Abstract
Objective
Cyclooxygenase-2 (COX-2) expression is associated with the pathogenesis of chronic inflammation and pain in osteoarthritis (OA). A study was undertaken to determine whether interleukin-1β (IL-1β)-mediated induction of COX-2 can be regulated by microRNAs (miRNAs) in OA.
Methods
Human chondrocytes were stimulated with IL-1β in vitro. Total RNA was prepared using Trizol reagent. Gene expression was quantified using TaqMan Assays and miRNA targets were identified using bioinformatics. Transfection with reporter construct and premiRNA and antimiRNA was employed to verify suppression of target mRNA. Expression of COX-2 proteins was determined by immunoblotting. The role of activated p38-MAPKs was evaluated using specific inhibitor.
Results
The 3′UTR of COX-2 mRNA contained the ‘seed-matched’ sequences for miR-199a* and miR-101_3. Increased expression of COX-2 correlated with the downregulation of miR-199a* and miR-101_3 in IL-1β-stimulated normal and OA chondrocytes. miR-199a* directly suppressed the luciferase activity of a COX-2 3′UTR reporter construct and inhibited the IL-1β-induced expression of COX-2 protein in OA chondrocytes. Modulation of miR-199a* expression also caused significant inhibition of IL-1β-induced upregulation of mPGES1 and prostaglandin E2 production in OA chondrocytes. Activation of p38-MAPK downregulated the expression of miR-199a* and induced COX-2 expression. Treatment with antimiR-101_3 increased COX-2 expression in IL-1β-stimulated chondrocytes, but overexpression of miR-101_3 had no significant effect on COX-2 protein expression.
Conclusions
miR-199a* is a direct regulator of COX-2 expression in OA chondrocytes. IL-1β-induced activation of p38-MAPK correlates inversely with miR199a* expression levels. miR-199a* may be an important regulator of human cartilage homeostasis and a new target for OA therapy.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous small (approximately 22 nucleotide) RNAs and mediate gene regulatory events by pairing with target mRNAs and suppressing their expression. Hundreds of miRNAs have been identified so far, many of which are conserved and predicted to regulate the expression of one-third of mammalian genes.1 In the last few years it has become clear that miRNAs play an important role in many human diseases including rheumatoid arthritis (RA) and osteoarthritis (OA).2–9 OA is a debilitating disease which probably evolves from a local inflammatory response to a chronic process with a variable degree of inflammation and degeneration of articular cartilage leading to the exposure of underlying bone, pain and disability.10,11 The role of miRNAs in maintaining cartilage homeostasis during development and their dysregulation in OA has also recently been shown.12–15 There is strong evidence for a key role of interleukin-1β (IL-1β) in the pathogenesis of OA,16 and the altered expression of miRNAs in OA and RA and in regulating the expression of matrix metalloproteinases (MMPs), ADAMTS-5, tumour necrosis factor α and insulin-like growth factor binding protein 5 (IGFBP-5) in OA has previously been reported.13,15,17–23 The expression of miR-146a was found to be induced by IL-1β and linked to pain-related pathology of OA; overexpression of miR-146a was found to be associated with upregulation of Aggrecan and COL2A1 expression in IL-1β-stimulated OA chondrocytes.22,24 Silencing of miR-34 was shown to reduce IL-1β-induced apoptosis in rat knee chondrocytes.25 The expression of miR-140 is high in normal cartilage but low in OA, and miR-140 knockout mice develop OA-like pathology with age.15,19 IL-1β-mediated overexpression of cyclooxygenase-2 (COX-2) strongly contributes to the inflammation and cartilage degeneration in OA via prostaglandin E2 (PGE2) production.26,27 As miRNAs are novel selective regulators of gene expression and probably have an important functional role in cartilage homeostasis, we determined whether the expression of COX-2 is regulated by specific miRNAs in human OA chondrocytes. We also determined the role of IL-1β and the activated signalling events in modulating the expression of COX-2 mRNA and the miRNAs that regulate COX-2 expression and PGE2 production. These results may be of value in the design of novel therapies for the treatment of OA.
METHODS
Clinical samples
OA was diagnosed according to the American College of Rheumatology criteria.28,29 OA cartilage samples were obtained from 46 patients with OA undergoing total joint arthroplasty at our hospital. It is important to note that these patients must have been treated with non-steroidal anti-inflammatory drugs (NSAIDs) but were unlikely to be on NSAIDs at the time of surgery since a 7–10-day washout period is required prior to surgery. Normal cartilage samples were taken from trauma patients with no known history of OA or RA (n=7).
Chondrocyte isolation and culture
Macroscopic cartilage degeneration was determined by staining with India ink.30 Portions of the cartilage with a smooth articular surface were used to prepare chondrocytes as previously described.17,31,32 Primary chondrocytes at 80% confluence were used for all the experiments described here.
Treatment of chondrocytes and preparation of microRNAs
OA chondrocytes were serum-starved overnight and then stimulated with IL-1β (5 ng/ml; R&D Systems, St Paul, Minnesota, USA) or p38-MAPK inhibitor (SB202190 100 μM; A G Scientific Inc, San Diego, California, USA) for the indicated time and total RNA was prepared using the TRIZOL reagent (Invitrogen, Carlsbad, California, USA). MicroRNAs were purified using the mirVANA kit according to the manufacturers’ instructions (Applied Biosystems, Foster City, California, USA).
Reverse transcription and quantitative RT-PCR analysis
Five hundred nanogram of genomic DNA-free total RNA was reverse-transcribed using QuantiTect Reverse Transcription Kit (Qiagen, Chatsworth, California, USA) according to the manufacturer’s protocol. The expression of COX-2 mRNA and mature miRNAs was quantified using the TaqMan Gene Expression Assays (Applied Biosystems). RNU6B/glyceraldehyde 3-phosphate dehydrogenase expression was used as an endogenous control. A threshold cycle (Ct) was observed in exponential phases of amplification and quantification of relative expression levels was determined by the ΔΔCt method. The value of each control sample was set at 1 and was used to calculate the fold change in mRNA/miRNA expression.
Luciferase reporter assay
A luciferase reporter vector containing the entire 3′UTR of COX-2 mRNA (NM_000963.2) was obtained commercially (SwitchGear Genomics, Menlo Park, California, USA). Empty vector containing only luciferase gene and its constitutively active promoter was used as control. OA chondrocytes were co-transfected with 100 ng reporter plasmid, 50 and 100 nM selected premiRNAs (Qiagen, Hilden, North Rhine-Westphalia, Germany) or negative control miRNAs (Ambion, Austin, Texas, USA) using HiPerfect Transfection Reagent (Qiagen) in a 96-well plate. Cell lysates were harvested 24 h after transfection and luciferase activity was assayed using a kit (Promega, Madison, Wisconsin, USA) and was measured using the EnSpire 2300 Multilabel Reader (Perkin Elmer, Waltham, Massachusetts, USA). The luciferase activity of COX-2 construct co-transfected with premiRNA was derived after subtracting the value of COX-2 construct co-transfected with negative control miRNA. Each experiment was performed in triplicate.
Modulation of COX-2 expression by premiRNAs and antimiRNAs in human OA chondrocytes
Human primary OA chondrocytes were transfected with selected premiRNAs and antimiRNAs (Qiagen) by nucleofection optimised for use with human articular chondrocytes using the Amaxa Human Chondrocytes Nucleofector kit (Lonza, Cologne, Germany).
Western blot analysis
Control and treated or transfected primary OA chondrocytes were washed with cold phosphate buffered saline and lysed using the cell lysis buffer and the blots were prepared as previously described.33 Membranes were blocked with blocking buffer containing non-fat dry milk powder in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) and probed with 1:1000 diluted antiCOX-2, anti-β-actin and 1:250 diluted ant-imPGES1 primary antibodies (Cell Signaling Technologies, Beverley, Massachusetts, USA; Santa Cruz Biotechnology, Santa Cruz, California, USA; Cayman Chemicals, Ann Arbor, Michigan, USA). After extensive washing, immunoreactive proteins were visualised by using horseradish peroxidase-linked secondary antibodies and enhanced chemiluminescence (GE Healthcare, Milwaukee, Wisconsin, USA). Images were captured using the AFP-Imaging System (Minimedical Series, Elms Ford, New York, USA) and analysed using the UN-SCAN-IT software (Silk Scientific Corporation, Idaho, Utah, USA). Each band was scanned three times with background correction and values were expressed as average pixel/band.
Measurement of PGE2 production
Levels of PGE2 in chondrocyte culture supernatant were quantified using a commercially available prostaglandin E2 EIA kit (Enzo Life Sciences, Farmingdale, New York, USA) according to the manufacturers’ instructions.
Statistical analysis
Comparisons were performed using Origin 6.1 software package (one paired two-tailed t test with one-way analysis of variance and Tukey post hoc analysis) and p<0.05 was considered significant.
RESULTS
Computer prediction of miRNAs targeting 3′UTR of COX-2 mRNA (NM_000963.2)
MIRANDA (http://www.microrna.org), TargetScan (http://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/) were used to identify the miRNAs targeting COX-2 3′UTR and four selected miRNAs (miR-199a*, miR-101_3, miR-26a and miR-26b) with seed-matched sequence in the 3′UTR of COX-2 mRNA for further studies. Predicted duplex of selected miRNAs with seed-matched sites in the 3′UTR of human COX-2 mRNA are shown in figure 1A–D. These findings suggest that the identified miRNAs might target the COX-2 mRNA by directly recognising the respective seed-matched sequences present in the 3′UTR.
Figure 1.
Seed sequences of selected miRNAs in 3′UTR of cyclooxygenase-2 (COX-2) mRNA. (A–D) Seed-matched sequences for miR-199a*, miR-101_3, miR-26a and miR-26b in the 3′UTR of COX-2 mRNA and conservation of their cross species was identified by computational algorithms and the data were used to predict the functional miRNA binding sites in human COX-2 mRNA. The sequences in red are the locations of the potential seed-matched sequence for the miRNAs studied.
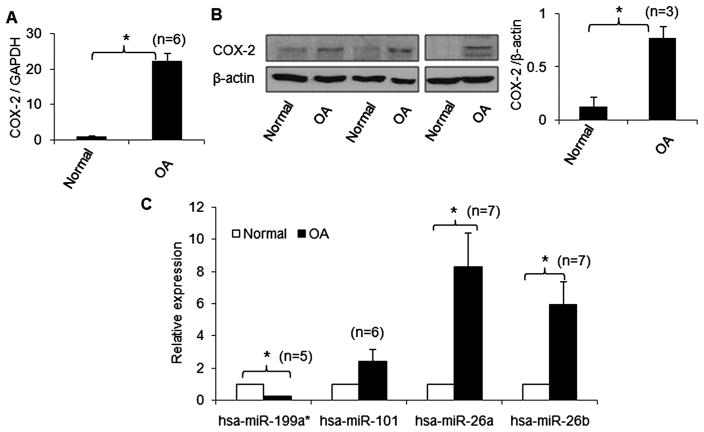
Basal expression levels of COX-2, miR-199a*, miR-101_3, miR-26a and miR-26b in OA and non-OA-derived chondrocytes
A high level of COX-2 expression has been found in the synovium and cartilage in arthritis and selective inhibition of COX-2 may result in amelioration of the disease.27 To determine if the expression levels of the selected miRNAs and COX-2 expression correlate in OA, expression of miRNAs and COX-2 was analysed by real-time PCR and western immunoblotting, respectively (figure 2). OA chondrocytes had higher levels of COX-2 mRNA and protein expression than chondrocytes from non-OA individuals (figure 2A, B). Interestingly, in chondrocytes derived from OA cartilage, the expression of miR-199a* was significantly lower (~4.5-fold; p<0.05) compared with normal chondrocytes (figure 2C). However, the expression levels of miR-26a and miR-26b were significantly higher (~8.2-fold and ~5.4-fold, respectively) in OA chondrocytes compared with normal chondrocytes (figure 2C; p<0.05). The expression level of miR-101_3 was not significantly different between OA and non-OA chondrocytes.
Figure 2.
Correlation between basal expression levels of cyclooxygenase-2 (COX-2), miR-199a*, miR-101_3, miR-26a and miR-26b in human chondrocytes derived from osteoarthritis (OA) and non-OA cartilage samples. (A, B) COX-2 mRNA and protein expression determined by TaqMan assay and western immunoblotting, respectively. (C) Basal levels of miR-199a*, miR-101_3, miR-26a and miR-26b in cartilage from OA and healthy individuals. Expression levels of miRNAs were determined by TaqMan assays. The expression level in non-OA samples was set as a control for comparison with diseased tissue and expression of RNU6B/GAPDH was used as an endogenous control. Each experiment was performed in duplicate with the indicated number of patient samples. *p<0.05.
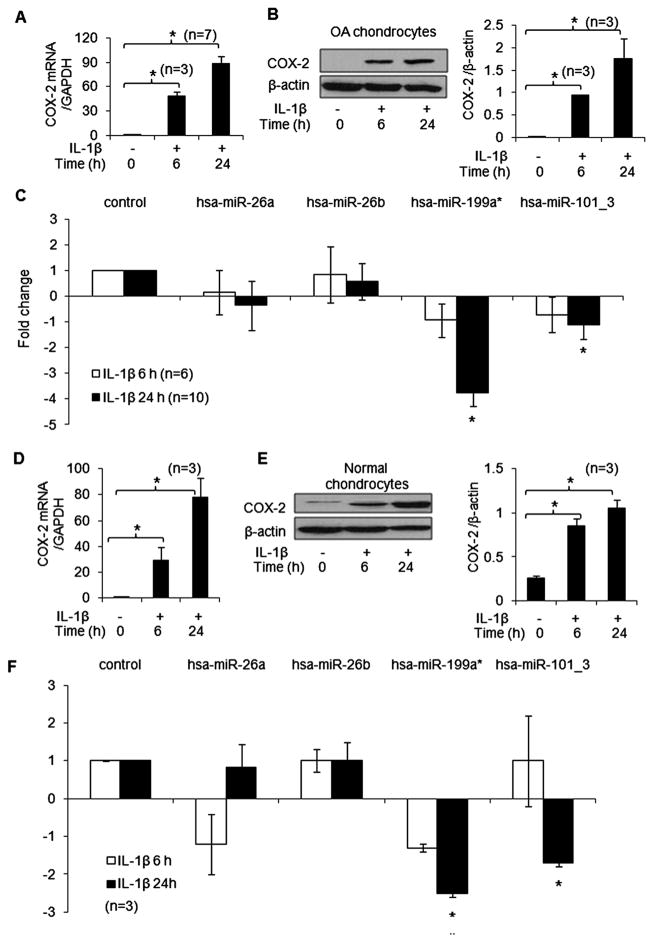
Correlation between IL-1β-stimulated COX-2 production and expression of selected miRNAs in normal and OA chondrocytes
IL-1β stimulation resulted in increased expression of COX-2 mRNA and protein in OA chondrocytes (figure 3A, B; p<0.05). Importantly, IL-1β-stimulated OA chondrocytes also showed a significant downregulation of miR-199a* expression 24 h after stimulation (~3.7-fold) but not at 6 h when COX-2 protein expression was lower (figure 3C; p<0.05). Similar inverse correlation between miR-199a* expression (~2.4-fold) and COX-2 protein was also observed when normal chondrocytes were stimulated with IL-1β (figure 3D–F; p<0.05). The expression of miR-101_3 was also significantly downregulated by IL-1β in both normal (~2.0-fold) and OA chondrocytes (~1.2-fold; n=19) and showed the same inverse correlation with COX-2 protein expression in these cells as was observed for miR-199a* (figure 3; p<0.05). No significant change in miR-26a and miR-26b expression was observed after stimulation with IL-1β in normal or OA chondrocytes (figure 3; p<0.05).
Figure 3.
Interleukin-1β (IL-1β) upregulates the expression of cyclooxygenase-2 (COX-2) and suppresses the expression of miR-199a*, miR-101_3 in osteoarthritis (OA) and normal chondrocytes. (A, D) COX-2 mRNA expression was determined by TaqMan assay. (B, E) COX-2 protein expression was determined by western immunoblotting. (C, F) Expression levels of miR-199a*, miR-101_3, miR-26a and miR-26b in IL-1β-stimulated OA and normal chondrocytes were determined by TaqMan assays. Unstimulated chondrocytes were used as controls and expression of RNU6B/glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control. Each experiment was performed in duplicate with the indicated number of patient samples. *p<0.05.
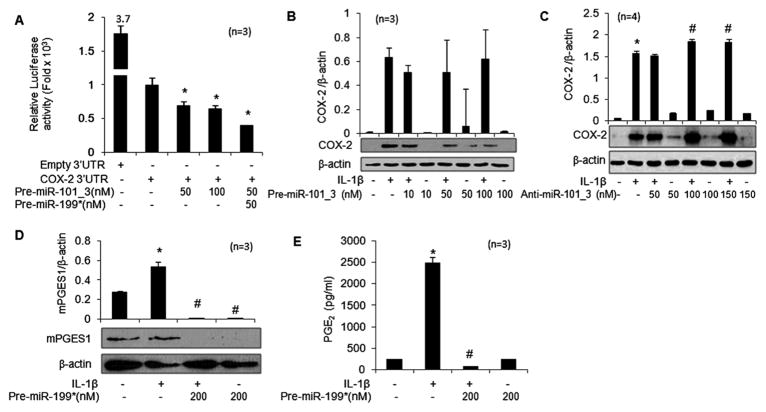
Overexpression of miR-199a* inhibits the activity of the reporter vector
To test whether the 3′UTR of COX-2 is a functional target of the computer predicted miRNAs, we used a COX-2 3′UTR luciferase reporter vector containing the seed-matched sites for the identified miRNAs. A marked concentration-dependent reduction in luciferase activity (50–60%) was observed in chondrocytes co-transfected with premiR-199a* at 24 h (figure 4A; p<0.05). Co-transfection of COX-2 reporter with premiR-101_3 also showed a decrease in luciferase activity of ~30% (figure 5A; p<0.05). In contrast, co-transfection of the reporter vector with either premiR-26a or premiR-26b had no significant effect on the luciferase activity (data not shown). The luciferase activity of COX-2 reporter was downregulated by ~60% when co-transfected with both miR-199a* and miR-101_3 (10 nM each), suggesting a synergistic effect (figure 5A; p<0.05). These results demonstrated that miR-199a* and miR-101_3 both bind the seed sequence present in the 3′UTR of human COX-2 mRNA, but the inhibitory effect on luciferase activity was more pronounced for miR-199a*.
Figure 4.
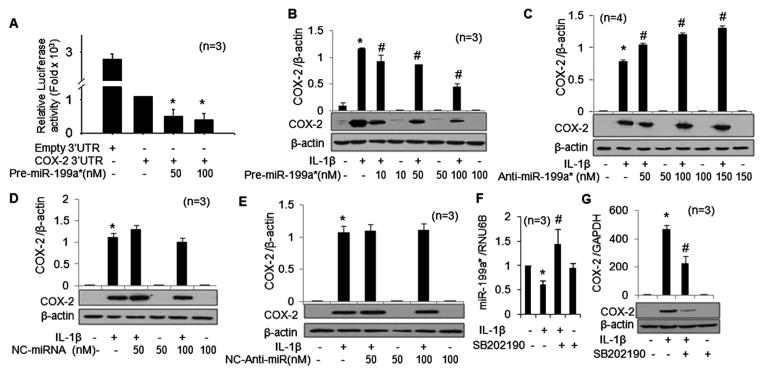
miR-199a* directly inhibits the expression of cyclooxygenase-2 (COX-2) in osteoarthritis chondrocytes. (A) Luciferase activity in chondrocytes transfected with the reporter vector and premiR-199a* (*p<0.05). (B, C) Expression of COX-2 protein in premiR-199a* or antimiR-199a* transfected chondrocytes after 72 h of stimulation with interleukin-1β (IL-1β). (D, E) Expression of COX-2 protein in negative control premiRNA or negative control-antimiRNA-transfected chondrocytes after 72 h of stimulation with IL-1β. (F, G) Expression levels of miR-199a* and COX-2 in chondrocytes treated with the p38-MAPK inhibitor (SB202190) and stimulated with IL-1β were determined by TaqMan assays. Gene expression in unstimulated chondrocytes was used as a control and expression of RNU6B/glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control. COX-2 protein expression was determined by western immunoblotting. Each experiment was performed in duplicate with samples from the indicated number of patients. *p<0.05 vs control; #p<0.05 vs IL-1β-stimulated chondrocytes.
Figure 5.
Regulation of cyclooxygenase-2 (COX-2) expression by miR-101_3 and effect of overexpression of miR-199a* on prostaglandin E2 production in OA chondrocytes. (A) miR-199a* and miR-101_3 synergistically inhibited luciferase activity in human OA chondrocytes transfected with a reporter vector containing the 3′UTR of human COX-2 mRNA; *p<0.05 vs COX-2 3′UTR. (B, C) Expression of COX-2 protein in premiR-101_3 or antimiR-101_3 transfected chondrocytes after 72 h of stimulation with interleukin-1β (IL-1β). (D, E) Overexpression of miR-199a* inhibited IL-1β-induced mPGES1 protein expression and prostaglandin E2 (PGE2) production in OA chondrocytes. COX-2 and mPGES1 protein expression was determined by western immunoblotting. PGE2 production in culture supernatants was analysed by ELISA. Each experiment was performed using indicated number of patients. *p<0.05 vs control; #p<0.05 vs IL-1β-stimulated chondrocytes.
miR-199a* directly and miR-101_3 indirectly regulate the expression of COX-2 protein in human chondrocytes
In these studies, transfection with premiR-199a* dramatically reduced COX-2 protein expression at 72 h up to ~62% (figure 4B; p<0.05), while transfection with miR-199a* antagomir showed an increase in COX-2 protein expression of 19–35% in IL-1β-stimulated OA chondrocytes (figure 4C; p<0.05). As expected, negative control premiRNA or negative control-antimiRNA had no effect on IL-1β-induced COX-2 protein expression in OA chondrocytes (figure 4D, E; p<0.05). IL-1β-stimulated OA chondrocytes transfected with premiR-101_3 showed a trend towards inhibition of COX-2 expression, but the level did not reach statistical significance (figure 5B; p>0.05). Interestingly, transfection of OA chondrocytes with the miR-101_3 inhibitor resulted in a dose-dependent increase in COX-2 protein expression upon IL-1β-stimulation (figure 5C; p<0.05). This suggests an indirect effect of miR-101_3 on the post-transcriptional regulation of COX-2 expression in OA chondrocytes.
Negative regulation of miR-199a* by p38-MAPKs in human OA chondrocytes
Studies have shown that the transcriptional response of the COX-2 gene to IL-1β is controlled by p38-MAPK but not by JNK or ERK.34 However, the role of MAPKs in regulating the expression of miRNAs in human OA chondrocytes has not yet been reported. In these studies, OA chondrocytes showed a significant increase in COX-2 mRNA (~470-fold) and protein expression upon IL-1β-stimulation (figure 4G; p<0.05). Importantly, OA chondrocytes pretreated with the p38-MAPK inhibitor SB202190 showed an increase of ~58% in miR-199a* expression (figure 4F; p<0.05) and a significant decrease in COX-2 mRNA and protein expression (figure 4G; p<0.05). These data suggest that IL-1β-induced activation of p38-MAPK negatively regulates the expression of miR-199a* which may be necessary for the unobstructed translation of COX-2 mRNA and expression of COX-2 protein in human OA chondrocytes.
Regulation of PGE2 production by miR-199a*
COX-2 converts arachidonic acid to PGH2 which is then converted to PGE2 by microsomal prostaglandin E synthase-1 (mPGES1). Both COX-2 and mPGES1 are co-induced in response to the proinflammatory cytokine IL-1β.27 As the expression of miR-199a* correlated with COX-2 protein expression, we determined whether this also affects PGE2 production in IL-1β-stimulated OA chondrocytes. OA chondrocytes were transfected with premiR-199a* and it was found that IL-1β stimulation of chondrocytes resulted in significant upregulation of mPGES1 expression and PGE2 production (figure 5D, E; p<0.05). However, overexpression of miR-199a* resulted in a significant decrease in mPGES1 protein expression and concomitant PGE2 production upon IL-1β stimulation in OA chondrocytes (figure 5D, E; p<0.05). Using TargetScanS (Version 5.2), no binding sites for miR-199a* were found in the 3′UTR of human mPGES1 mRNA (NM_004878.4). These data suggest that miR-199a* indirectly regulates the expression of mPGES1 in OA chondrocytes.
DISCUSSION
OA is a progressive degenerative joint disorder characterised by gradual degeneration of articular cartilage, periarticular bone changes and secondary synovitis.35 Pain and inflammation of the affected joints is the most important symptom in patients with OA.36 Proinflammatory cytokine IL-1β-induced COX-2 expression contributes to inflammation and pain via PGE2 production.36 COX-2 is the rate-limiting enzyme in PGE2 production and its inhibition is sufficient to achieve analgesic and anti-inflammatory efficacy.37 Recent studies have shown that miRNAs play a crucial role in human disease and can be a potential new therapeutic target.3,5,6,9 Since miRNAs provide quantitative regulation of genes rather than on/off signals, they can be thought of as molecules that fine tune the response of a cell to different stimuli.1 Consequently, manipulation of miRNA levels could lead to novel therapeutic strategies to combat OA. Miyaki et al19 showed that IL-1β treatment suppresses miR-140 expression, and overexpression of miR-140 can downregulate IL-1β-induced ADAMTS-5 expression in chondrocytes. Furthermore, they also showed that miR-140−/− mice develop age-related OA-like changes characterised by proteoglycan loss and fibrillation of articular cartilage.15 Yamasaki et al24 found an association between the decreased expression of miR-146a and increased expression of MMP-13 in OA cartilage. Moreover, the detection of miRNAs in plasma and synovial fluid from patients with RA and OA has been postulated to be a useful biomarker for monitoring disease activity.38 Overexpression of miR-155 has been linked to repression of MMP-3 expression in synovial fibroblasts from patients with RA, and miR-140 has been reported to be expressed in cartilage during limb development and to inhibit histone deacetylase 4 expression.20,39 miR-124a has been reported to regulate the proliferation of RA synovial fibroblasts by suppressing the production of the cyclin-dependent kinase 2 and monocyte chemoattractant protein 1 proteins.40 These studies point to the importance of miRNAs in maintaining cartilage homeostasis as their deregulation correlated strongly with the disease.
This is the first study to identify the regulation of COX-2 by miRNAs in OA chondrocytes. We found that COX-2 expression was regulated directly by miR-199a* and indirectly by miR-101_3 in human chondrocytes, and provide the data linking miR-199a* and miR-101_3 to OA by demonstrating their presence and modulation by IL-1β in human chondrocytes derived from normal and OA cartilage. After identifying miRNAs with seed-matched sequence in the COX-2 mRNA 3′UTR region (miR-199a*, miR-101_3, miR-26a and miR-26b), their expression was analysed in chondrocytes derived from OA cartilage. Expression analysis showed that only miR-199a* had an inverse correlation with COX-2 protein expression in chondrocytes derived from normal and OA cartilage samples. Downregulation of miR-199a* expression by IL-1β and enhanced production of COX-2 protein was observed in human chondrocytes, suggesting a direct link between the expression of miR-199a* and the expression of COX-2 protein. We also showed that, although miR-26a and miR-26b were predicted to target the 3′UTR of COX-2 mRNA and their expression was significantly higher in patients with OA, no change in expression of these miRNAs was seen upon IL-1β stimulation in normal and OA chondrocytes, suggesting that these are not IL-1β-responsive genes. Additionally, these miRNAs do not bind the seed sequences in 3′UTR as no significant effect on luciferase activity was observed in transfected chondrocytes (data not shown). Thus, these miRNAs may not be involved in post-transcriptional regulation of COX-2 expression in human chondrocytes. Next we tested whether the 3′UTR of COX-2 is a direct target of miR-199a* or miR-101_3 using the luciferase reporter assay. A significant reduction was observed in the luciferase activity (50–60%; p<0.05) in chondrocytes with overexpression of miR-199a*. Co-transfection of the reporter vector with premiR-101_3 also showed a decrease in luciferase activity, but the reduction was not as pronounced as with miR-199a*. These data confirm the direct binding between miR-199a* and the seed sequence present in COX-2 3′UTR in OA chondrocytes. Furthermore, transfection of mature miR-199a* or its inhibitor induced significant silencing or marked upregulation, respectively, of COX-2 protein expression in IL-1β-stimulated OA chondrocytes. Silencing of COX-2 protein expression by mmu-miR-199a* and mmu-miR-101a has been shown during embryonic implantation in mice.41 Regulation of COX-2 expression by miR-101a in other cell types has also been shown previously.42,43 The sequences of miR-101a and miR-101_3 are the same, and both of these miRNAs target the same seed region (1735–1741 bp) in COX-2 mRNA. In our studies, transfection of chondrocytes with antimiR-101_3 upregulated the expression of COX-2 protein in IL-1β-stimulated OA chondrocytes, but we did not find a statistically significant change in the levels of COX-2 protein in chondrocytes from different patients with overexpression of miR-101_3 (figure 5B, C; p<0.05). This suggests that, in human chondrocytes, miR-101_3 may be an indirect regulator of COX-2 expression. p38-MAPK has been implicated in the regulation of COX-2 expression in IL-1β-stimulated chondrocytes.44,45 We found that activation of p38-MAPK acts as a negative regulator of miR-199a* expression as, in human OA chondrocytes treated with p38-MAPK inhibitor (SB202190), the expression of miR-199a* was enhanced but that of COX-2 mRNA and protein expression was reduced. IL-1β is one of the most prominent inducers of mediators of cartilage degradation and joint inflammation.46 miR-199a* and miR-101_3 appear to be components of a novel additional mechanism which maintains cartilage homeostasis that is suppressed by IL-1β in chondrocytes in order to allow the full manifestation of the inflammatory response in OA.
The synovial fluid from patients with OA contains high levels of PGE2.47 COX-2 acts on arachidonic acid to form PGH2. The prostanoid synthase mPGES1 produces PGE2 from PGH2. Several recent studies have shown that mPGES1 is critical for the inflammatory response in vivo and its expression can be induced by IL-1β in chondrocytes.48,49 As we found that miR-199a* transfected chondrocytes produce very little PGE2, we determined whether miR-199a* also regulates mPGES1 expression in IL-1β-stimulated OA chondrocytes. IL-1β stimulation of chondrocytes resulted in significant upregulation of mPGES1 expression and PGE2 production which was significantly inhibited by overexpression of miR-199a*. Although several miR-NAs have been predicted to target multiple mRNA transcripts involved in PGE2 production, using TargetScanS (Version 5.2) we could not find any binding site of miR-199a* within the 3′UTR of mPGES1.50 This could be an indirect effect of miR-199a* on mPGES1 expression in OA chondrocytes.
In conclusion, we have shown that miR-199a* can directly target COX-2 mRNA in human OA chondrocytes through binding a conserved site in the 3′UTR. Furthermore, the levels of miR-199a* are inversely correlated with COX-2 mRNA and protein levels in IL-1β-stimulated human chondrocytes. Our results suggest that expression of miR-199a* and possibly of miR-101_3 can be exploited in novel therapeutic strategies for the treatment of OA.
Acknowledgments
The authors thank Dr Brendan Patterson (Department of Orthopaedics, MetroHealth Medical Center) for providing human OA cartilage samples.
Funding This work was supported in part by National Institute of Health/National Centre for Complementary and Alternative Medicine grants (RO1 AT-003267; RO1-AT-005520, R21-AT504615) and funds from the MetroHealth Medical Center, Cleveland, Ohio, USA.
Footnotes
Competing interests None.
Ethics approval Permission to use de-identified and discarded human cartilage was obtained from the Institutional Review Board, MetroHealth Medical Centre, prior to the initiation of the studies. These studies were approved as ‘Exempt’ and no informed consent was required.
Provenance and peer review Not commissioned; externally peer reviewed.
Contributors NA carried out the experimental work, collection and interpretation of data and manuscript drafting. TMH conceived the study, its design, coordination, data interpretation and manuscript drafting and editing.
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–31. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 3.Mishra PK, Tyagi N, Kumar M, et al. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009;13:778–89. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Q, Gao Z, Alarcon RM, et al. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276:2348–58. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheedy FJ, O’Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008;67(Suppl 3):iii50–5. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- 6.Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–46. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y, Luo X, Cui H, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 9.Nakasa T, Nagata Y, Yamasaki K, et al. A mini-review: microRNA in arthritis. Physiol Genomics. 2011;43:566–70. doi: 10.1152/physiolgenomics.00142.2010. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann M. Pathogenesis of arthritis: recent research progress. Nat Immunol. 2001;2:771–3. doi: 10.1038/ni0901-771. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence RC, Felson DT, Helmick CG, et al. National Arthritis Data Workgroup. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos D, Malizos KN, Oikonomou P, et al. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr Cartil. 2009;17:464–72. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949–54. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blom AB, van der Kraan PM, van den Berg WB. Cytokine targeting in osteoarthritis. Curr Drug Targets. 2007;8:283–92. doi: 10.2174/138945007779940179. [DOI] [PubMed] [Google Scholar]
- 17.Akhtar N, Rasheed Z, Ramamurthy S, et al. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62:1361–71. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanczyk J, Ospelt C, Karouzakis E, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63:373–81. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyaki S, Nakasa T, Otsuki S, et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–30. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–9. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 21.Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–92. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Gibson G, Kim JS, et al. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011;480:34–41. doi: 10.1016/j.gene.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardif G, Hum D, Pelletier JP, et al. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamasaki K, Nakasa T, Miyaki S, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–41. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abouheif MM, Nakasa T, Shibuya H, et al. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49:2054–60. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 26.Laine L, White WB, Rostom A, et al. COX-2 selective inhibitors in the treatment of osteoarthritis. Semin Arthritis Rheum. 2008;38:165–87. doi: 10.1016/j.semarthrit.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Martel-Pelletier J, Pelletier JP, Fahmi H. Cyclooxygenase-2 and prostaglandins in articular tissues. Semin Arthritis Rheum. 2003;33:155–67. doi: 10.1016/s0049-0172(03)00134-3. [DOI] [PubMed] [Google Scholar]
- 28.Altman R, Alarcón G, Appelrouth D, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hip. Arthritis Rheum. 1991;34:505–14. doi: 10.1002/art.1780340502. [DOI] [PubMed] [Google Scholar]
- 29.Altman R, Asch E, Bloch D, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986;29:1039–49. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong CG, Mow VC. Variations in the intrinsic mechanical properties of human articular cartilage with age, degeneration, and water content. J Bone Joint Surg Am. 1982;64:88–94. [PubMed] [Google Scholar]
- 31.Singh R, Ahmed S, Islam N, et al. Epigallocatechin-3-gallate inhibits interleukin-1beta-induced expression of nitric oxide synthase and production of nitric oxide in human chondrocytes: suppression of nuclear factor kappaB activation by degradation of the inhibitor of nuclear factor kappaB. Arthritis Rheum. 2002;46:2079–86. doi: 10.1002/art.10443. [DOI] [PubMed] [Google Scholar]
- 32.Shukla M, Gupta K, Rasheed Z, et al. Bioavailable constituents/metabolites of pomegranate (Punica granatum L) preferentially inhibit COX2 activity ex vivo and IL-1beta-induced PGE2 production in human chondrocytes in vitro. J Inflamm (Lond) 2008;5:9. doi: 10.1186/1476-9255-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasheed Z, Akhtar N, Haqqi TM. Advanced glycation end products induce the expression of interleukin-6 and interleukin-8 by receptor for advanced glycation end product-mediated activation of mitogen-activated protein kinases and nuclear factor-κB in human osteoarthritis chondrocytes. Rheumatology (Oxford) 2011;50:838–51. doi: 10.1093/rheumatology/keq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulivi V, Giannoni P, Gentili C, et al. p38/NF-kB-dependent expression of COX-2 during differentiation and inflammatory response of chondrocytes. J Cell Biochem. 2008;104:1393–406. doi: 10.1002/jcb.21717. [DOI] [PubMed] [Google Scholar]
- 35.Flugge LA, Miller-Deist LA, Petillo PA. Towards a molecular understanding of arthritis. Chem Biol. 1999;6:R157–66. doi: 10.1016/S1074-5521(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 36.Dubois RN, Abramson SB, Crofford L, et al. Cyclooxygenase in biology and disease. FASEB J. 1998;12:1063–73. [PubMed] [Google Scholar]
- 37.Lipsky PE, Isakson PC. Outcome of specific COX-2 inhibition in rheumatoid arthritis. J Rheumatol Suppl. 1997;49:9–14. [PubMed] [Google Scholar]
- 38.Murata K, Yoshitomi H, Tanida S, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuddenham L, Wheeler G, Ntounia-Fousara S, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–7. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 40.Nakamachi Y, Kawano S, Takenokuchi M, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 41.Chakrabarty A, Tranguch S, Daikoku T, et al. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA. 2007;104:15144–9. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strillacci A, Griffoni C, Sansone P, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–47. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka T, Haneda S, Imakawa K, et al. A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation. 2009;77:181–7. doi: 10.1016/j.diff.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Faour WH, He Y, He QW, et al. Prostaglandin E(2) regulates the level and stability of cyclooxygenase-2 mRNA through activation of p38 mitogen-activated protein kinase in interleukin-1 beta-treated human synovial fibroblasts. J Biol Chem. 2001;276:31720–31. doi: 10.1074/jbc.M104036200. [DOI] [PubMed] [Google Scholar]
- 45.Thomas B, Thirion S, Humbert L, et al. Differentiation regulates interleukin-1beta-induced cyclo-oxygenase-2 in human articular chondrocytes: role of p38 mitogen-activated protein kinase. Biochem J. 2002;362(Pt 2):367–73. doi: 10.1042/0264-6021:3620367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daheshia M, Yao JQ. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. J Rheumatol. 2008;35:2306–12. doi: 10.3899/jrheum.080346. [DOI] [PubMed] [Google Scholar]
- 47.Notoya K, Jovanovic DV, Reboul P, et al. The induction of cell death in human osteoarthritis chondrocytes by nitric oxide is related to the production of prostaglandin E2 via the induction of cyclooxygenase-2. J Immunol. 2000;165:3402–10. doi: 10.4049/jimmunol.165.6.3402. [DOI] [PubMed] [Google Scholar]
- 48.Trebino CE, Stock JL, Gibbons CP, et al. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci USA. 2003;100:9044–9. doi: 10.1073/pnas.1332766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuko-Hongo K, Berenbaum F, Humbert L, et al. Up-regulation of microsomal prostaglandin E synthase 1 in osteoarthritic human cartilage: critical roles of the ERK-1/2 and p38 signaling pathways. Arthritis Rheum. 2004;50:2829–38. doi: 10.1002/art.20437. [DOI] [PubMed] [Google Scholar]
- 50.Moore AE, Young LE, Dixon DA. MicroRNA and AU-rich element regulation of prostaglandin synthesis. Cancer Metastasis Rev. 2011;30:419–35. doi: 10.1007/s10555-011-9300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]