal women having hormone therapy (Primary Endocrine Response Study) Edinburgh: 2011. [Google Scholar]
- 5.Smith I. A trial of a short course of hormone therapy before and after surgery for early breast cancer (POETIC) Multiple cities, UK: 2011. [Google Scholar]
- 6.Kagawa S, Matsuo A, Yagi Y, Ikematsu K, Tsuda R, Nakasono I. The time-course analysis of gene expression during wound healing in mouse skin. Leg Med (Tokyo) 2009;11:70–75. doi: 10.1016/j.legalmed.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 8.Mello-Grand M, Singh V, Ghimenti C, Scatolini M, Regolo L, Grosso E, Zambelli A, Da Prada GA, Villani L, Fregoni V, Baiardi P, Marsoni S, Miller WR, Costa A, Chiorino G. Gene expression profiling and prediction of response to hormonal neoadjuvant treatment with anastrozole in surgically resectable breast cancer. Breast Cancer Res Treat. 2010;121:399–411. doi: 10.1007/s10549-010-0887-y. [DOI] [PubMed] [Google Scholar]
- 9.Mackay A, Urruticoechea A, Dixon JM, Dexter T, Fenwick K, Ashworth A, Drury S, Larionov A, Young O, White S, Miller WR, Evans DB, Dowsett M. Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res. 2007;9:R37. doi: 10.1186/bcr1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper L, Johnson C, Burslem F, Martin P. Wound healing and inflammation genes revealed by array analysis of ‘macrophageless’ PU.1 null mice. Genome Biol. 2005;6:R5. doi: 10.1186/gb-2004-6-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorski DH, Walsh K. Control of vascular cell differentiation by homeobox transcription factors. Trends Cardiovasc Med. 2003;13:213–220. doi: 10.1016/s1050-1738(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 12.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008;19:133–144. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Yang J, Dai C, Wu C, Liu Y. Role for integrin-linked kinase in mediating tubular epithelial to mesenchymal transition and renal interstitial fibrogenesis. J Clin Invest. 2003;112:503–516. doi: 10.1172/JCI17913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Q, Sui W, Xie S, Chen H, Zou G, Guo J, Zou H. Expression and role of integrin-linked kinase and collagen IV in human renal allografts with interstitial fibrosis and tubular atrophy. Transpl Immunol. doi: 10.1016/j.trim.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Peroukides S, Bravou V, Varakis J, Alexopoulos A, Kalofonos H, Papadaki H. ILK overexpression in human hepatocellular carcinoma and liver cirrhosis correlates with activation of Akt. Oncol Rep. 2008;20:1337–1344. [PubMed] [Google Scholar]
- 16.Sobolewski C, Cerella C, Dicato M, Ghibelli L, Diederich M. The role of cyclooxygenase-2 in cell proliferation and cell death in human malignancies. Int J Cell Biol. 2010;2010:215158. doi: 10.1155/2010/215158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nassar A, Radhakrishnan A, Cabrero IA, Cotsonis G, Cohen C. COX-2 expression in invasive breast cancer: correlation with prognostic parameters and outcome. Appl Immunohistochem Mol Morphol. 2007;15:255–259. doi: 10.1097/01.pai.0000213130.63417.b3. [DOI] [PubMed] [Google Scholar]
- 18.Ristimaki A, Sivula A, Lundin J, Lundin M, Salminen T, Haglund C, Joensuu H, Isola J. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–635. [PubMed] [Google Scholar]
- 19.Schmitz KJ, Callies R, Wohlschlaeger J, Kimmig R, Otterbach F, Bohr J, Lee HS, Takeda A, Schmid KW, Baba HA. Overexpression of cyclo-oxygenase-2 is an independent predictor of unfavourable outcome in node-negative breast cancer, but is not associated with protein kinase B (Akt) and mitogen-activated protein kinase (ERK1/2, p38) activation or with Her-2/neu signalling pathways. J Clin Pathol. 2006;59:685–691. doi: 10.1136/jcp.2005.030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller WR, Larionov AA, Renshaw L, Anderson TJ, White S, Murray J, Murray E, Hampton G, Walker JR, Ho S, Krause A, Evans DB, Dixon JM. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet Genomics. 2007;17:813–826. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]
- 21.Midwood KS, Williams LV, Schwarzbauer JE. Tissue repair and the dynamics of the extracellular matrix. Int J Biochem Cell Biol. 2004;36:1031–1037. doi: 10.1016/j.biocel.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Retsky M, Demicheli R, Hrushesky W. Wounding from biopsy and breast-cancer progression. Lancet. 2001;357:1048. doi: 10.1016/S0140-6736(05)71626-3. [DOI] [PubMed] [Google Scholar]
- 23.Retsky M, Demicheli R, Hrushesky WJ. Does surgery induce angiogenesis in breast cancer? Indirect evidence from relapse pattern and mammography paradox. Int J Surg. 2005;3:179–187. doi: 10.1016/j.ijsu.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Miller WR, White S, Dixon JM, Murray J, Renshaw L, Anderson TJ. Proliferation, steroid receptors and clinical/pathological response in breast cancer treated with letrozole. Br J Cancer. 2006;94:1051–1056. doi: 10.1038/sj.bjc.6603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cvoro A, Tatomer D, Tee MK, Zogovic T, Harris HA, Leitman DC. Selective estrogen receptor-beta agonists repress transcription of proinflammatory genes. J Immunol. 2008;180:630–636. doi: 10.4049/jimmunol.180.1.630. [DOI] [PubMed] [Google Scholar]
- 26.Clarke RB, Laidlaw IJ, Jones LJ, Howell A, Anderson E. Effect of tamoxifen on Ki67 labelling index in human breast tumours and its relationship to oestrogen and progesterone receptor status. Br J Cancer. 1993;67:606–611. doi: 10.1038/bjc.1993.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, Griffith C, Boeddinghaus I, Salter J, Detre S, Hills M, Ashley S, Francis S, Walsh G. Short-term changes in Ki-67 during neoadjuvant treatment of primary breast cancer with anastrozole or tamoxifen alone or combined correlate with recurrence-free survival. Clin Cancer Res. 2005;11:951s–958s. [PubMed] [Google Scholar]
- 28.Ellis MJ, Coop A, Singh B, Tao Y, Llombart-Cussac A, Janicke F, Mauriac L, Quebe-Fehling E, Chaudri-Ross HA, Evans DB, Miller WR. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 2003;63:6523–6531. [PubMed] [Google Scholar]
- 29.Dowsett M, Ebbs SR, Dixon JM, Skene A, Griffith C, Boeddinghaus I, Salter J, Detre S, Hills M, Ashley S, Francis S, Walsh G, Smith IE. Biomarker changes during neoadjuvant anastrozole, tamoxifen, or the combination: influence of hormonal status and HER-2 in breast cancer--a study from the IMPACT trialists. J Clin Oncol. 2005;23:2477–2492. doi: 10.1200/JCO.2005.07.559. [DOI] [PubMed] [Google Scholar]
- 30.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R, Salter J, Detre S, Hills M, Walsh G. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99:167–170. doi: 10.1093/jnci/djk020. [DOI] [PubMed] [Google Scholar]
- 31.Ellis MJ, Tao Y, Luo J, A’Hern R, Evans DB, Bhatnagar AS, Chaudri Ross HA, von Kameke A, Miller WR, Smith I, Eiermann W, Dowsett M. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100:1380–1388. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]