SUMMARY
The melanoma antigen (MAGE) family consists of more than 60 genes, many of which are cancer-testis antigens that are highly expressed in cancer and play a critical role in tumorigenesis. However, the biochemical and cellular functions of this enigmatic family of proteins have remained elusive. Here, we identify really interesting new gene (RING) domain proteins as binding partners for MAGE family proteins. Multiple MAGE family proteins bind E3 RING ubiquitin ligases with specificity. The crystal structure of one of these MAGE-RING complexes, MAGE-G1-NSE1, reveals structural insights into MAGE family proteins and their interaction with E3 RING ubiquitin ligases. Biochemical and cellular assays demonstrate that MAGE proteins enhance the ubiquitin ligase activity of RING domain proteins. For example, MAGE-C2-TRIM28 is shown to target p53 for degradation in a proteasome-dependent manner, consistent with its tumorigenic functions. These findings define a biochemical and cellular function for the MAGE protein family.
INTRODUCTION
Cancer-testis antigens are genes whose expression is typically restricted to cells of the germline and trophoblast lineage but are aberrantly expressed in a wide variety of human tumors (Simpson et al., 2005). The list of cancer-testis antigens has grown during the past two decades to include more than 100 different genes, with additional genes continuing to be discovered (Scanlan et al., 2004). The majority of these genes are on the X chromosome as multigene families that share a common genomic locus (Scanlan et al., 2002). These include the melanoma antigen (MAGE), G antigen (GAGE), and X chromosome antigen (XAGE) multigene families. Evolutionarily, these gene families have recently undergone expansion and share high sequence homology between family members (Scanlan et al., 2002). In addition, they are often under similar transcriptional regulation, resulting in their coexpression, typically in the highly proliferative spermatogonia germ cells (Jungbluth et al., 2000). The function of cancer-testis antigens within germ cells is unknown.
The first cancer-testis antigen, MZ2-E (now known as MAGE-A1), was discovered in 1991 as a cell surface antigen presented by major histocompatibility complex (MHC) molecules mediating cytotoxic T cell recognition of melanoma cells (van der Bruggen et al., 1991). Additional MAGE family genes were later identified by sequence homology. The MAGE family comprises more than 60 genes in humans, some of which are predicted pseudogenes (Chomez et al., 2001). The MAGE family of proteins is divided into two classes based on their expression pattern, the type I cancer-testis antigen and the type II ubiquitous MAGEs (Barker and Salehi, 2002). Type I MAGEs comprise more than 45 genes encoded in three clusters on the X chromosome, including 12 MAGE-A, 18 MAGE-B, and seven MAGE-C genes (Chomez et al., 2001). Type II MAGEs consist of 15 genes whose genomic loci are not restricted to the X chromosome, including four MAGE-D, three MAGE-E, MAGE-F1, MAGE-G1, MAGE-H1, two MAGE-I, MAGE-K1, MAGE-L2, and NECDIN genes (Chomez et al., 2001). Unlike the type I MAGE cancer-testis antigens, type II MAGE genes are not restricted to germline expression and are expressed in a variety of tissues (Ohman Forslund and Nordqvist, 2001). Type II MAGEs have also been implicated in human pathologies. For example, the maternally imprinted type II MAGEs, MAGE-L2 and Necdin, are implicated in the development of the neurobehavioral disorder known as Prader-Willi syndrome (Gérard et al., 1999; Jay et al., 1997; Kozlov et al., 2007).
Both type I and type II MAGE proteins share a conserved domain known as the MAGE homology domain (MHD). The MHD is ~170 amino acids long and can be evolutionarily traced back to protozoa (Barker and Salehi, 2002; López-Sánchez et al., 2007). In general, MHDs are quite similar to one another, even between type I and type II MAGEs. On average, all human MHDs are 46% conserved at the amino acid level (Figure S1A available online). The MHDs in specific subfamilies are even more conserved, such as 75% conservation between the four human MAGE-D MHDs and 70% between the twelve human MAGE-A MHDs (Figure S1A). Even though these domains are common and conserved, the biochemical function of the MHD has remained unknown.
The specific expression of type I MAGE genes in tumor, but not normal, somatic cells has been extensively investigated. DNA methylation of type I MAGE gene promoters prevents their expression in normal somatic cells (De Smet et al., 1995; De Smet et al., 1999). However, type I MAGE genes are hypomethylated and highly expressed in a variety of cancer types, including brain, breast, liver, lung, ovarian, prostate, skin, testicular, and thyroid. The expression of several type I MAGEs is highest in more advanced diseases of several cancer types and correlates with poor prognosis in patients (Bolli et al., 2002; Brasseur et al., 1995; Brichard and Lejeune, 2007; Dhodapkar et al., 2003; Patard et al., 1995). In addition, recent studies in mice have suggested an active role of MAGE proteins in tumor formation. In an orthotopic xenograft model of thyroid cancer, overexpression of MAGE-A3 promotes increased cell proliferation and primary tumor size, as well as increased number and size of lung metastatic foci (Liu et al., 2008). Furthermore, knockdown of type I MAGE-B genes suppressed the growth of melanomas in xenograft mouse models (Yang et al., 2007). In addition, MAGE and other cancer-testis antigens have been shown to facilitate the malignant phenotype by conferring resistance to chemotherapeutic agents, such as paclitaxel, doxorubicin, and tumor necrosis factor (Cilensek et al., 2002; Duan et al., 2003; Park et al., 2002). These results suggest that type I MAGE cancer-testis antigens display oncogenic properties that actively facilitate cancer cell survival, proliferation, tumor formation, and metastasis.
Type I MAGE proteins are ideal targets for cancer immunotherapies due to their specific expression and antigen presentation on a wide range of tumor, but not normal, cells. Extensive efforts have gone into developing anti-tumor antibodies and cancer vaccines toward type I MAGE cancer-testis antigens (Brichard and Lejeune, 2007; Goldman and DeFrancesco, 2009). However, the biochemical function of cancer antigens, including MAGE proteins, within tumor and germ cells has been relatively understudied and remains a mystery. To discover the biochemical function of MAGE proteins within cells, we took an unbiased approach to identify binding partners of nine different MAGE family proteins, utilizing tandem affinity purification (TAP) coupled to mass spectrometry (TAP-MS). Using this approach, we identify E3 RING ubiquitin ligases as binding partners of MAGE proteins. Through structural and biochemical methods, we show that MAGE and RING domain proteins form active E3 ubiquitin ligases. These results define the biochemical function of MAGE proteins within cells.
RESULTS
MAGE Proteins Form Complexes with E3 RING Ubiquitin Ligases
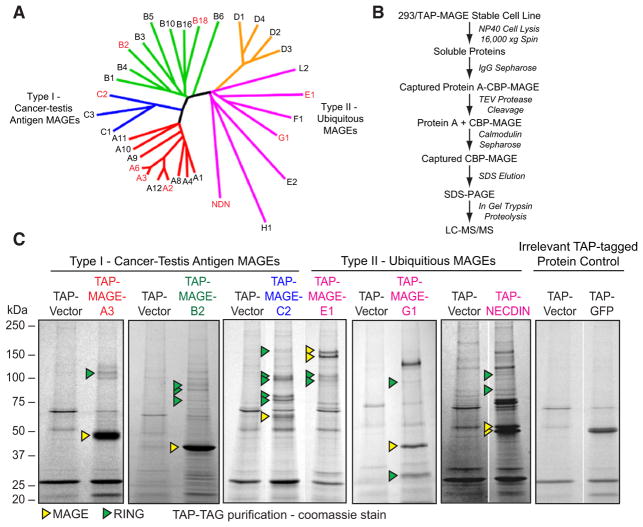
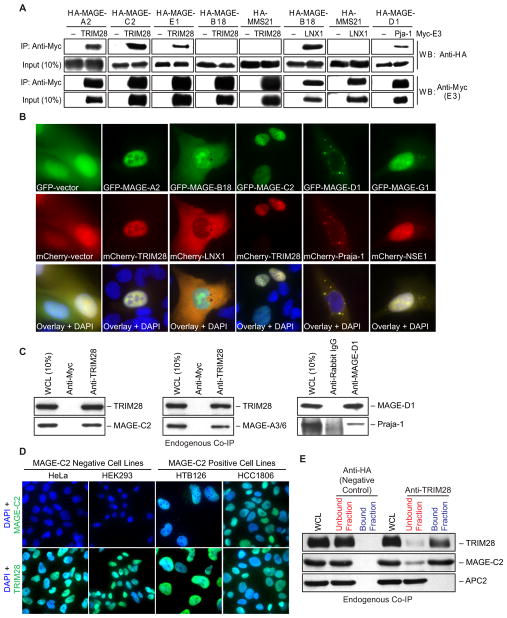
To elucidate the cellular functions of the MAGE family, we examined the binding partners of six type I and three type II MAGE proteins (Figure 1A) by TAP-MS. Full-length MAGE-A2, -A3, -A6, -B2, -B18, -C2, -E1, -G1, and Necdin were stably expressed as fusion proteins containing an N-terminal TAP tag (protein A-TEV-CBP) in HEK293 cells that express relatively few endogenous MAGE proteins (Figure S1B), and two-step purification was performed (Figure 1B). Those proteins present in the TAP-MAGE purifications (Figure 1C and data not shown) but absent in the control TAP vector alone or irrelevant TAP-GFP purifications were sequenced by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Remarkably, in all nine TAP-MAGE purifications, but not in TAP-vector purifications, we found proteins containing really interesting new gene RING domains (Figure 1C, Table S1, and data not shown). RING domains are cysteine-rich domains that form a cross-brace structure that typically coordinates two zinc ions (Borden, 2000). Many RING domain proteins function in the ubiquitylation cascade as E3 ubiquitin ligases by binding to and localizing E2 ubiquitin-conjugating enzymes to substrates for ubiquitylation (Lorick et al., 1999). To confirm those MAGE-RING complexes found by TAP-MS experiments, we performed coimmunoprecipitation (co-IP) and colocalization studies. First, we coexpressed Myc-tagged E3 RINGs (LNX1, Praja-1, or TRIM28/KAP1) and HA-tagged MAGEs (MAGE-A2, -B18, -C2, -D1, or -E1) and performed co-IP studies from cells. Indeed, those MAGEs identified to bind specific RINGs by TAP-MS also bound specific RING proteins by coexpression and co-IP (Figure 2A). For example, MAGE-B18 bound to LNX1, but not TRIM28 (Figure 2A). Next, we examined whether MAGEs colocalize with their RING binding partners in cells. We coexpressed and imaged GFP-tagged MAGEs (MAGE-A2, -B18, -C2, -D1, or -G1) and mCherry-tagged RINGs (LNX1, NSE1, or TRIM28) in U2OS cells due to their optimal imaging properties. Strikingly, those MAGE and RING proteins identified as binding partners by TAP-MS and co-IP also colocalized within cells (Figure 2B). Of interest, these MAGE-RING complexes localized to various subcellular compartments, such as MAGE-B18-LNX1 in the cytoplasm, MAGE-A2/C2-TRIM28 and MAGE-G1-NSE1 in the nucleus, and MAGE-D1-Praja-1 at undefined cytosolic puncti and the nuclear rim (Figure 2B). A portion of GFP-MAGE-B18 localized to the nucleus where mCherry-LNX1 was absent, suggesting that MAGE-B18 may bind another nuclear RING protein or that the nuclear MAGE-B18 is an artifact of transient expression of a GFP-tagged MAGE-B18.
Figure 1. Tandem Affinity Purification of MAGE Proteins.
(A) Dendrogram tree of MAGE protein family. Type I MAGEs include MAGE-A, MAGE-B, and MAGE-C families. Type II MAGEs include MAGE-D, MAGE-E, MAGE-F, MAGE-G, MAGE-H, MAGE-L2, and Nec-din families. Tandem affinity purification (TAP) was performed on the MAGE proteins indicated in red.
(B) TAP scheme to identify MAGE-interacting proteins.
(C) Colloidal Coomassie-stained SDS-PAGE gels of the indicated TAP-MAGEs isolated from HEK293 stable cell lines. See Table S1 for LC-MS/MS identification of indicated Coomassie-stained bands. TAP-GFP was used as an irrelevant negative control protein.
Figure 2. MAGEs Bind E3 RING Ubiquitin Ligases.
(A) Interactions between MAGE and RING domain proteins were validated by coimmunoprecipitation (IP). The indicated Myc- and HA-tagged proteins were expressed in HeLa cells, Myc-RING proteins were IPed with an anti-Myc antibody, and HA-MAGE proteins were detected by anti-HA western blot (WB). HA-MMS21 was included as an irrelevant HA-tagged negative control protein.
(B) MAGEs and their cognate E3 RINGs colocalize in specific subcellular compartments. The indicated GFP-tagged MAGEs and mCherry-tagged RING domain proteins were coexpressed and imaged in U2OS cells.
(C) Endogenous MAGE-RING ligase complexes. Cell lysates were subjected to immunoprecipitation and western blotting with the indicated antibodies.
(D) Endogenous MAGE-C2 and TRIM28 localize in the nucleus. MAGE-C2-negative (HEK293 and HeLa) and MAGE-C2-positive (HTB126 and HCC1806) cells were immunostained with mouse monoclonal MAGE-C2 (green) or mouse monoclonal TRIM28 (green). Nuclei were defined by DAPI staining (blue).
(E) The majority of MAGE-C2 associates with TRIM28 in cells. Cell lysates were subjected to immunoprecipitation with anti-HA (negative control) or anti-TRIM28 antibodies. The amount of MAGE-C2, TRIM28, and APC2 (negative control) in the cell lysate before immunoprecipitation (WCL), after immunoprecipitation (unbound fraction), and bound to the antibody beads (bound fraction) is shown.
Consistent with the formation of MAGE-RING complexes in cells, MAGE-C2-TRIM28, MAGE-A3/6-TRIM28, and MAGE-D1-Praja-1 interactions were all confirmed by endogenous co-IP (Figure 2C). Furthermore, both endogenous MAGE-C2 and TRIM28 localized to the nucleus of cells in a similar manner as the GFP- and mCherry-tagged proteins (Figure 2D). Finally, we determined what fraction of endogenous MAGE-C2 bound to endogenous TRIM28 in cells. By comparing the amount of MAGE-C2 in cell lysates before and after TRIM28 immunoprecipitation, we found that the majority of endogenous MAGE-C2 binds TRIM28 in cells (Figure 2E). Taken together, these results suggest that a widespread property of MAGE family proteins is their ability to form complexes with RING domain proteins in cells.
Characterization of MAGE-RING Binding and Specificity
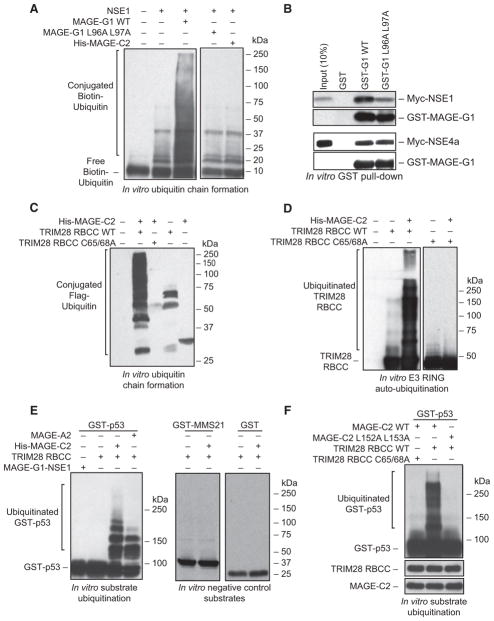
To determine whether MAGE proteins directly bind RING domain proteins, we purified recombinant His-TRIM28, NSE1, MAGE-B18, MAGE-C2, and GST-MAGE-G1 from bacteria (Figure S2A) and tested their binding in vitro using GST and Ni-NTA pull-down assays. We observed direct binding of MAGE-C2 to His-TRIM28 (Figure S2B) and GST-MAGE-G1 to NSE1 (Figure S2C). In addition, when coexpressed in bacteria, MAGE-C2-TRIM28 and MAGE-G1-NSE1 form tight protein complexes (Figures S2D and S2E). These results suggest that MAGE proteins directly bind RING domain proteins.
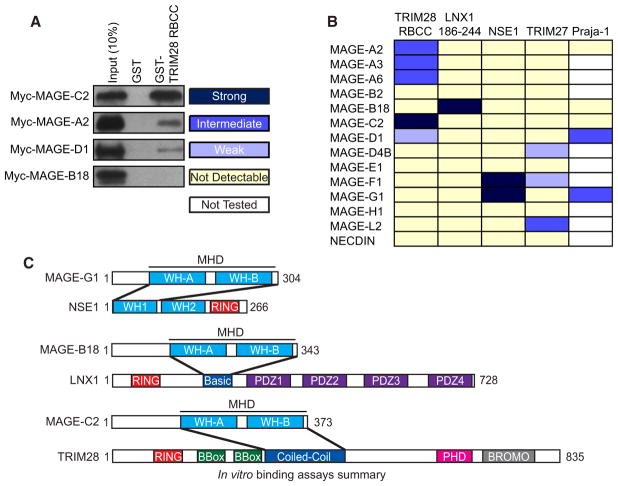
Given the ability of MAGEs to directly bind RING proteins and the large number of MAGE-RING complexes identified by TAP-MS and co-IP (Figures 1 and 2 and Table S1), we next examined whether specific MAGE proteins bind specific RING domain proteins. We systematically tested the ability of 14 different MAGEs to bind five different RING domain proteins by GST pull-down assays (Figures 3A, 3B, and S3A). MAGEs preferentially bound one specific RING domain protein, with the exception of MAGE-G1 and MAGE-F1, which bound two different RING domain proteins (Figures 3B and S3A). Furthermore, similar MAGEs (Figure 1A) tended to bind the same RING protein, such as MAGE-A2, MAGE-A3, MAGE-A6, and MAGE-C2 binding TRIM28 and MAGE-F1 and MAGE-G1 binding NSE1 (Figures 3B and S3A). Unlike MAGEs, RING domain proteins tended to have less specificity and typically bound at least two different MAGEs, with TRIM28 binding five of the 14 tested MAGEs (Figures 3B and S3A). These results establish that MAGE proteins directly bind RING domain proteins with specificity. Furthermore, these results identify a large number of MAGE-RING complexes (Table S1).
Figure 3. MAGE Proteins Directly Bind E3 RING Ubiquitin Ligases with Specificity.
(A) Representative examples of strong, intermediate, weak, and not detectable in vitro binding from Figure S3A.
(B) The relative binding affinities of the indicated MAGE and E3 RING ubiquitin ligases.
(C) Summary of in vitro binding assays from Figures S3B–S3M mapping the interacting regions of MAGE-B18-LNX1, MAGE-C2-TRIM28, and MAGE-G1-NSE1. MHD denotes the MAGE homology domain (MHD) comprising WH-A and WH-B motifs.
To gain insights into how specificity is achieved between MAGE and RING proteins, we mapped the minimal regions on MAGEs and RINGs required for binding. We performed this analysis on three different MAGE-RING complexes, MAGE-B18-LNX1, MAGE-C2-TRIM28, and MAGE-G1-NSE1, to gain insights into the similarities and differences among the various MAGE family proteins (summarized in Figure 3C). In all three cases, MAGE proteins bound RING domain-containing proteins through the MHD (Figures S3B–S3G) composed of two winged-helix motifs (WH-A and WH-B; see Figure 4). The region on which MAGEs bound RING domain-containing proteins was variable but did not require the RING domain (Figures S3H–S3M). MAGE-C2 bound the coiled-coil region on TRIM28 (Figures S3H and S3I). MAGE-B18 bound a basic region between the RING and first PDZ domain of LNX1 (Figures S3J and S3K). MAGE-G1 bound the WH-A motif within the N terminus of NSE1 (Figures S3L and S3M). These results suggest that MAGE family proteins do not recognize a common motif on their RING partners. The specificity for which RING proteins a given MAGE binds is determined by the MHD recognizing unique regions on their RING partner.
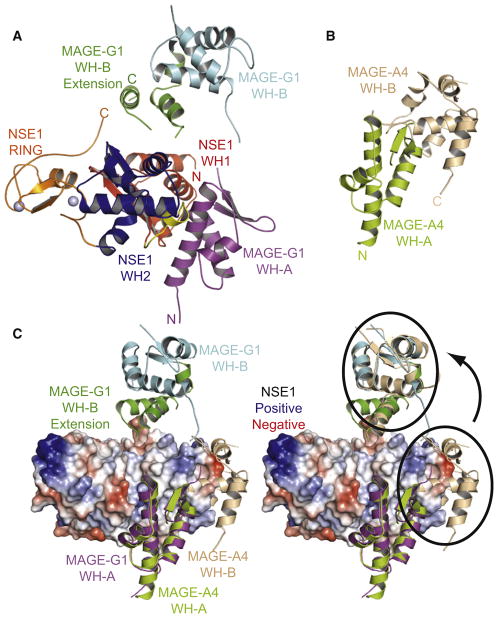
Figure 4. MAGE-G1-NSE1 Crystal Structure.
(A) Crystal structure of MAGE-G1-NSE1 complex. MAGE-G1 MHD forms a tandem winged helix (WH-A, purple; WH-B, cyan). The two zinc ions coordinated by the NSE1 RING domain are shown as gray spheres.
(B) Crystal structure of the MAGE-A4 MAGE homology domain (PDB: 2WA0) consisting of two winged helix motifs (WH-A, yellow-green; WH-B, cream).
(C) Orientation of MAGE WH-A to WH-B differs between MAGE-G1 and MAGE-A4. (Left) MAGE-A4 aligned to MAGE-G1-NSE1 based on WH-A motif. (Right) MAGE-A4 WH-B aligns with MAGE-G1 WH-B after rotation, as indicated by the arrow.
MAGE-G1-NSE1 Crystal Structure
To gain clues into the biochemical and structural properties of MAGE proteins, we coexpressed and purified full-length MAGE-G1 and its cognate NSE1 RING domain protein from bacteria and solved the crystal structure of the MAGE-G1-NSE1 protein complex at 2.75 Å(Table 1). The MAGE-G1 MHD (amino acids 78–295) formed two winged-helix motifs (referred to as WH-A and WH-B; Figure 4A). In addition, MAGE-G1 WH-B contained three extra α helices formed by the extreme C-terminal residues of the protein that are predicted to be conserved throughout the MAGE family (Figures 4A and S4A). We could not observe the N-terminal region of MAGE-G1 (amino acids 1–77), presumably due to structural flexibility. Within the crystal structure, MAGE-G1 mainly contacts NSE1 through its WH-A motif (Figure 4A). Of interest, NSE1 also contains two winged-helix motifs (WH1/2) and a RING domain that forms a cross-brace structure coordinating two zinc ions (Figure 4A). Consistent with our in vitro mapping experiments (Figure 3C), the NSE1 RING domain did not interact with MAGE-G1 but, rather, interacted with both NSE1 WH1 and WH2 motifs through a series of hydrogen bonds and with WH2 through a hydrophobic core (Figure S4B). E77 of WH2 formed three ionic bonds with the side chain of H212 and the main chain of L198, I199, and Q200 in the RING domain (Figure S4B). V188, L197, and I199 in the RING domain formed a hydrophobic core that interacts with the hydrophobic surface of I117, T167, I173, and I180 in the WH2 motif (Figure S4B). Importantly, these contacts between NSE1’s RING and WH1/2 motifs are not predicted to sterically impede binding of NSE1’s RING to E2 ubiquitin-conjugating enzymes (Figure S4C).
Table 1.
Statistics of Data Collection and Refinement
| Data Collection | Se-SAD | Native |
|---|---|---|
| Diffraction beam | BSRF | BL38B1, Spring-8 |
| Space group | P21212 | P21212 |
| Unit cell (Å) | 88.48, 158.65, 54.94 | 86.89, 154.7, 53.54 |
| Number of molecules in ASU | 2 | 2 |
| Wavelength (Å) | 0.979 | 1.000 |
| Resolution (Å) | 30 – 3.2 (3.31 – 3.2) | 40 – 2.75 (2.85 – 2.75) |
| Rsym (%) | 11.2 (69) | 4.1 (40.8) |
| I/σ | 20.85 (2.69) | 27.95 (1.54) |
| Completeness (%) | 99.3 (100) | 95.8 (66.0) |
| Number of measured reflections | 90,304 | 60,512 |
| Number of unique reflections | 12,927 | 17,756 |
| Redundancy | 7.0 (7.3) | 3.4 (1.7) |
| Wilson B factor (Å2) | 204.6 | 111.1 |
| SAD Phasing | ||
| Anomalous scatterers | 10 Se + 2 Zn2+ | 2 Zn2+ |
| Figure of merit (FOM) | 0.391 | 0.217 |
| FOM after DM | 0.689 | 0.747 |
| FOM after phase combination | 0.789 | |
| Refinement | ||
| R factor | 0.2195 | |
| Rfree | 0.2713 | |
| No. atoms | 3516 protein atoms + 2 Zn2+ + 1 SO42− | |
| B factors | ||
| Overall | 110.33 | |
| Main chain | 107.32 | |
| Side chain | 113.11 | |
| Rmsd bond lengths (Å) | 0.011 | |
| Rmsd bond angles (°) | 1.523 | |
| Ramachandran favored (%) | 87.8 | |
| Ramachandran outliers (%) | 3.59 | |
Values in parentheses are for the highest-resolution shell. Rsym=ΣhΣi|Ih,I –Ih|/ΣhΣiIh,i, wherein Ih is the mean intensity of the i observations of symmetry-related reflections of h. R=Σ|Fobs – Fcalc|/ΣFobs, wherein Fcalc is the calculated protein structure factor from the atomic model. (Rfree was calculated with 5% of the reflections.)
While we were preparing this manuscript, the structure of the free MAGE-A4 MHD was solved by structural genomics efforts (Figure 4B; PDB: 2WA0). Consistent with our structure, the MAGE-A4 MHD also contains WH-A and extended WH-B motifs (Figure 4B). The MAGE-G1 and MAGE-A4 WH-A and WH-B motifs are very similar, with rmsd values of 1.05 Åand 1.07 Å, respectively. Of interest, the relative orientation of WH-A to WH-B is different in the free MAGE-A4 and NSE1-bound MAGE-G1 structures (Figure 4C). This difference could reflect a conformational change in MAGE-G1’s MHD upon binding the NSE1 RING domain protein. These results structurally define the conserved MHD in the MAGE family of proteins.
MAGE Proteins Enhance the Ubiquitin Ligase Activity of RING Domain Proteins
We next examined the functional consequences of MAGE family proteins on the ubiquitin ligase activity of their cognate RING domain proteins. We first screened a panel of E2 ubiquitin-conjugating enzymes for those that potentially function with NSE1 in vitro. Consistent with previous reports (Pebernard et al., 2008), NSE1 on its own had weak in vitro ubiquitin ligase activity in the presence of the UbcH13/Mms2 E2 ubiquitin-conjugating enzyme (Figure 5A and data not shown). However, the activity of NSE1 was significantly enhanced upon addition of MAGE-G1, but not MAGE-C2 that does not bind NSE1 (Figure 5A and data not shown). To determine whether MAGE-G1-induced ubiquitin chain formation required binding to NSE1, we examined the MAGE-G1-NSE1 crystal structure for conserved residues important for MAGE-RING complex formation. MAGE-G1 L96 and L97 contribute to NSE1 interaction and form part of the hydrophobic core of MAGE-G1 WH-A motif (Figures S5A and S5B). Furthermore, this dileucine motif is conserved within the MAGE family (Figure S5C). Mutation of MAGE-G1 L96 and L97 to alanines significantly impaired MAGE-G1 binding to its cognate NSE1 RING protein, but not its non-RING binding partner NSE4a, and did not significantly alter its subcellular localization (Figures 5B, S5D, and S5G). More importantly, MAGE-G1 L96A L97A did not promote ubiquitin chain formation by NSE1 (Figure 5A). Therefore, these results suggest that MAGE-G1 directly enhances NSE1 ubiquitin ligase activity in vitro through binding NSE1.
Figure 5. MAGEs Enhance the Ubiquitin Ligase Activity of E3 RING Proteins.
(A) MAGE-G1 enhances NSE1 ubiquitin ligase activity in vitro. Biotin-ubiquitin, ubiquitin E1, UbcH13/MMS2 E2, Mg-ATP, and the indicated MAGE-RING proteins were incubated for 60 min at 37°C. Biotin-ubiquitin was detected by Streptavidin-HRP.
(B) Mutation of the dileucine motif within MAGE-G1 WH-A disrupts binding to the NSE1 RING protein, but not NSE4a. The indicated GST-tagged MAGE-G1 proteins were incubated with in vitro translated Myc-NSE1 or Myc-NSE4a. Proteins were detected by anti-Myc and anti-GST immunoblotting.
(C) MAGE-C2 enhances TRIM28 ubiquitin ligase activity in vitro. FLAG-ubiquitin, ubiquitin E1, UbcH2 E2, Mg-ATP, and the indicated MAGE-RING proteins were incubated for 60 min at 37°C. FLAG-ubiquitin was detected by anti-FLAG immunoblotting.
(D) MAGE-C2 enhances TRIM28 autoubiquitylation in vitro. Reactions were performed as in (C) except TRIM28 was detected by anti-TRIM28 immunoblotting.
(E) MAGE-A2 and -C2 enhance p53 ubiquitylation by TRIM28 in vitro. Reactions were performed as in (A) except substrate (GST-p53 or negative controls GST-MMS21 AA1-165 or free GST) was added and detected by anti-GST immunoblotting. MAGE-G1-NSE1 complex was included as an irrelevant MAGE-RING ligase that does not target p53.
(F) MAGE-C2-induced p53 ubiquitylation requires a functional TRIM28 RING domain and the ability to bind TRIM28. Reactions were performed as described in (E).
Next, we wanted to determine whether the ability of MAGE proteins to enhance E3 ubiquitin ligase activity is a general property of the MAGE family proteins by examining MAGE-C2 enhancement of TRIM28 ubiquitin ligase activity in vitro. We first screened a panel of E2 ubiquitin-conjugating enzymes for those that function with MAGE-C2-TRIM28 in vitro and found that UbcH2 was the optimal E2 enzyme for MAGE-C2-TRIM28 (Figure S5H). Next, we determined whether MAGE-C2 enhances TRIM28 ubiquitin ligase activity in a similar manner to MAGE-G1-NSE1. Indeed, MAGE-C2 enhanced TRIM28 ubiquitin ligase activity (Figures 5C and S5H). Importantly, MAGE-C2-induced ubiquitin ligase activity required an intact TRIM28 RING domain, as MAGE-C2 failed to enhance ubiquitin chain formation in the presence of the TRIM28 RING mutant C65A C68A or when incubated in the absence of TRIM28 (Figure 5C). Similar results were observed when measuring ubiquitin ligase activity as a function of TRIM28 autoubiquitylation (Figure 5D). These findings suggest that multiple MAGE proteins facilitate efficient ubiquitin ligase activity of RING domain proteins.
Next, we examined whether MAGE-C2 also enhances substrate ubiquitylation by TRIM28. Previous work has suggested that TRIM28 can ubiquitinate the tumor suppressor p53 (Wang et al., 2005). Therefore, we addressed the ability of MAGE-C2 to promote TRIM28-dependent ubiquitylation of p53 in vitro. Strikingly, MAGE-C2 enhanced TRIM28-induced p53 ubiquitylation in a RING-dependent manner in in vitro ubiquitylation reactions in which TRIM28 was limiting (Figures 5E and 5F). Consistent with MAGE-C2-TRIM28 ubiquitin chain formation, MAGE-C2-TRIM28 p53 ubiquitylation was maximal with the UbcH2 E2 ubiquitin-conjugating enzyme (Figure S5I). Importantly, MAGE-C2-TRIM28 promoted the ubiquitylation of GST-p53 and His-p53, but not the irrelevant control proteins GST or GST-MMS21 (Figures 5E and S5J). In addition, the related TRIM28-associated MAGE, MAGE-A2, also enhanced TRIM28-mediated p53 ubiquitylation (Figure 5E), although to a slightly less degree than MAGE-C2, possibly due to its slightly weaker binding affinity (Figure 3A). However, the ubiquitylation of p53 was specific to MAGE-TRIM28, as MAGE-G1-NSE1 failed to ubiquitinate GST-p53 in vitro (Figure 5E).
To determine whether MAGE-C2 activates TRIM28 in an analogous manner to MAGE-G1 activation of NSE1, we mutated the corresponding dileucine motif in MAGE-C2 L152 L153 that we previously found by structure function analysis to be important for MAGE-G1 binding and activation of NSE1. Consistent with the MAGE-G1 dileucine motif mutant, MAGE-C2 L152A L153A failed to bind TRIM28 (Figures S5E and S5F) and induce p53 ubiquitylation in vitro (Figure 5F). These results suggest that a conserved feature of MAGE proteins is their ability to bind and enhance the ubiquitin ligase activity of E3 RING proteins in vitro.
MAGE-TRIM28 Complexes Regulate p53 Protein Stability in Cells
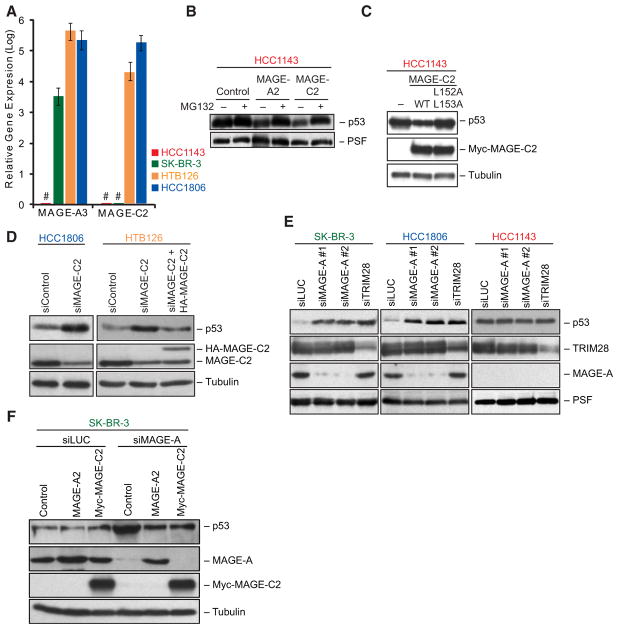
Finally, we examined whether MAGEs enhance the ubiquitin ligase activity of E3 RING proteins in cells. To do so, we examined whether MAGE-TRIM28 complexes are both necessary and sufficient to regulate p53 protein stability in cells. First, we identified breast cancer cell lines that express all of the TRIM28-associated MAGEs (MAGE-A2, -A3, -A6, and -C2) (HTB126 and HCC1806); only MAGE-A2, -A3, and -A6 (SK-BR-3); or none of the TRIM28-associated MAGEs (HCC1143) (Figures 6A and S1B). Then, we examined whether expression of MAGE-A2 or MAGE-C2 in HCC1143 MAGE-negative cells was sufficient to reduce p53 protein levels. Indeed, expression of either MAGE-A2 or MAGE-C2 reduced p53 protein levels in a proteasome-dependent manner in HCC1143 cells (Figure 6B). In addition, expression of the MAGE-C2 L152A L153A mutant that is incapable of binding TRIM28 (Figures S5E–S5G) did not promote p53 instability in MAGE-negative HCC1143 cells (Figure 6C), suggesting that MAGE-C2 regulates p53 protein levels through TRIM28.
Figure 6. MAGE-TRIM28 Ligases Promote p53 Degradation in Cells.
(A) RT-QPCR analysis of MAGE-A3 and MAGE-C2 expression in HCC1143, HCC1806, HTB126, and SK-BR-3 cells. Results were normalized to GAPDH expression. Data are represented as the mean ± standard deviation. # denotes no signal above background detected by QPCR. MAGE-A3 expression is representative of MAGE-A2 and MAGE-A6 (See Figure S1B).
(B) Expression of MAGE-A2 or -C2 in HCC1143 MAGE-negative breast cancer cells results in decreased p53 protein levels in a proteasome-dependent manner. Twenty-four hours after transfection, HCC1143 cells were treated with 10 μM MG132 for 16 hr. Cell lysates were immunoblotted with the indicated antibodies.
(C) MAGE-C2-induced p53 degradation requires TRIM28 binding. HCC1143 cells were transfected with wild-type or L152A L153A Myc-MAGE-C2 followed by immunoblotting with the indicated antibodies 48 hr later.
(D) MAGE-C2-RNAi increases p53 protein levels in HCC1806 and HTB126 breast cancer cells. MAGE-C2-RNAi was rescued by a siRNA-resistant HA-MAGE-C2 construct in HTB126 cells. Cell lysates were collected 72 hr after transfection and immunoblotted with the indicated antibodies.
(E) MAGE-A- and TRIM28-RNAi increase p53 protein levels in MAGE-positive HCC1806 and SK-BR-3 cells, but not in MAGE-negative HCC1143 cells. p53 levels were determined as described in (D).
(F) MAGE-A2 and MAGE-C2 share similar activities in regulating p53. SK-BR-3 MAGE-A-positive, MAGE-C2-negative cells were treated with the indicated siRNAs for 24 hr before transfection with the indicated expression constructs. Cell lysates were collected 48 hr later and immunoblotted with the indicated antibodies.
Next, we tested whether endogenous MAGE-TRIM28 ubiquitin ligases actually function to reduce p53 protein levels in cells using MAGE-C2, pan-MAGE-A, or TRIM28 siRNAs (Figures 6D and 6E). Consistent with the overexpression studies, knockdown of endogenous MAGE-C2 in HTB126 or HCC1806 breast cancer cells or MAGE-As in HCC1806 or SK-BR-3 cells substantially increased p53 protein levels (Figures 6D and 6E). These effects were not due to off-target effects, as expression of an RNAi-resistant MAGE-A2 or MAGE-C2 could rescue MAGE-A or MAGE-C2 RNAi, respectively (Figures 6D and 6F). In addition, knockdown of TRIM28 stabilized p53 in breast cancer cells expressing MAGE-A2, -A3, -A6, or -C2 (HCC1806 and SK-BR-3), but not in cells negative for these MAGEs (HCC1143) (Figure 6E). Previous studies have reported that TRIM28 can collaborate with the p53 E3 ligase MDM2 to regulate p53 protein stability in cells (Okamoto et al., 2006; Wang et al., 2005). However, we found that MAGE-C2-TRIM28 regulation of p53 was independent of MDM2 (Figures S6A–S6E). These results suggest that MAGE-A and MAGE-C2 proteins bind to and promote the activity of TRIM28 to promote p53 degradation in cells.
To directly address whether MAGE-A and MAGE-C2 proteins share similar activities to promote TRIM28 ubiquitin ligase activity in cells, we examined the ability of MAGE-C2 to rescue MAGE-A-RNAi-induced accumulation of p53 in SK-BR-3 cells that normally do not express endogenous MAGE-C2 (Figure 6A). Expression of MAGE-A2 or MAGE-C2 in MAGE-A-RNAi cells rescued p53 protein levels back to baseline (Figure 6F). Therefore, activation of TRIM28 ubiquitin ligase activity is conserved between the TRIM28-associated MAGEs. In sum, these results suggest that a defining characteristic of MAGE proteins is their ability to bind RING proteins and enhance their ubiquitin ligase activity in vitro and in cells.
DISCUSSION
Recent evidence suggests that type I MAGE cancer-testis antigens play an active role in tumorigenesis and cancer cell viability (Cilensek et al., 2002; Duan et al., 2003; Liu et al., 2008; Yang et al., 2007). Furthermore, type II ubiquitous MAGE proteins are increasingly implicated in a number of important biological and pathological processes (Barker and Salehi, 2002; Gérard et al., 1999; Jay et al., 1997; Kozlov et al., 2007; Ohman Forslund and Nordqvist, 2001; Pebernard et al., 2004). In spite of this, the biochemical and cellular mechanisms by which MAGE family proteins function in tumor, normal, and germ cells have remained elusive.
Through in vitro biochemical and cellular assays, we show that a general property conserved throughout the MAGE protein family is their binding to E3 RING proteins (Figures 1–3). Of interest, several MAGEs bound members of the TRIM family E3 RING ubiquitin ligases, including TRIM27 and TRIM28 (Figures 1–3). The TRIM family of proteins is a large and relatively poorly understood family of proteins implicated in a number of biological processes, including cell proliferation, development, apoptosis, and innate immunity (Ozato et al., 2008). Whether additional TRIM family proteins also form MAGE-RING complexes will need to be investigated. Through in vitro binding assays, we show that MAGE-B18, MAGE-C2, and MAGE-G1 all bind E3 RING proteins through their MAGE homology domain (MHD) consisting of two winged-helix motifs (Figure 3C). However, the region on the E3 protein in which MAGEs bound was variable in amino acid composition, secondary structure, and distance from the RING domain (Figure 3C). Therefore, there may not be a common type of contact between the MHD of MAGE proteins and their cognate E3 proteins.
We further show that a general feature of both type I and type II MAGE proteins is not only binding to E3 RING ligases, but also enhancing their ubiquitin ligase activity (Figures 5 and 6). The mechanisms behind this enhancement are currently not fully understood. One possible mechanism is that MAGE binding induces a conformational change of the RING protein, resulting in increased activity. However, this does not seem to be the case for MAGE-G1-NSE1, as MAGE-G1 binding does not alter NSE1 structure (M.Y. and P.R.P., unpublished data). A second possible mechanism would involve MAGEs facilitating substrate binding to the E2–E3 ubiquitin ligase complex. However, in the case of MAGE-TRIM28, MAGE-A2, -A3, -A6, and -C2 do not show appreciable direct binding to the p53 substrate or enhance TRIM28 binding to p53 (Figures S6F–S6H). A third possibility is that MAGEs stimulate E2 charging. However, MAGE-C2 did not affect the charging of the UbcH2 E2 ubiquitin-conjugating enzyme by the ubiquitin E1 (Figure S6K). Finally, we examined a fourth possible mechanism whereby MAGEs bind to and thereby help recruit E2 ubiquitin-conjugating enzymes to the E3-substrate complex. We observed that MAGE-A2 and MAGE-C2 did specifically bind the UbcH2, but not UbcH5b, E2 ubiquitin-conjugating enzyme in cells and in vitro (Figures S6I and S6J). These results suggest that MAGEs may enhance ubiquitin ligase activity through recruitment and/or stabilization of the E2 ubiquitin-conjugating enzymes at the E3-substrate complex. Several important questions arise from these findings. First, can MAGE-A2/C2 bind both UbcH2 and TRIM28 at the same time, or are they mutually exclusive? Second, does MAGE-A2/C2 binding to UbcH2 contribute to increased TRIM28 ubiquitin ligase activity? Finally, is the binding of MAGEs to E2 ubiquitin-conjugating enzymes conserved throughout the MAGE family, or do different MAGEs enhance E3 ubiquitin ligase activity by distinct mechanisms?
MAGE-G1 and NSE1 are part of the larger SMC5/6 complex that promotes homologous recombination-mediated DNA repair (Potts, 2009; Potts et al., 2006). The biochemical function of MAGE-G1 (NSE3 in yeast) and NSE1 in DNA repair has been relatively enigmatic. We show that MAGE-G1 promotes the activity of NSE1 in the presence of the UbcH13/MMS2 E2 ubiquitin-conjugating enzyme in vitro (Figure 5A and data not shown). Of interest, UbcH13/MMS2 specifically catalyzes the formation of K63-linked polyubiquitin chains and has also been implicated in homologous recombination repair (Zhao et al., 2007). It will be interesting in the future to examine the cofunctionality of MAGE-G1-NSE1 with UbcH13/MMS2 in cells and to determine their relevant ubiquitylation substrates contributing to homologous recombination-mediated double-strand break repair.
MAGE-A2, -A3, -A6, and -C2 are cancer-testis antigens specifically expressed in the germline and aberrantly expressed in a broad spectrum of tumors due to DNA hypomethylation (Zhuang et al., 2006). We and others found MAGE-A2, -A3, -A6, and -C2 to interact with the TRIM28 E3 RING ubiquitin ligase (Table S1) (Yang et al., 2007). Of interest, TRIM28 has recently been reported to be overexpressed in gastric cancer and, similar to MAGE-A3, correlates with poorer patient survival (Yokoe et al., 2010). We show here that one possible mechanism by which MAGE-A2, -A3, -A6, -C2, and TRIM28 contribute to tumorigenesis is through the ubiquitylation and degradation of p53 (Figure 6). Consistent with our findings, Bonus, the Drosophila ortholog of the TIF1 family (including TRIM24, TRIM28, and TRIM33) also regulates Dmp53 (Allton et al., 2009). Whether the Drosophila MAGE also regulates Bonus activity and therefore Dmp53 stability will need to be investigated. The importance of MAGE-RING ligase-mediated p53 inactivation in tumorigenesis is likely highlighted in tumors that do not frequently mutate p53, such as the case in melanomas (Flørenes et al., 1994). In these cases, MAGE-RING ligases are ideal, cancer-specific drug targets.
In summary, our work has shed light on the mysterious structure and function of MAGE proteins. We show that both type I cancer-testis antigen and type II ubiquitous MAGEs bind to and activate RING E3 ubiquitin ligases. The identification of MAGE-RING ubiquitin ligases opens the door to understanding the biological and pathological function of both type I and type II MAGE proteins.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and siRNAs
HCC1143, HCC1806, HEK293, HeLa Tet-ON, HTB126, and SK-BR-3 were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 2 mM L-glutamine (Invitrogen), 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 0.25 μg/ml amphotericin B (Invitrogen). Plasmid transfection was performed with either Effectene (QIAGEN) or Lipofectamine 2000 (Invitrogen) reagents according to the manufacturer’s protocol. siRNAs transfection was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol for reverse transfection. The siLuciferase (5′-CUUACGCUGAGUACUUCGAdTdT-3′), pan-siMAGE-A #1 (5′-GAUGGUUGAAUGAGCGUCAdTdT-3′), pan-siMAGE-A #2 (5′-GGUAA AGAUCAGUGGAGGAdTdT-3′), siTRIM28 (5′-GCAUGAACCCCUUGUGCU GdTdT-3′), and siMAGE-C2 SmartPool oligonucleotides were purchased from Dharmacon.
Tandem Affinity Purification and Mass Spectrometry
Tandem affinity purification (TAP) was performed using 293/TAP-MAGE stable cell lines. Detailed methods on TAP and LC/MS-MS are described in Supplemental Experimental Procedures. In brief, ten 15 mm2 dishes of 293/TAP-MAGE cells were lysed, bound to IgG Sepharose beads (GE Amersham), cleaved off the beads with TEV protease, collected on Calmodulin Sepharose beads (GE Amersham), eluted with SDS sample buffer, separated by SDS-PAGE, and stained with colloidal Coomassie blue (Pierce). Unique protein bands were excised, in-gel proteolyzed, and identified by LC/MS-MS.
Immunoprecipitation, Immunoblotting, and Antibodies
Immunoprecipitation and immunoblotting were performed as previously described (Potts and Yu, 2007). Commercial antibodies used in this study were as follows: anti-hemagglutinin (Roche; 12CA5), anti-MAGE-D1 (Millipore; 07-394), anti-MDM2 (Santa Cruz; SMP14), anti-Myc (Roche; 9E10), anti-p53 (Santa Cruz; DO-1), anti-PSF (Sigma, P2860), anti-TRIM28/KAP1 (Abcam; 20C1), and anti-tubulin (DM1; Sigma). The production of polyclonal anti-GST antibody was previously described (Potts and Yu, 2005). A rabbit polyclonal MAGE-A3/A6 antibody was generated against full-length, recombinant MAGE-A3 protein (Yenzyme Inc.). Crude antibody sera were affinity-purified against full-length, recombinant MAGE-A3 and tested for specificity by immunoblotting cells treated with control or MAGE-A3 siRNAs. Anti-MAGE-C2 was generously provided by Dr. Lloyd Old, Ludwig Institute for Cancer Research (Zhuang et al., 2006). Anti-Praja-1 was generously provided by Dr. Lopra Mishra, Georgetown University (Mishra et al., 1997).
Protein Purification and In Vitro Binding Assays
GST-tagged MAGE proteins were produced in BL21(DE3) cells by overnight induction at 16°C with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Proteins were purified from bacterial lysates with glutathione Sepharose (GE Amersham) and eluted with 10 mM glutathione. The His-MAGE-C2/TRIM28 RBCC complex was purified from BL21(DE3) cells coexpressing full-length His-MAGE-C2 and GST-TRIM28 RBCC fragment (amino acids 22–418) using the pRSF-Duet system (Novagen). Cells were induced as above in the presence of 1 mM ZnCl2. Cleared cell lysates were first applied to Ni-NTA beads (QIAGEN), and bound proteins were subsequently eluted with 250 mM imidazole. The eluted proteins were then bound to glutathione Sepharose, cleaved on-column by PreScission Protease, and subsequently subjected to ion exchange chromatography (Resource Q).
For in vitro binding assays, Myc-tagged proteins were in vitro translated using the SP6-TNT Quick rabbit reticulocyte lysate system (Promega) according to manufacturer’s instructions. In vitro binding assays were performed by incubating 15 μg of purified GST-tagged protein with glutathione Sepharose beads (Amersham) for 1 hr in binding buffer (25 mM Tris [pH 8.0], 2.7 mM KCl, 137 mM NaCl, 0.05% Tween-20, and 10 mM 2-mercaptoethanol). Bound beads were blocked for 1 hr in binding buffer containing 5% milk powder. In vitro translated proteins (5 μl) were then incubated with the bound beads for 1 hr in binding buffer containing 5% milk powder. After four washes in binding buffer, the proteins were eluted in SDS-sample buffer, boiled, subjected to SDS-PAGE, and blotted with anti-Myc or anti-GST as a loading control. For quantitation of in vitro binding assays, films were scanned and subjected to densitometry analysis. Background binding to GST was subtracted from the binding to the relevant GST-tagged proteins and expressed as a percentage of the input protein.
Crystallization, Data Collection, and Structure Determination
The MAGE-G1-NSE1 crystal structure was solved by coexpression and purification of full-length proteins from bacteria. Crystals were grown by hanging-drop method in 22% PEG500, Sodium Citrate (pH 5.5) and 2% 1,1,1,3,3,3-Hexafluoro-2-Propanol reservoir solution. Native and Se-SAD data were used to solve the structure. Detailed methods on protein purification, crystallization, and structure determination are described in the Supplemental Experimental Procedures.
In Vitro Ubiquitylation Assays
In vitro ubiquitylation assays were performed using 100 nM His-Ube1 (Biomol), 2.5 μM E2 (His-UbcH2 or His-Ubc13/MMS2; Biomol), 5 μM E3 (NSE1, MAGE-G1-NSE1, TRIM28 RBCC, or MAGE-C2-TRIM28 RBCC), 2.5 μM biotinylated ubiquitin (Biomol), 0.5 mM Mg-ATP (Biomol), 20 units/ml inorganic pyrophosphatase (Sigma), and 1 mM DTT in 1 ×ubiquitylation buffer (Biomol). For those reactions examining substrate ubiquitylation, 1 μM GST-p53 (Biomol) was added. Negative control reactions (–Mg-ATP) were set up as described above except Mg-ATP was replaced with 5 mM EDTA. Reactions were stopped by the addition of SDS sample buffer after incubation at 37°C for 60 min and subjected to SDS-PAGE and immunoblotting for ubiquitin chain formation (Streptavidin-HRP; Vector Laboratories), substrate ubiquitylation (anti-p53; Santa Cruz DO-1), or E3 autoubiquitylation (anti-TRIM28/KAP1; Abcam 20C1).
Supplementary Material
Acknowledgments
We would like to thank Hongtao Yu (HHMI and UT Southwestern Medical Center) for support during the early stages of this work and Drs. Lloyd Old (Ludwig Institute for Cancer Research) and Lopra Mishra (Georgetown University) for generously providing the MAGE-C2 and Praja-1 antibodies, respectively. We would also like to thank Jianhua He (Shanghai Sychrotron Radiation Facility BL17U), S. Baba (Spring-8 beamline BL38B1), Yuhui Dong (Beijing Synchrotron Radiation Facility), Na Wang (Tsinghua University), and Fangfang Gao (Tsinghua University) for technical assistance. Finally, we would like to thank Steven McKnight, John Minna, Matthew Porteus, Malia Potts, Michael Rosen, Yigong Shi, and Michael White for helpful discussion and critical reading of the manuscript. This work was supported by Tsinghua University Startup Funds (M.Y.), National Natural Science Foundation of China, National Laboratory Special Fund 2060204 (M.Y.), and the Sara and Frank McKnight Fellowship (P.R.P.).
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors of the MAGE-G1-NSE1 complex described here have been deposited in the Protein Data Bank under ID 3NW0.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at doi:10. 1016/j.molcel.2010.08.029.
References
- Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, Barton MC. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci USA. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- Bolli M, Kocher T, Adamina M, Guller U, Dalquen P, Haas P, Mirlacher M, Gambazzi F, Harder F, Heberer M, et al. Tissue microarray evaluation of Melanoma antigen E (MAGE) tumor-associated antigen expression: potential indications for specific immunotherapy and prognostic relevance in squamous cell lung carcinoma. Ann Surg. 2002;236:785–793. doi: 10.1097/01.SLA.0000036266.09823.6C. discussion 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Brasseur F, Rimoldi D, Liénard D, Lethé B, Carrel S, Arienti F, Suter L, Vanwijck R, Bourlond A, Humblet Y, et al. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int J Cancer. 1995;63:375–380. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- Brichard VG, Lejeune D. GSK’s antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25(Suppl 2):B61–B71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- Cilensek ZM, Yehiely F, Kular RK, Deiss LP. A member of the GAGE family of tumor antigens is an anti-apoptotic gene that confers resistance to Fas/CD95/APO-1, Interferon-gamma, taxol and gamma-irradiation. Cancer Biol Ther. 2002;1:380–387. [PubMed] [Google Scholar]
- De Smet C, Courtois SJ, Faraoni I, Lurquin C, Szikora JP, De Backer O, Boon T. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics. 1995;42:282–290. doi: 10.1007/BF00176446. [DOI] [PubMed] [Google Scholar]
- De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV, Iversen K, Kolb D, Geller MD, Hassoun H, Kewalramani T, et al. Expression of cancer/testis (CT) antigens MAGE-A1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun. 2003;3:9. [PubMed] [Google Scholar]
- Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, Seiden MV. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin Cancer Res. 2003;9:2778–2785. [PubMed] [Google Scholar]
- Flørenes VA, Oyjord T, Holm R, Skrede M, Børresen AL, Nesland JM, Fodstad O. TP53 allele loss, mutations and expression in malignant melanoma. Br J Cancer. 1994;69:253–259. doi: 10.1038/bjc.1994.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- Goldman B, DeFrancesco L. The cancer vaccine roller coaster. Nat Biotechnol. 2009;27:129–139. doi: 10.1038/nbt0209-129. [DOI] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei MG, Malzac P, Roëckel N, Taviaux S, Lefranc JL, Cau P, et al. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- Jungbluth AA, Stockert E, Chen YT, Kolb D, Iversen K, Coplan K, Williamson B, Altorki N, Busam KJ, Old LJ. Monoclonal antibody MA454 reveals a heterogeneous expression pattern of MAGE-1 antigen in formalin-fixed paraffin embedded lung tumours. Br J Cancer. 2000;83:493–497. doi: 10.1054/bjoc.2000.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, Hogenesch JB, Muglia LJ, Van Gelder RN, Herzog ED, Stewart CL. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- López-Sánchez N, González-Fernández Z, Niinobe M, Yoshikawa K, Frade JM. Single mage gene in the chicken genome encodes CMage, a protein with functional similarities to mammalian type II Mage proteins. Physiol Genomics. 2007;30:156–171. doi: 10.1152/physiolgenomics.00249.2006. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L, Tully RE, Monga SP, Yu P, Cai T, Makalowski W, Mezey E, Pavan WJ, Mishra B. Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene. 1997;15:2361–2368. doi: 10.1038/sj.onc.1201405. [DOI] [PubMed] [Google Scholar]
- Ohman Forslund K, Nordqvist K. The melanoma antigen genes—any clues to their functions in normal tissues? Exp Cell Res. 2001;265:185–194. doi: 10.1006/excr.2001.5173. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kitabayashi I, Taya Y. KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction. Biochem Biophys Res Commun. 2006;351:216–222. doi: 10.1016/j.bbrc.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kong GH, Lee SW. hMAGE-A1 overexpression reduces TNF-alpha cytotoxicity in ME-180 cells. Mol Cells. 2002;14:122–129. [PubMed] [Google Scholar]
- Patard JJ, Brasseur F, Gil-Diez S, Radvanyi F, Marchand M, François P, Abi-Aad A, Van Cangh P, Abbou CC, Chopin D, et al. Expression of MAGE genes in transitional-cell carcinomas of the urinary bladder. Int J Cancer. 1995;64:60–64. doi: 10.1002/ijc.2910640112. [DOI] [PubMed] [Google Scholar]
- Pebernard S, McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Perry JJ, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol Biol Cell. 2008;19:4099–4109. doi: 10.1091/mbc.E08-02-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 2009;8:499–506. doi: 10.1016/j.dnarep.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, III, Chen J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- Yokoe T, Toiyama Y, Okugawa Y, Tanaka K, Ohi M, Inoue Y, Mohri Y, Miki C, Kusunoki M. KAP1 is associated with peritoneal carcinomatosis in gastric cancer. Ann Surg Oncol. 2010;17:821–828. doi: 10.1245/s10434-009-0795-8. [DOI] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Zhuang R, Zhu Y, Fang L, Liu XS, Tian Y, Chen LH, Ouyang WM, Xu XG, Jian JL, Güre AO, et al. Generation of monoclonal antibodies to cancer/testis (CT) antigen CT10/MAGE-C2. Cancer Immun. 2006;6:7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.