Abstract
Whole exome sequencing (WES) and RNA sequencing (RNA-Seq) are two main platforms used for next-generation sequencing (NGS). While WES is primarily for DNA variant discovery and RNA-Seq is mainly for measurement of gene expression, both can be used for detection of genetic variants, especially single nucleotide variants (SNVs). How consistently variants can be detected from WES and RNA-Seq has not been systematically evaluated. In this study, we examined the technical and biological inconsistencies in SNV detection using WES and RNA-Seq data from 27 pairs of tumor and matched normal samples. We analyzed SNVs in three categories: WES unique - those only detected in WES, RNA-Seq unique - those only detected in RNA-Seq, and shared – those detected in both. We found a small overlap (average ∼14%) between the SNVs called in WES and RNA-Seq. The WES unique SNVs were mainly due to low coverage, low expression, or their location on the non-transcribed strand in RNA-Seq data, while the RNA-Seq unique SNVs were primarily due to their location out of the WES-capture boundary regions (accounting ∼71%), as well as low coverage of the regions, low coverage of the mutant alleles or RNA-editing. The shared SNVs had high locus-specific coverage in both WES and RNA-Seq and high gene expression levels. Additionally, WES unique and RNA-Seq unique SNVs showed different nucleotide substitution patterns, e.g., ∼55% of RNA-Seq unique variants were A:T→G:C, a hallmark of RNA editing. This study provides an important evaluation on the inconsistencies of somatic SNVs called in WES and RNA-Seq data.
Keywords: Single nucleotide variants, whole exome sequencing, RNA-Seq, somatic mutations, allele frequency, RNA editing
1. Introduction
Single nucleotide variants (SNVs) are the most abundant form of genetic variation in genome sequences and somatic SNVs play critical roles in disease [1]. The discovery of many driver SNVs has led to new targets for therapeutic treatments and preventive measures. Examples include vemurafenib for the BRAF V600 mutations in melanoma [2, 3] and gefitinib, erlotonib, and afatinib for EGFR mutations in lung cancer [4]. The recent advances in next-generation sequencing (NGS) technologies, especially whole exome sequencing (WES) and whole transcriptome sequencing (RNA-Seq), have helped investigators generate a massive amount of NGS data, from which genetic variants, including SNVs, are detected. Many tools are now available for the detection of somatic SNVs from NGS data [5].
Both whole genome sequencing (WGS) and WES have been applied to detect SNVs in large scale cancer studies. While WGS can detect the full spectrum of variants (SNVs, insertions/deletions (indels), copy number variations (CNVs), and structural variants (SVs) across the whole cancer genome, WES is more cost-effective in detecting SNVs and indels located in the 1–2% of the genome that encodes for functional proteins [6]. There is good evidence that SNVs within the exome are responsible for many diseases, so WES has been applied extensively in research and clinically [6–8]. RNA-Seq is commonly used for the measurement of gene expression levels, detection of gene fusions, and identification of splicing events. Because RNA-Seq is based on direct sequencing of cDNA, the product of the mRNA through reverse transcription, it is practically feasible to detect SNVs from RNA-Seq data [9, 10]. This is a unique feature that is different from the traditional microarray-based gene expression. RNA-Seq also has the ability to detect RNA editing, which is a post-transcriptional process that modifies RNA transcripts. One of the most common mechanisms of RNA editing is the deamination of adenosine to inosine by the protein Adenosine Deaminase Acting on RNA (ADAR). The inosine is interpreted in a similar way to guanosine and, thus, results in an adenosine to guanine (A → G) change [11].
RNA-Seq has been extensively applied to genomic and transcriptomic studies, including cancer. For example, a large-scale RNA-Seq study of lung adenocarcinoma identified several cancer driver genes [12], indicating its utility in a transcriptome analysis of cancer samples. This study demonstrated that in addition to identifying fusion genes and differential gene expression, RNA-Seq could detect well-known cancer driver genes. RNA-Seq has also been combined with WGS to better understand the mutational landscape of lung cancer [13, 14]. These studies, in addition to showing the standard applications of RNA-Seq in gene expression analysis, highlight its usefulness as a technology platform for SNV detection, though challenges remain [15]. Large consortia such as The Cancer Genome Atlas (TCGA), have applied both WES and RNA-Seq, as well as other platforms, to comprehensively catalog the cancer genome landscape [16]. The combined WES and RNA-Seq of the same tumor samples allow for large-scale examinations of somatic mutations in both the DNA and RNA. By applying these two types of technology together, one can improve the detection of various mutations, including those in the expressed genes with different splicing and expression levels, and those in non-transcribed regions. However, sequencing the same tumor using both platforms is rarely used in real projects due to the cost and analysis issues.
A detailed comparison of SNVs called from WES and RNA-Seq data using the same samples can not only reveal the technical differences of these two technologies, but also help us better understand the underlying biological processes that lead to the ambiguous observations of SNVs at the DNA and RNA levels, respectively. Such a comparison can provide guidance on the utility of WES and RNA-Seq in SNV detection. So far, there have been only a few attempts to unveil the advantages and disadvantages of WES and RNA-Seq in SNV detection. For example, Cirulli et al. [17] recently compared WGS with RNA-Seq in detecting SNVs using peripheral blood mononuclear cells from the same subjects. They highlighted many important aspects for SNV detection such as expression levels and read depth, but its conclusions are yet to be validated due to the limited sample size. Another recent review compared WES and RNA-Seq [18], but it only discussed several global features without a systematic comparison of many detailed features.
In this study, we compared the features of SNVs from WES and RNA-Seq using a collection of 27 lung tumor and matched normal samples from the same patients. Through our systematic analyses, we attempted to unveil the unique features of SNVs from each platform and determined why variants are missed between these platforms. Because of the high false calling rate of indels, we only focused on SNVs. We observed only a small overlap of SNVs between WES and RNA-Seq, and identified multiple technological and biological reasons leading to discrepancies in SNV calling.
2. Materials and methods
2.1 Samples and sequencing
Twenty-seven paired tumor and normal lung cancer samples from patients undergoing lung cancer surgery at Massachusetts General Hospital were used for this analysis. For all 27 paired tumor and normal lung cancer samples, we performed both WES and RNA-Seq experiments. All participants provided written informed consent. Tumor content was assessed with an average of 60% across samples. The exome regions were captured using the Agilent SureSelect Human All Exon kit and then sequenced on an Illumina HiSeq 2000 platform (paired end, 100 bp) in a MGH core. We obtained a total of 3,677,811,274 paired-end reads with an average sequencing depth of 121×. For RNA-Seq, Illumina Tru-Seq v2 RNA-Seq kit was used for enrichment of mRNA, cDNA synthesis, and library construction. Then, RNA sequencing was performed on an Illumina HiSeq 2000 platform in the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core (paired end, 100 bp). We obtained a total of 4,778,766,598 paired end reads with an average of 88,495,678 paired end reads per sample. We used FASTQC to check the quality of reads of all samples (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
2.2 WES data analysis
We mapped the WES reads to the human reference genome hg19 (GRCh37) using BWA (version 0.5.9c) [19]. In order to further process the data, we used Picard (version 1.95) [20] to mark duplicate reads and used GATK (version 1.0.3825) to perform local realignment and recalibration [21, 22]. After post-alignment processing of the data, we called SNVs with MuTect (version 1.1.4). To generate mpileup files for each tumor and normal sample, we used the “mpileup” function in Samtools (version 0.1.19) [23]. Read count values were obtained from the mpileup files using VarScan2 (version 2.3.5) [24] with the “readcounts” function. Read count values were split up into categories of values: not covered (NA), single read (1), low coverage (2–7) and high coverage (≥ 8).
2.3 RNA-Seq data analysis
We used TopHat2 (version 2.0.0) [25] to map RNA-Seq reads to the human reference transcriptome and genome (hg19). TopHat2 firstly attempts to map reads to the reference transcriptome and then for the unmapped reads from the initial transcriptome, it attempts to map them to the human genome reference. As we did for WES data, we called SNVs using MuTect (version 1.1.4). Specifically, we generated mpileup files using Samtools and obtained read count values using VarScan2. We used Cufflinks (version 2.1.1) [26] to obtain gene-based FPKM (Fragments Per Kilobase of exon per Million fragments Mapped) values for all samples. FPKM values corresponding to degrees of expression were as follows: not covered (NA), no expression (FPKM < 1), very low expression (FPKM 1–5), low to moderate expression (FPKM 5–20), and high expression (FPKM > 20).
2.4 Read counting for the RNA-Seq SNVs covered by the WES capture kit
We used Bedtools (version 2.17.0) to determine whether the SNVs identified from RNA-Seq were covered by the WES capture kit using the “-intersectBed” function. SNVs were categorized into four groups by read count values as was done for the aforementioned read count analysis: not covered (NA), single read (1), low coverage (2–7) and high coverage (≥ 8).
2.5 Mutation pattern categorization for all SNVs
We categorized SNVs into six groups according to their nucleotide changes: A:T→C:G, A:T→T:A, A:T→G:C, C:G→A:T, C:G→G:C, and C:G→T:A. The number of each mutation type was further counted in four groups: WES unique SNVs, RNA-Seq unique SNVs, overlap from RNA-Seq, and RNA-Seq unique SNVs covered by the WES capture regions.
2.6 Allele frequency analysis for RNA-Seq SNVs
We determined the allele frequency for RNA-Seq unique SNVs covered by the WES capture regions using the read count values generated from VarScan2 [24]. An allele frequency of 0.2 was used as a threshold.
The computational tools that we used were summarized in Supplemental Table 1.
3. Results
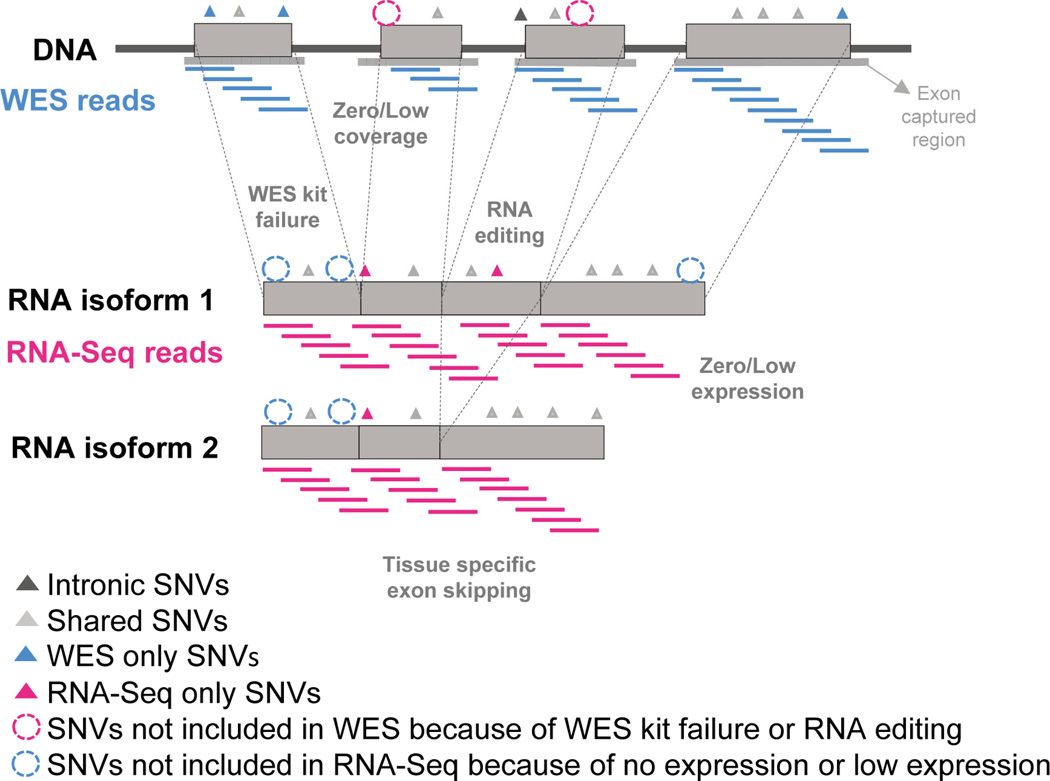
Fig. 1 illustrates the concept of our SNV comparison from WES and RNA-Seq data. There are several factors that may cause the difference in detecting SNVs from WES and RNA-Seq data, even from the same samples. First, the two sequencing technologies and their sequencing strategy will have variation in the enrichment of sequence regions. Second, at the biological level, SNVs detected from DNA-Seq (i.e., WES) may not be detectable by RNA-Seq due to low coverage, or tissue-specific expression and alternative splicing. In contrast, SNVs in the transcriptome may not be detected in WES because of low coverage, RNA editing, or their location outside of the WES capture regions. With these factors, we performed an in-depth comparison between SNVs detected by the two sequencing techniques.
Fig. 1.
Comparison between WES data and RNA-Seq data. This image shows the motivation and the concept behind our study. WES reads will be generated on the exon captured regions. RNA-Seq reads will be generated on the content of gene expression conditions. SNVs may exist in various locations of the genome including introns adjacent to exons in the DNA, and for locations within the transcriptome. SNVs for the intronic, WES and RNA-Seq shared, WES only, RNA-Seq only are colored with dark grey, light grey, blue and pink, respectively. SNVs not included in WES by the low coverage or WES kit failure or RNA editing are represented with pink dotted circles. SNVs not included in RNA-Seq by the low expression or coverage are represented with blue dotted circles.
3.1 Poor concordance for SNVs called in WES and RNA-Seq data
We obtained WES and RNA-Seq data for 27 lung cancer tumor samples and their matched normal samples. We applied a standard pipeline to analyze the samples and detect somatic SNVs (Supplemental Fig. 1). We refer to those SNVs that were uniquely detected in WES but not in RNA-Seq data as “WES unique SNVs,” those SNVs that were uniquely detected in RNA-Seq but not in WES data as “RNA-Seq unique SNVs,” and those observed in both WES and RNA-Seq as “WES shared SNVs” or “RNA-Seq shared SNVs.” Note that although the WES shared SNVs and the RNA-Seq shared SNVs have the same genomic coordinates, they may have different alternative allele frequencies, or even different alternative alleles, in the WES data and in the RNA-Seq data. Thus, we referred to them separately as WES shared SNVs and RNA-Seq shared SNVs. Overall, we identified 15,662 SNVs from the WES data, with an average of 580 ± 517 SNVs per sample, and 15,473 SNVs from the RNA-Seq data, with an average of 573 ± 332 SNVs per sample. Surprisingly, only ∼14% (2150) of these SNVs were detected by both WES and RNA-Seq (Table 1).
Table 1.
Summary of all SNVs detected in RNA-Seq and WES by MuTect.
| Sample ID | RNA-Seq | WES | Overlap | Overlap with RNA-Seq (%) |
Overlap with WES (%) |
|---|---|---|---|---|---|
| 1 | 452 | 388 | 52 | 11.5 | 13.4 |
| 2 | 1082 | 1206 | 263 | 24.3 | 21.8 |
| 3 | 731 | 902 | 175 | 23.9 | 19.4 |
| 4 | 531 | 62 | 9 | 1.7 | 14.5 |
| 5 | 572 | 83 | 4 | 0.7 | 4.8 |
| 6 | 640 | 619 | 92 | 14.4 | 14.9 |
| 7 | 317 | 94 | 8 | 2.5 | 8.5 |
| 8 | 220 | 85 | 5 | 2.3 | 5.9 |
| 9 | 659 | 168 | 23 | 3.5 | 13.7 |
| 10 | 524 | 78 | 8 | 1.5 | 10.3 |
| 11 | 529 | 447 | 36 | 6.8 | 8.1 |
| 12 | 597 | 773 | 112 | 18.8 | 14.5 |
| 13 | 432 | 335 | 54 | 12.5 | 16.1 |
| 14 | 533 | 1360 | 124 | 23.3 | 9.1 |
| 15 | 403 | 540 | 30 | 7.4 | 5.6 |
| 16 | 768 | 892 | 143 | 18.6 | 16.0 |
| 17 | 590 | 296 | 56 | 9.5 | 18.9 |
| 18 | 753 | 172 | 26 | 3.5 | 15.1 |
| 19 | 313 | 1060 | 42 | 13.4 | 4.0 |
| 20 | 422 | 1017 | 125 | 29.6 | 12.3 |
| 21 | 188 | 716 | 9 | 4.8 | 1.3 |
| 22 | 1425 | 901 | 158 | 11.1 | 17.5 |
| 23 | 348 | 309 | 19 | 5.5 | 6.1 |
| 24 | 310 | 227 | 45 | 14.5 | 19.8 |
| 25 | 97 | 66 | 1 | 1.0 | 1.5 |
| 26 | 508 | 577 | 58 | 11.4 | 10.1 |
| 27 | 1529 | 2289 | 473 | 30.9 | 20.7 |
| Mean ± SD | 573 ± 332 | 580 ± 517 | 80 ± 102 | 13.9 ± 9.0 | 13.7 ± 6.0 |
| Total | 15473 | 15662 | 2150 | ||
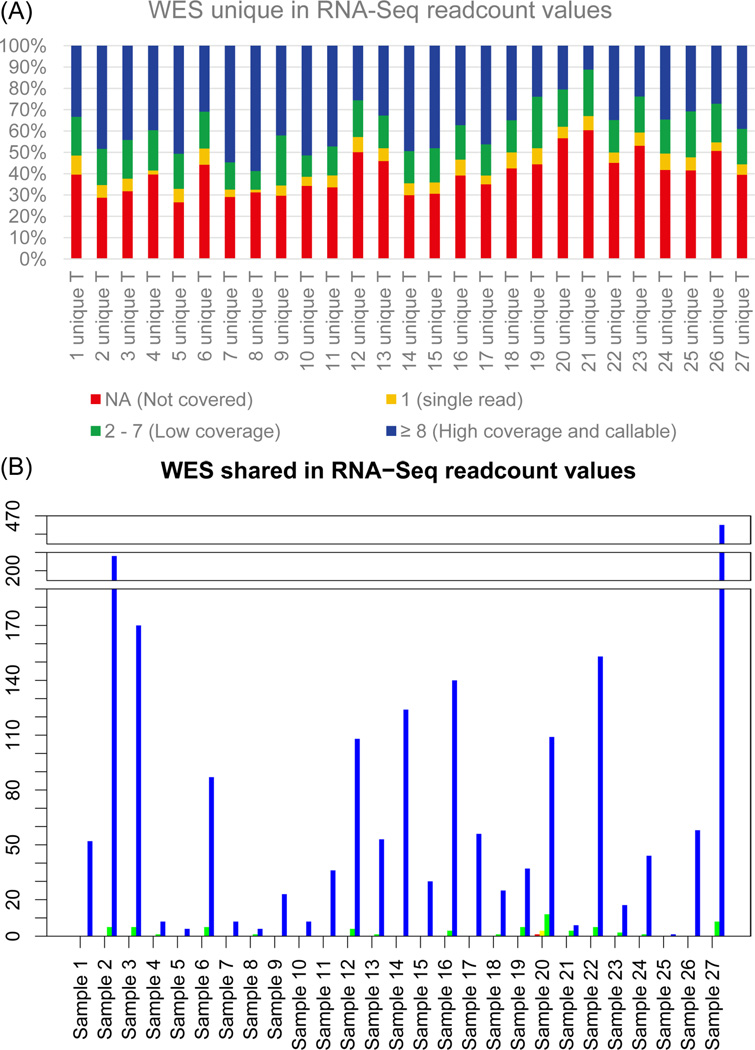
We explored the reasons why such a small portion of WES SNVs was detected in the RNA-Seq data. One possibility is that the positions of the WES SNVs are not well covered in RNA-Seq. Table 2 shows a summary of the RNA-Seq read counts for SNVs detected in WES using VarScan2. A large proportion of the WES unique SNVs (41.0%) are not covered in RNA-Seq. However, the majority (96.9%) of the WES shared SNVs have at least eight RNA-Seq reads mapped to their position (Fig. 2). There is a small proportion of WES unique and WES shared SNVs moderately covered in RNA-Seq (2–7 reads), 8.8 – 24.2% and 0 – 33.3% respectively. Interestingly, 11.2 – 58.8% of the WES unique SNVs have a high number (≥8) of RNA-Seq reads aligned to their position. However, these are still undetected in RNA-Seq. As we expected, the number of WES shared SNVs in positions that are not covered (NA) is low – on average it is less than 0.1% (in a range of 0–0.8%). We hypothesized that some of the WES unique SNVs may be located in genes which are not expressed, or have very low expression levels, and therefore are undetected by RNA-Seq.
Table 2.
Summary of read counta values from RNA-Seq for SNVs detected by WES.
| NAb | 1 | 2 – 7 | ≥ 8 | Total | |
|---|---|---|---|---|---|
| WES unique SNVsc | |||||
| Mean ± SD | 205 ± 179 | 30 ± 25 | 87 ± 77 | 179 ± 175 | 500 ± 429 |
| Range | 21 – 717 | 1 – 89 | 7 – 302 | 20 – 708 | 53 – 1816 |
| Range of % | 26.6 – 60.4 % | 1.3 – 8.9 % | 8.8 – 24.2 % | 11.2 – 58.8 % | |
| WES shared SNVsd | |||||
| Mean ± SD | 0 ± 0 | 0 ± 1 | 2 ± 3 | 77 ± 100 | 80 ± 102 |
| Range | 0 – 1 | 0 – 3 | 0 – 12 | 1 – 465 | 1 – 473 |
| Range of % | 0 – 0.8 % | 0 – 2.4 % | 0 – 33.3 % | 66.7 – 100 % |
Read count values were generated from RNA-Seq data using chromosomal locations of SNVs detected by WES.
NA: SNV positions from WES that are not covered at the nucleotide level in RNA-Seq.
WES unique SNVs: SNVs detected only in WES.
WES shared SNVs: SNVs detected in both WES and RNA-Seq.
Fig. 2.
VarScan2 read count values determine why WES unique SNVs are not called by RNA-Seq. (A) Stacked column graph showing read counts results in RNA-Seq for WES unique SNVs. (B) Barplot showing read counts results in RNA-Seq for WES shared SNVs. Red represents read counts NA (not covered), yellow represents readcounts 1, green represents read counts 2–7, and blue represents read counts ≥ 8. Most WES unique SNVs are not covered in RNA-Seq.
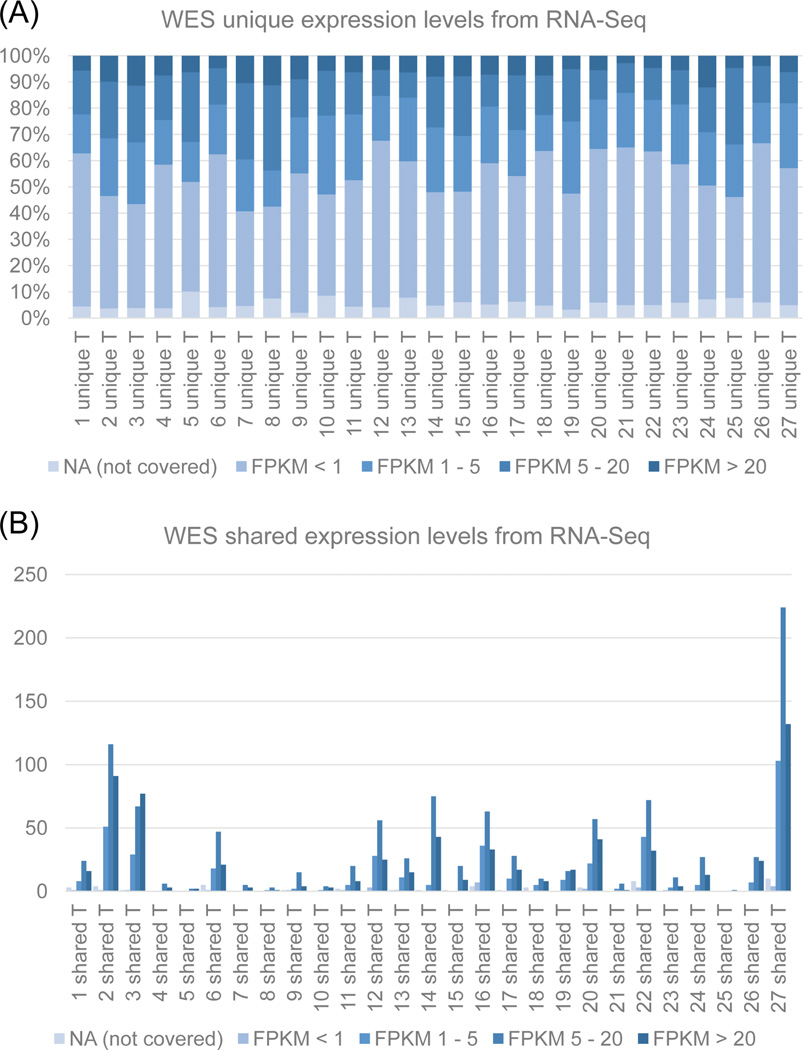
We further explored the features of WES unique SNVs regarding their gene expression levels. We used Cufflinks to generate FPKM values from RNA-Seq data for the chromosomal loci of WES SNVs (Table 3). We categorized FPKM values as not covered (NA), not expressed (< 1 FPKM), low expression (1–5 FPKM), low to moderate expression (5 – 20 FPKM) and high expression (> 20 FPKM) (Fig. 3). Many of the WES unique SNVs are located in genes that are not expressed (51.0%). In contrast, 77.7% of WES shared SNVs are located in genes with FPKM > 5, including 0 – 66.7% of WES shared SNVs located in genes with low to moderate expression (FPKM 5–20), and 11.1 – 100% WES shared SNVs located in genes with high expression levels (> 20 FPKM). It is interesting to note that 1.3% of the shared SNVs are located in genes which are not expressed in RNA-Seq.
Table 3.
Summary of FPKMa levels from RNA-Seq for SNVs detected by WES.
| NAb | < 1 | 1 – 5 | 5 – 20 | > 20 | Total | |
|---|---|---|---|---|---|---|
| WES unique SNVsc | ||||||
| Mean ± SD | 24 ± 20 | 255 ± 220 | 109 ± 106 | 79 ± 68 | 33 ± 31 | 500 ± 429 |
| Range | 2 – 90 | 25 – 948 | 9 – 449 | 9 – 240 | 3 – 114 | 53 – 1816 |
| Range of % | 2.1 – 10.1% | 35.0 – 63.5% | 13.7 – 30.0% | 9.6 – 32.5% | 2.8 – 12.1% | |
| WES shared SNVsd | ||||||
| Mean ± SD | 2 ± 3 | 1 ± 2 | 15 ± 23 | 38 ± 47 | 24 ± 31 | 80 ± 102 |
| Range | 0 – 10 | 0 – 7 | 0 – 103 | 0 – 224 | 1 – 132 | 1 – 473 |
| Range of % | 0 – 11.5% | 0 – 5.3% | 0 – 27.2% | 0 – 66.7% | 11.1 – 100% |
FPKM: Fragments Per Kilobase of transcript per Million mapped reads. FPKM gene expression values were generated by Cufflinks.
NA: SNV positions from WES that are not covered at the gene level in RNA-Seq.
WES unique SNVs: SNVs detected only in WES.
WES shared SNVs: SNVs detected in both WES and RNA-Seq.
Fig. 3.
Cufflinks analysis to determine gene expression levels of WES unique SNVs in RNA-Seq. (A) FPKM values for WES unique SNVs. (B) FPKM values for SNVs shared between WES and RNA-Seq. Most WES unique SNVs are located within genes which are not expressed in RNA-Seq. FPKM NA: not covered, FPKM < 1: not detected; FPKM 1–5: not expressed; FPKM 5 −20: low to moderate expression; and FPKM > 20: high expression.
3.2 Strand analysis of WES unique SNVs
As mentioned in the previous paragraphs, most of the WES unique SNVs are located in regions of genes with low expression. Interestingly, some of the WES unique SNVs are located in genes with FPKM > 20 (2.8 – 12.1%), but are not detected by MuTect. There may be several explanations for this. For example, the mutant (non-reference) alleles of the WES unique SNVs happen to be located on the untranscribed strand and were not transcribed at the mRNA level; or the SNV indeed occurred on the transcribed strand but its mutant allele frequency was too low to be detected due to allele specific expression. We checked the strand information of the mutant alleles. We used Oncotator [27] (http://www.broadinstitute.org/oncotator/) to generate annotation information for the WES SNVs. This annotation includes cDNA base-pair changes, and the strand information of the genes on which the SNVs are located. We examined the WES unique SNVs to determine whether they occur on the transcribed or non-transcribed strand. For the 13,512 WES unique SNVs, 10,897 (80.6%) had annotated cDNA information available, due to their location within the coding region. We found that 216 (2.0%) of these SNVs were located within annotated splice sites, and we excluded these SNVs from the strand level analysis. For the remaining 10,681 SNVs, 5271 (49.3%) were located on the non-transcribed strand (Table 4). Among the 5271 SNVs on the non-transcribed strand, some are located in regions of high expression in RNA-Seq (FPKM > 20), but the proportion is small, i.e., only 5.0%.
Table 4.
Strand-specific location of WES unique SNVs.
| Sample ID | Total # of WES unique SNVs |
# of SNVs with cDNA annotationa |
# SNVs (%) on complimentary strandb |
|---|---|---|---|
| 1 | 336 | 280 | 160 (57.1) |
| 2 | 943 | 735 | 375 (51.0) |
| 3 | 727 | 575 | 273 (47.5) |
| 4 | 53 | 41 | 22 (53.7) |
| 5 | 79 | 59 | 27 (45.8) |
| 6 | 527 | 425 | 201 (47.3) |
| 7 | 86 | 72 | 34 (47.2) |
| 8 | 80 | 64 | 24 (37.5) |
| 9 | 145 | 121 | 52 (43.0) |
| 10 | 70 | 59 | 26 (44.1) |
| 11 | 411 | 318 | 155 (48.7) |
| 12 | 661 | 503 | 242 (48.1) |
| 13 | 281 | 199 | 92 (46.2) |
| 14 | 1236 | 999 | 487 (48.7) |
| 15 | 510 | 426 | 229 (53.8) |
| 16 | 749 | 570 | 268 (47.0) |
| 17 | 240 | 180 | 82 (45.6) |
| 18 | 146 | 115 | 59 (51.3) |
| 19 | 1018 | 845 | 414 (49.0) |
| 20 | 892 | 721 | 366 (50.8) |
| 21 | 707 | 567 | 283 (49.9) |
| 22 | 743 | 589 | 280 (47.5) |
| 23 | 290 | 232 | 112 (48.3) |
| 24 | 182 | 137 | 64 (46.7) |
| 25 | 65 | 50 | 33 (66.0) |
| 26 | 519 | 396 | 207 (52.3) |
| 27 | 1816 | 1403 | 704 (50.2) |
| Total | 13512 | 10681 | 5271 (49.3) |
| Mean | 500 | 396 | 195 (49.3) |
Excluding SNVs within known annotated splice sites.
Complimentary strand: non-transcribed strand.
3.3 Feature analysis of RNA-Seq unique SNVs
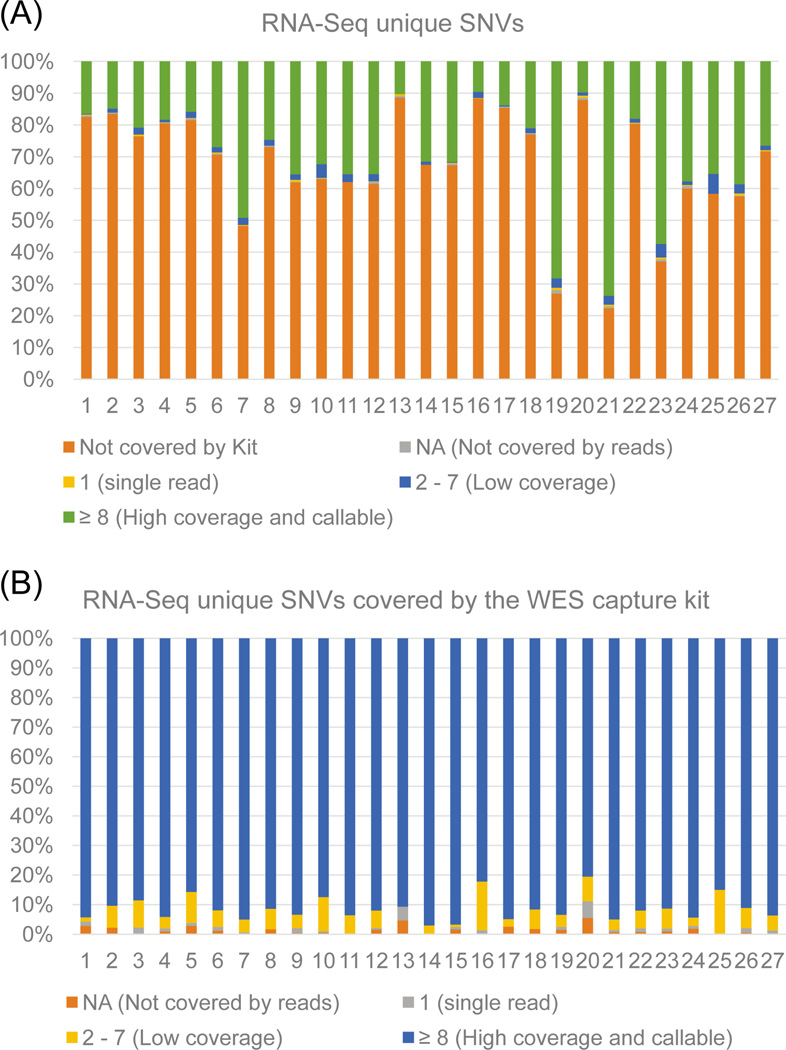
We then examined the features of RNA-Seq unique variants. We first explored RNA-Seq unique SNVs that may be located outside of the WES capture regions. RNA-Seq does not contain a specific exome capture step, so the variants detected are not constrained to the specific 1–2% of the genome sequenced by WES, and are only limited to the genomic regions that are being transcribed. We first explored the proportion of RNA-Seq unique SNVs that lie outside of the WES capture region. We used the”-intersectBed” command in Bedtools to identify RNA-Seq unique SNVs that are not covered by the WES capture region. For the 13,323 RNA-Seq unique SNVs, 9,513 (71.4%) are located outside of the WES capture regions (Fig. 4). We used VarScan2 to identify the read count values for the positions that are covered by the WES capture kit. We discovered that for the RNA-Seq unique SNVs covered by the kit, an average of ∼93% (82.2 – 98.3%) are in locations that are highly covered (≥ 8 reads) (Table 5). This is an interesting observation - it means that only approximately 7.0% of the SNVs uniquely called in RNA-Seq are potentially missed in WES due to low coverage of sequencing. Thus, the remaining SNVs are not missed due to technical issues, but due to biological issues.
Fig. 4.
RNA-Seq unique SNVs not covered by the WES kit and coverage levels. (A) Barplot shows the percentage of RNA-Seq unique SNVs within each sample that are not covered by the WES capture kit. Also included are VarScan2 read count values for covered positions. Figure 4A shows that most SNVs are not covered by the WES kit. Here, ‘not covered by kit’ represents RNA-Seq SNVs outside of the capture kit region, read counts values represented by NA, 1, 2 – 7, and ≥ 8. (B) Barplot containing VarScan2 read counts values for only the positions covered by the WES kit. Most SNVs covered by the WES kit have high coverage. Read counts values represented by NA, 1, 2 – 7, and ≥ 8.
Table 5.
Summary of WES coverage for RNA-Seq SNVs that are covered by the WES capture kit.
| NA | 1 | 2 – 7 | ≥ 8 | Total in kit | |
|---|---|---|---|---|---|
| Mean | 1 ± 1 | 1 ± 1 | 8 ± 5 | 130 ± 64 | 140 ± 68 |
| Range | 0 – 3 | 0 – 4 | 0 – 22 | 29 – 280 | 34 – 299 |
| Range of % | 0 – 5.9% | 0 – 5.9% | 0 – 16.4% | 82.2 – 98.3% |
We hypothesized that low frequency of the mutant alleles is a biological factor leading to the observation that many RNA-Seq SNVs are undetected in WES. We searched the allele frequency values for all RNA-Seq unique SNVs covered by the whole exome capture kit. MuTect calls variants having an allele fraction of 0.2 with 99.9% sensitivity at coverage rates of 50× [28]. Therefore, we used 0.2 as a threshold to determine the number of RNA-Seq unique SNVs covered by the WES capture kit. We observed that only 3.0% of these SNVs have allele frequency values ≥ 0.2. This suggests that a large proportion of highly covered RNA-Seq SNVs are not detected in WES because not many reads are mapped to the mutant allele.
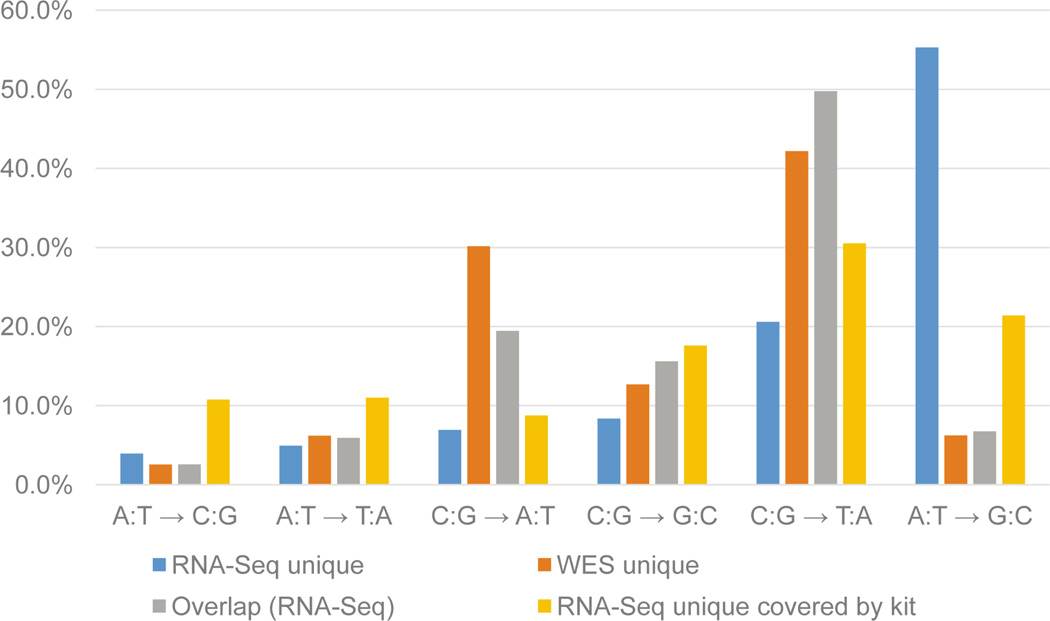
We hypothesized that RNA editing is another factor leading to the RNA-Seq SNVs being undetected in WES. Although there are known difficulties detecting RNA editing in NGS data [29–31], we explored this mechanism as a potential reason for inconsistencies in mutation calling between WES and RNA-Seq. We used the results from MuTect to analyze the base-pair mutation pattern across all SNVs for signatures of RNA editing. Interestingly, the most common mutation pattern for the RNA-Seq unique SNVs was the A:T→G:C mutation pattern, occurring in 55.3% of SNVs (Fig. 5). Another interesting finding was that 21.4% of the RNA-Seq unique SNVs that were covered by the WES capture kit (but not detected in WES) also shared this same mutation pattern. In comparison, only 6.7% of the total number of overlapping SNVs called in both WES and RNA-Seq had this mutation pattern. The A→G mutation is a common RNA-editing mechanism arising from A→I editing acted upon by Adenosine Deaminase Acting on RNA (ADARs) [11].
Fig. 5.
Mutation pattern for all SNVs. Mutation pattern was determined for all categories of SNVs and percentages plotted. Several patterns are more highly enriched than others, such as the A:T→G:C mutation in RNA-Seq.
4. Discussion
Few studies have examined mutation detection from both WES and RNA-Seq data of the same samples. However, such information is critical in assessing the mutations at different biological stages as well as their effects on disease. In this study, our comparison of WES and RNA-Seq data from the 27 pairs of tumor and matched normal samples revealed that on average only ∼14% of SNVs overlap. This value is quite low considering that the samples are identical. Thus, we explored possible reasons that cause this small overlap. We found that many of the WES unique SNVs are not called in RNA-Seq because they are poorly covered. This information is important for using a SNV-calling software tool like MuTect, where the coverage limitations allowed to call an SNV is at least 14 reads in the tumor and at least 8 in the normal.
We noticed that although low coverage levels explained why most WES unique SNVs were not detected in RNA-Seq, many other SNVs had high read counts values but were still missed. We decided to interrogate gene expression levels to determine if this may explain why some SNVs are not detected in RNA-Seq. We used FPKM values, and found that the majority of WES unique SNVs are located in genes which are not expressed. In contrast, the SNVs that were shared between sequencing methods were found to have moderate to high expression levels. This is an important finding, because many studies use WES as the single method for somatic mutation detection in cancer, and this analysis demonstrates that it is important to measure expression levels when trying to determine deleterious variants. Many SNVs may be called in WES, but may not have an impact at the biological level because the variant is located within a non-expressed gene. Expression levels were able to help explain why many SNVs were not called in RNA-Seq, but many more still remained highly expressed and non-callable in RNA-Seq. We explored why these still are not detected by analyzing the strand specific expression of these SNVs.
We used Oncotator to annotate the WES samples to determine if some SNVs were located on the non-transcribed strand. One limitation of this analysis is that we were limited to variants that were within the coding regions. We found that about half of all the detected SNVs with cDNA information available in WES are located on the non-transcribed strand. In order to determine if this is why SNVs within expressed genes are not called in RNA-Seq, we further examined if some of these SNVs located on the non-transcribed strand are in locations that harbor highly expressed genes. We found that ∼5% of the total WES SNVs located on the non-transcribed strand are highly expressed in RNA-Seq, but would unlikely be called due to their location on the non-transcribed strand. These results are informative, because it implies that many of the variants detected by WES may not be causative or damaging due to their location on the DNA strand. After determining these potential causes for the WES unique SNVs not being called in RNA-Seq, we next focused on the reasons why RNA-Seq unique SNVs were missed by WES.
We first thought that many of the RNA-Seq unique SNVs may be missed by WES because they fall outside of the WES capture regions. This is an important aspect to consider, because while RNA-Seq covers the whole transcriptome, WES is limited to detecting variants in the exons and their flanking regions. Currently, many exon capture kits are designed to have their probes covering well-annotated coding genes using representative gene models like CCDS and RefSeq. And the capture method using target-probe hybridization has the limitation of GC-content bias. To compare the regions covered by the kit with the RNA-Seq, we used Bedtools and found that the majority of the RNA-Seq unique SNVs are not covered in WES. This means that many potentially important SNVs not located in exome regions would be missed if WES were applied. This is becoming more important as ENCODE data has determined that many non-exonic regions in the genome are expressed, and that they may be playing important roles in gene regulation [32]. It also implies that only performing WES on a tumor sample may miss potential variants that may be of important function. However, coverage levels are high for the RNA-Seq unique SNVs that are detected by the kit. We hypothesized that allele frequency may help explain why these variants are being missed in WES. We found that most of the SNVs detected in RNA-Seq had very low variant allele frequency values in WES. This may have important consequences, because if there is a variant at a low allele frequency in WES, it may be preferentially expressed over the reference allele. This may lead to deleterious effects that would have been missed if only looking at the WES data.
Another biological reason why RNA-Seq unique SNVs may be missed in WES is due to RNA editing. We used the output from MuTect to generate the mutation pattern for all samples. An interesting mutation pattern in RNA-Seq unique SNVs was A:T→G:C. This pattern is indicative of RNA editing occurring by deamination of the adenosine to inosine, which gets interpreted as a guanine, editing in RNA achieved by the Adenosine Deaminase Acting on RNA proteins [11]. This result has two implications for tumor sequencing. First, there may be a defect in the RNA editing machinery that leads to over-editing occurring in loci that normally do not get edited. Studies have shown that increased and decreased levels of RNA-editing may occur in different types of cancer [33, 34]. This editing may give rise to new functions, or lose functions of important proteins in the tissue of interest. These mutations would be completely missed if sequencing were only focused on the whole genome or whole exome. Second, these mutations edited at RNA level are not expected to be detected by WES or WGS; therefore, their potential causative or deleterious effects will remain hidden. A list of factors that may lead to inconsistencies in detecting SNVs in RNA-Seq versus WES is summarized in Table 6.
Table 6.
Summary of factors that may lead to inconsistencies in detecting SNVs in WES versus RNA-Seq.
| Factors causing RNA- Seq unique SNVs |
Observation | Factors causing WES unique SNVs |
Observation |
|---|---|---|---|
| SNVs outside of the WES capture regions |
71.4% of RNA-Seq unique SNVs |
Low coverage of SNVs in RNA-Seq |
41.0% of WES- unique SNVs have no RNA-Seq coverage |
| Low coverage of SNVs in WES |
8.0% of RNA-Seq unique SNVs that are within the WES regions, have low or no WES coverage |
SNVs located in non- expressed genes (< 1 FPKM) |
51.0% of WES- unique SNVs |
| Low mutant allele frequency of RNA- Seq SNVs within WES regions |
97.0% of RNA-Seq SNVs within WES regions had mutant allele frequency < 0.2 |
SNVs on the non- transcribed strand |
49.3% of WES- unique SNVs with cDNA information available |
| RNA-editing | 55.3% of RNA-Seq unique SNVs were A:T→G:C mutations |
SNVs potentially edited in RNA-Seq |
55.3% of RNA-Seq unique SNVs were A:T→G:C mutations |
Although we discovered many important differences between variants detected in WES versus RNA-Seq, there are some limitations to the interpretation of the results. Our samples were exclusively from tumor material, so it will be interesting to see if these results are similar for non-tumor tissue and germline mutations. We only used a total of 27 pairs of samples, and while this is large number and adequate for this analysis, it may miss some important conclusions. Furthermore, while the number of reads per sample in our RNA-Seq is large, it is not sufficient enough for RNA splicing analysis. Finally, the SNVs called in each tumor type and sequencing type vary widely, so a pan-cancer study may identify additional reasons for the small overlap of variants detected in WES versus RNA-Seq.
5. Conclusions
In conclusion, our systematic comparison of SNVs from WES and RNA-Seq data revealed a low overlap. We pinpointed multiple reasons for the inconsistencies in SNV detection with RNA-Seq and WES. It was discovered that most WES SNVs were undetected by RNA-Seq because of low coverage, low expression levels, or their strand-specific location on DNA. We found that most SNVs detected by RNA-Seq were missed in WES because they are located outside the boundary of the WES capture regions. It was also discovered that RNA-Seq SNVs that are highly covered by the WES capture kits may still be undetected due to low allele frequency of variants. Lastly, we found that many SNVs detected by RNA-Seq had a mutational signature of RNA editing. This analysis serves as an important resource to investigators regarding the strengths and limitations on SNV calls using WES and RNA-Seq, especially in a tumor genomic study.
Supplementary Material
Highlights.
We compared SNVs called from WES versus RNA-Seq of the same samples
We found a low overlap of ∼14% between SNVs called in WES and RNA-Seq
Low coverage and expression levels explain why some SNVs are missed in RNA-Seq
Location of SNVs outside of WES capture kit explain why some are missed in WES
Acknowledgements
This study was supported by a grant from LUNGevity Foundation and Upstage Lung Cancer. We also thank the financial support from US National Institutes of Health grants (R01LM011177, P50CA095103, P50CA098131, P30CA068485, and T32GM080178), a Vanderbilt Breast SPORE pilot grant, and Ingram Professorship Funds (ZZ). TO was supported by a National Institute of General Medical Sciences Training Grant (T32GM080178). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest
The authors claim no competing interest.
Contributor Information
Timothy D. O’Brien, Email: timothy.obrien@vanderbilt.edu.
Peilin Jia, Email: peilin.jia@vanderbilt.edu.
Junfeng Xia, Email: jfxia@ahu.edu.cn.
Uma Saxena, Email: stqa8350@gmail.com.
Hailing Jin, Email: hailing.jin@Vanderbilt.Edu.
Huy Vuong, Email: huy.vuong@gmail.com.
Pora Kim, Email: pora.kim@vanderbilt.edu.
Qingguo Wang, Email: josephw10000@gmail.com.
Martin J Aryee, Email: Aryee.Martin@mgh.harvard.edu.
Mari Mino-Kenudson, Email: MMINOKENUDSON@partners.org.
Jeffrey Engelman, Email: JENGELMAN@partners.org.
Long P. Le, Email: LPLE@partners.org.
A. John Iafrate, Email: AIAFRATE@partners.org.
Rebecca S. Heist, Email: RHEIST@partners.org.
William Pao, Email: william.pao@Vanderbilt.Edu.
Zhongming Zhao, Email: zhongming.zhao@vanderbilt.edu.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Jia P, Li F, Chen H, Ji H, Hucks D, Dahlman KB, Pao W, Zhao Z. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? J Med Genet. 2011;48:580–589. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- 7.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 8.Jia P, Jin H, Meador CB, Xia J, Ohashi K, Liu L, Pirazzoli V, Dahlman KB, Politi K, Michor F, et al. Next-generation sequencing of paired tyrosine kinase inhibitor-sensitive and -resistant EGFR mutant lung cancer cell lines identifies spectrum of DNA changes associated with drug resistance. Genome Res. 2013;23:1434–1445. doi: 10.1101/gr.152322.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greif PA, Eck SH, Konstandin NP, Benet-Pages A, Ksienzyk B, Dufour A, Vetter AT, Popp HD, Lorenz-Depiereux B, Meitinger T, et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25:821–827. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- 11.Maas S. Posttranscriptional recoding by RNA editing. Adv Protein Chem Struct Biol. 2012;86:193–224. doi: 10.1016/B978-0-12-386497-0.00006-2. [DOI] [PubMed] [Google Scholar]
- 12.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012;22:2315–2327. doi: 10.1101/gr.140988.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkerson MD, Cabanski CR, Sun W, Hoadley KA, Walter V, Mose LE, Troester MA, Hammerman PS, Parker JS, Perou CM, Hayes DN. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic Acids Res. 2014;42:e107. doi: 10.1093/nar/gku489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirulli ET, Singh A, Shianna KV, Ge D, Smith JP, Maia JM, Heinzen EL, Goedert JJ, Goldstein DB. Center for HIVAVI: Screening the human exome: a comparison of whole genome and whole transcriptome sequencing. Genome Biol. 2010;11:R57. doi: 10.1186/gb-2010-11-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ku CS, Wu M, Cooper DN, Naidoo N, Pawitan Y, Pang B, Iacopetta B, Soong R. Exome versus transcriptome sequencing in identifying coding region variants. Expert Rev Mol Diagn. 2012;12:241–251. doi: 10.1586/erm.12.10. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picard Web Site [Google Scholar]
- 21.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. [ http://www.broadinstitute.org/oncotator/]
- 28.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 30.Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin W, Piskol R, Tan MH, Li JB. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210419. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 32.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.