Abstract
Interferon-γ (IFN-γ) primes macrophages for enhanced inflammatory activation by Toll-like receptors (TLRs) and microbial killing, but little is known about the regulation of cell metabolism or mRNA translation during priming. We found that IFN-γ regulates human macrophage metabolism and translation by targeting the kinases mTORC1 and MNK that both converge on the selective regulator of translation initiation eIF4E. Physiological downregulation of mTORC1 by IFN-γ was associated with autophagy and translational suppression of repressors of inflammation such as HES1. Genome-wide ribosome profiling in TLR2-stimulated macrophages revealed that IFN-γ selectively modulates the macrophage translatome to promote inflammation, further reprogram metabolic pathways, and modulate protein synthesis. These results add IFN-γ-mediated metabolic reprogramming and translational regulation as key components of classical inflammatory macrophage activation.
Interferon-γ (IFN-γ) activates innate responses by augmenting inflammatory cytokine and chemokine production, microbial killing, and antigen presentation by mononuclear phagocytes such as macrophages1. Immune cell activation by IFN-γ is entirely dependent on its activation of the transcription factor STAT1, which binds to and activates transcription of interferon-stimulated genes (ISGs)2. Direct and rapid activation of ISGs plays a key role in IFN-γ-mediated functions2. It has become clear that many IFN-γ activities can not be explained by the direct effector functions of ISGs, and that important IFN-γ functions are mediated by crossregulation of distinct signaling pathways or reprogramming of cell states to alter their responses to extracellular stimuli1. For example, IFN-γ- augments macrophage cytokine production in response to inflammatory stimuli such as TLR ligands1 by attenuating signaling via the suppressive transcription factor STAT33, and by inhibiting expression of the TLR-induced Notch-dependent transcriptional repressors HES1 and HEY14. In parallel, IFN-γ reprograms the ‘epigenetic landscape’ of macrophages by inducing and priming enhancers to increase transcriptional output in response to TLR signaling5, 6. Whether IFN-γ can reprogram macrophage metabolism to alter cell function remains to be elucidated.
The importance of translational control of immune responses is increasingly appreciated7. Increased translation of select cytokine, chemokine and transcription factor mRNAs has been observed after TLR stimulation8, 9, and key immune regulators such as I-κBα and IRF7 are under translational control10, 11. Selective translational regulation of mRNA transcripts typically occurs at the level of initiation and can be achieved by specific RNA-binding proteins, microRNAs, and by modulation of the activity of 5’ cap-binding eukaryotic initiation factor 4E (eIF4E)7. Although eIF4E is a general translation initiation factor, changes in its activity do not globally regulate translation but instead selectively affect translation of a subset of transcripts, including inefficiently translated transcripts with long 5’ untranslated regions (UTRs)7, 11, 12. eIF4E activity is regulated by MNK kinases and mechanistic target of rapamycin complex 1 (mTORC1)7, and thus is responsive to upstream signals that activate MAPK signaling or mTORC1 activity. Type I IFNs suppress translation13 by inactivating translation factor eIF2α14 and inducing ISGs that translationally silence viral RNAs15. IFNs can promote translation of ISGs by various mechanisms16, and IFN-γ suppresses translation of a small set of mRNAs17. Little is known about regulation of translation by IFN-γ in immune cells.
The mechanistic target of rapamycin (mTOR) serine threonine kinase is a component of distinct mTORC1 and mTORC2 complexes, of which mTORC1 is an important regulator of mRNA translation18. mTORC1 senses and coordinates cellular responses to the nutrient status of extracellular and intracellular microenvironments, and is regulated by growth factors, oxygen, stress, and intracellular amino acid and energy levels. mTORC1 activation requires binary inputs from growth factor-induced Akt-mediated signaling that activates mTOR, and repletion of intracellular amino acids, which enables translocation of mTORC1 to lysosomal membranes where mTOR activation occurs18. Under nutrient replete conditions, mTORC1 promotes anabolic, biosynthetic and proliferative pathways, including protein, lipid and nucleotide synthesis that are required for cell growth18. mTORC1 promotes protein synthesis by phosphorylating and inactivating negative regulators of eIF4E termed 4E-BPs, and by activating kinase p70S6K that phosphorylates ribosome proteins7. Understanding of the role of mTORC1 in innate immunity is limited19, 20.
In this study we investigated how IFN-γ alters macrophage cell state to potentiate TLR responses. To maximize physiological relevance for human inflammatory conditions, we used primary human monocytes and macrophages that play a key role in human inflammatory diseases. We found that IFN-γ regulates TLR2 responses in human macrophages by suppressing MNKs and mTORC1 and modulating mRNA translation. A genome-wide ribosome profiling approach revealed that translational regulation selectively affected pathways important for cytokine expression, protein synthesis and cell metabolism. Our findings reveal an unrecognized function of IFN-γ to reprogram macrophage metabolism to alter inflammatory responses.
RESULTS
Inhibition of HES1 mRNA translation by IFN-γ
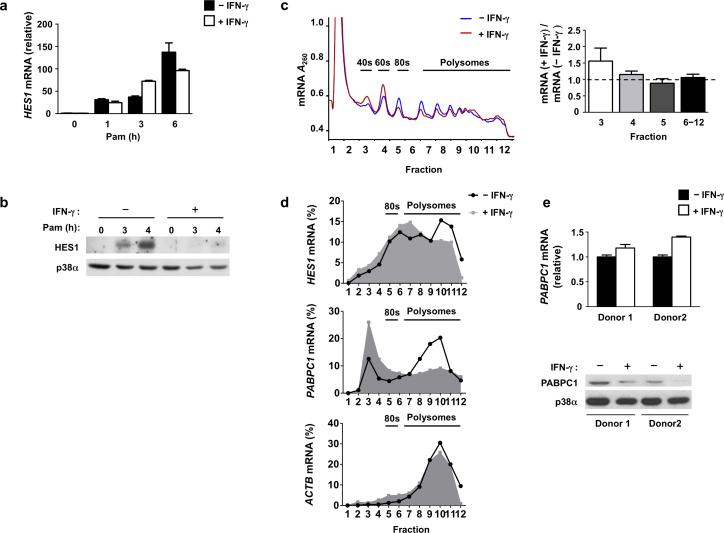
It was previously shown that IFN-γ suppresses TLR4-induced expression of transcriptional repressors HES1 and HEY1, thereby disrupting a feedback inhibitory loop and augmenting production of the inflammatory cytokines IL-6 and IL-124. We tested whether IFN-γ could also repress induction of HES1 mRNA by TLR2 in primary human macrophages. Stimulation with the TLR2 ligand Pam3CSK4 resulted in HES1 mRNA expression within 1 h, and its abundance increased in a time-dependent manner (Fig. 1a). In contrast to 10-fold repression of TLR4 (lipolysacharride, LPS)-induced HES1 mRNA by IFN-γ (Supplementary Fig. 1a), TLR2-induced HES1 transcript amounts were minimally suppressed by IFN-γ (Fig. 1a). This observation was consistent among more than 20 human blood donors tested, and no statistically significant difference was apparent when we pooled data from 23 different donors (Supplementary Fig. 1b). In striking contrast to mRNA regulation, TLR2-induced HES1 protein expression was almost completely abrogated by IFN-γ (Fig. 1b; HES1 was suppressed by 85.5% in 6 independent experiments, p <0.0001). The inhibitory effect of IFN-γ on TLR2-induced HES1 protein expression became apparent with 4 h of IFN-γ pre-treatment and was clearly established after 6 h of priming (data not shown), which is consistent with previous studies of IFN-γ priming21. The prominent discrepancy between HES1 mRNA and protein expression suggests that IFN-γ negatively regulates HES1 expression at the protein level.
Figure 1. IFN-γ suppresses HES1 translation.
(a) Quantitative PCR (qPCR) analysis of HES1 mRNA in human primary macrophages cultured with or without IFN-γ (100 U/ml) for 24 h and then stimulated with Pam3CSK4 (10 ng/ml) for 0-6 h; results are normalized relative to GAPDH mRNA; Pam= Pam3CSK4. Data are shown as means + SD of triplicate determinants and are representative of more than 20 independent experiments (cumulative results, Supplementary Fig. 1b). (b) Immunoblot analysis of HES1 in control or IFN-γ-treated macrophages stimulated with Pam3CSK4 (10 ng/ml) for 0-4 h as indicated; p38α serves as a loading control; Pam= Pam3CSK4. Data are representative of 23 independent experiments. (c) Representative polysome profiles (left panel) of control (blue) or IFN-γ-treated (red) macrophages stimulated with Pam3CSK4 (10 ng/ml) for 4 h. Quantitation (right panel) of ratio of RNA amounts in IFN-γ-treated relative to control conditions in the indicated fractions from three independent experiments (error bars, s.d.). Data are presented as mean ± SD and were analyzed using one way ANOVA, p = 0.1383. (d) Polysome shift qPCR analysis of HES1, PABPC1 and ACTB mRNA in polysome fractions from (c), depicted as the percentage of mRNA in each fraction compared to total mRNA of all fractions (fractions 1-12). Data are representative of three independent experiments. (e) qPCR analysis (upper panel) of PABPC1 mRNA in control or IFN-γ-treated macrophages from two independent donors (error bars, s.d.); data are shown as means + SD of triplicate determinants. Immunoblot analysis (lower panel) of PABPC1 from parallel samples in the same experiments; p38α serves as loading control.
IFN-γ could downregulate HES1 protein either by suppressing its synthesis or increasing its degradation. To distinguish between these possibilities, we first monitored HES1 protein half life in control and IFN-γ-treated macrophages, and found that HES1 protein decay rates were comparable in both conditions (Supplementary Fig. 1c). We also found that neither the proteasome inhibitor MG132 nor autophagosome-lysosome fusion inhibitor Bafilomycin A1 reversed the downregulation of HES1 protein amounts by IFN-γ (Supplementary Fig. 1d-e), implying that accelerated protein degradation was not responsible for the reduced HES1 protein abundance in IFN-γ-treated cells. This suggested that IFN-γ suppressed translation of HES1 mRNA. We directly tested this hypothesis by performing sucrose gradient centrifugation to track the ribosomal distribution of mRNAs in cytosolic extracts from control and IFN-γ-treated macrophages. The frequency of ribosome binding to an mRNA corresponds to the translation efficiency of each mRNA molecule, and actively translated mRNAs are found in the polysome fractions. IFN-γ had no effect on total mRNA amounts associated with heavy polysome fractions 9-12 and a minimal suppressive effect on light polysome fractions 6-8, indicating that IFN-γ did not globally and nonspecifically suppress translation (Fig. 1c). However, IFN-γ slightly reduced monomeric 80S ribosomes (fraction 5) and correspondingly increased mRNA amounts associated with individual ribosome 40S and 60S subunits (fractions 3 and 4) (Fig. 1c), implying that IFN-γ might suppress translation initiation, which is associated with 80S ribosome assembly, of select mRNA transcripts.
A shift in individual mRNAs from heavy polysome to light polysome or monosome fractions indicates decreased translation efficiency. Polysome shift analysis of mRNAs encoded by specific genes showed that in control TLR2-stimulated macrophages a substantial fraction of HES1 mRNA resided in heavy polysome fractions (Fig. 1d, top panel, fractions 10-12), indicating efficient translation. In IFN-γ-treated cells the HES1 mRNA peak shifted to light polysome fractions (Fig. 1d, top panel, fractions 6-8), indicating decreased translational efficiency. As a positive control for the polysome shift analysis, we found that mRNA encoded by PABPC1, which is known to be sensitive to translational regulation, was strongly shifted to the light polysome and monosome fractions in IFN-γ-treated macrophages (Fig. 1d, middle panel). Accordingly, IFN-γ suppressed PABPC1 protein amounts without affecting mRNA levels (Fig. 1e, representative data; PABPC1 was suppressed by 68.75% in pooled data from 4 independent experiments, p = 0.01). As a specificity control, we found that ACTB mRNA was present in the heavy polysome fractions, and thus actively translated, regardless of IFN-γ treatment (Fig. 1d, bottom panel). Collectively, the results demonstrate that IFN-γ exerts selective effects on translation efficiency of distinct mRNAs, and that IFN-γ inhibits translation of PABPC1 and HES1 mRNA in primary human macrophages.
IFN-γ attenuates TLR2-induced MAPK-MNK-eIF4E signaling
We wished to test whether IFN-γ inhibits translation by targeting the key regulator of translation initiation efficiency eIF4E, which is activated following phosphorylation by mitogen activated protein kinase (MAPK) interacting kinases (MNKs)22 (Supplementary Fig. 2a). TLR2 stimulation induced activating phosphorylation of MNK1 and eIF4E, which was substantially but not completely inhibited by IFN-γ at all time points tested (Fig. 2a) (p-eIF4E was suppressed by 69% in 6 independent experiments, p = 0.0009; p-MNK1 by 66% in 3 independent experiments, p = 0.02). These results show that IFN-γ negatively regulates activation of TLR2-induced MNK-eIF4E signaling; the functional importance of this suppression was supported by IFN-γ-mediated suppression of translation of the signaling inhibitor I-κBα and the transcription factor IRF8, which was previously shown to be dependent on the MNK-eIF4E pathway9, 10 (Fig. 2b and Supplementary Fig. 2b-d). These results suggest that MNKs play a role in promoting translation of HES1. We tested this notion by inhibiting MNKs or knocking down their expression using siRNAs and measuring HES1 expression by immunoblot. The MNK inhibitor CGP57380 strongly suppressed HES1 protein expression in a dose-dependent manner (Fig. 2c) without affecting HES1 mRNA induction (Fig. 2d). Furthermore siRNA-mediated knockdown of MNKs (Supplementary Fig. 2e) resulted in diminished phosphorylation of eIF4E and decreased expression of HES1 (Fig. 2e) without decreasing HES1 mRNA expression (Supplementary Fig. 2f). These data indicate that efficient HES1 translation requires MNK-mediated phosphorlyation of eIF4E. To gain insight into how IFN-γ suppresses TLR2-induced MNK phosphorylation, we examined upstream MAPK activation. IFN-γ attenuated TLR2-induced phosphorylation of p38 and ERK MAPKs in primary human macrophages (Fig. 2f), which is consistent with our previous results3. These MAPKs are dephosphorylated and inactivated by dual specificity phosphatases (DUSPs); accordingly IFN-γ increased baseline and TLR2-induced expression of DUSP 1, 2, 4, 8 and 16 (Fig. 2g). A role for phosphatases in attenuation of MAPK signaling was further supported by reversal of IFN-γ-mediated dampening of p38 activation by the phosphatase inhibitor okadaic acid (Supplementary Fig. 2g). Taken together, our results suggest that optimal translation of HES1 mRNA requires phosphorylation of 5’ mRNA cap-binding protein eIF4E, and that IFN-γ suppresses HES1 translation in part by attenuating TLR2-induced activation of MAPK-MNK-eIF4E signaling.
Figure 2. IFN-γ inhibits TLR2-induced activation of MAPK-MNK-eIF4E axis.
(a) Immunoblot analysis of phosphorylated (p-) eIF4E and p-MNK1 in control or IFN-γ-primed macrophages treated with Pam3CSK4 (10 ng/ml) for 0-60 min; p38α serves as loading control. (b) Polysome shift analysis of NFKBIA mRNA. (c) Immunoblot analysis of HES1 in human primary macrophages pretreated for 30 min with the vehicle control dimethyl sulfoxide (DMSO) or increasing concentrations of MNK inhibitor CGP57380, then stimulated for 0, 2 or 4h with Pam3CSK4 (10 ng/ml); p38α serves as loading control. (d) qPCR analysis of HES1 mRNA in human primary macrophages (error bars, s.d.). UT=untreated; Pam= Pam3CSK4. Data are shown as means + SD of triplicate determinants and are normalized relative to GAPDH mRNA. (e) Immunoblot analysis of HES1 and phosphorylated (p-) eIF4E in human primary macrophages transfected with scrambled control small interfering RNA (siRNA) or siRNA specific for both MNK1 and MNK2 for 72h, and then stimulated for 0-4 h with Pam3CSK4 (10 ng/ml); p38α serves as loading control. (f) Immunoblot analysis of phosphorylated (p-) p38 and p-ERK in control or IFN-γ-primed macrophages treated with Pam3CSK4 (10 ng/ml) for 0-60 min; p38α and ERK serve as loading controls. (g) qPCR analysis of DUSP1, DUSP2, DUSP4, DUSP8 and DUSP16 mRNA in control or IFN-γ-primed macrophages treated with or without Pam3CSK4 for 4h (error bars, s.d.). Data are shown as means + SD of triplicate determinants and are normalized relative to GAPDH mRNA. Data are representative of at least three independent experiments (a-g).
IFN-γ suppresses mTORC1 activation and downstream function
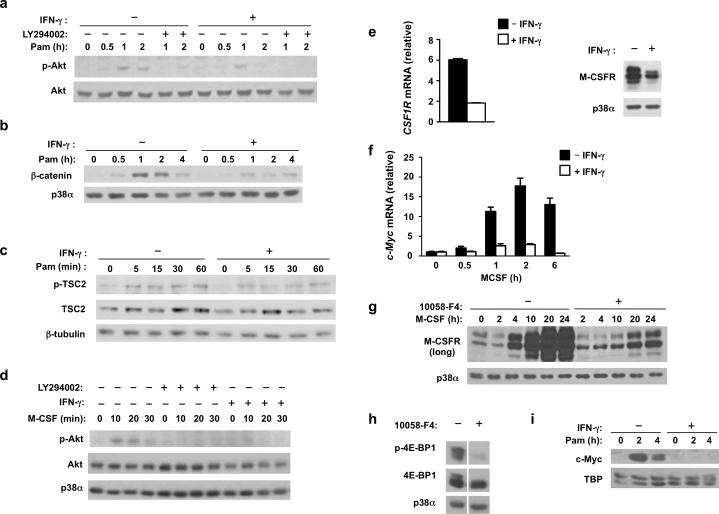
Given the partial attenuation of MNK activation by IFN-γ, but near-complete suppression of HES1, we hypothesized an involvement of additional signaling pathways in the process. A major positive regulator of translation is the mTORC1 complex, which phosphorylates eIF4E binding proteins (4E-BPs) to release them from eIF4E and thereby promote translation (Supplementary Fig. 2a). Therefore, we tested whether IFN-γ suppressed mTORC1 activity. mTORC1 activity is typically assessed by measuring phosphorylation of its downstream substrates 4E-BP and p70S6K. As expected, 4E-BP1 was phosphorylated on Thr37 and/or Thr46 in cultured macrophages, consistent with mTORC1 activity maintained by serum growth factors (Fig. 3a). IFN-γ suppressed basal 4E-BP1 phosphorylation (Fig. 3a, lane 6 vs. lane 1; 52% mean suppression in 7 independent experiments, p = 0.0036) and diminished 4E-BP1 phosphorylation was maintained throughout the time course of TLR2 stimulation in IFN-γ-treated macrophages (Fig. 3a). IFN-γ also suppressed 4E-BP1 phosphorylation in TLR-stimulated human monocyte-derived dendritic cells (Supplementary Fig. 3a). In macrophages IFN-γ suppressed 4E-BP1 phosphorylation comparably or slightly more strongly than the well established allosteric inhibitor of mTORC1 rapamycin (Fig. 3b), which only partially inhibits mTORC1 but has strong biological effects related to suppression of mTORC1 function23. The ATP-competitive inhibitors of mTOR kinase activity, Torin1 and PP242, more strongly suppressed 4E-BP1 phosphorylation, as expected. IFN-γ also suppressed phosphorylation of p70S6K, another substrate of mTORC1 that is involved in translation regulation (Fig. 3c, lane 6 vs. lane 1; 50.5% mean suppression in 4 independent experiments, p=0.0031). These data show that IFN-γ suppresses phosphorylation of mTORC1 substrates important in regulation of translation, and support a role for IFN-γ in repressing mTORC1 activity.
Figure 3. IFN-γ suppresses mTORC1 activation and downstream functions.
(a-c) Immunoblot analysis of whole cell lysates from control or IFN-γ-treated macrophages stimulated with Pam3CSK4 (10 ng/ml) for the indicated times and probed with antibodies against p-4E-BP1 (a, b) or p-p70S6K (c). In (b), mTOR inhibitors PP242 (50 nM ), Torin1 (50 nM) or Rapamycin (500 nM) were added for 30 min. (d) Immunoblot analysis of LC3A and LC3B in control or IFN-γ-primed macrophages. (e) Upper: Immunofluorescence images of LAMP1 (red) and mTOR (green) co-staining in control or IFN-γ-primed macrophages stimulated with Pam3CSK4 (10 ng/ml) for 4 h; nuclei were stained with DAPI (blue). Quantitation of co-localization (lower panel) between LAMP1 and mTOR; data are presented as mean ± SEM of the percentage of co-localized cells from 600 cells analyzed in three independent experiments; * p = 0.0001 by unpaired student t test. (f) Immunoblot analysis of HES1 in human primary macrophages pretreated for 30min with vehicle control DMSO or increasing concentrations of Rapamycin (0 nM, 500 nM, 1 μM), and then stimulated with Pam3CSK4 (10 ng/ml) for 0, 2, or 4h; p38α serves as loading control. Data are representative of at least three independent experiments (a-e).
mTORC1 suppresses autophagy, a critical cellular process that degrades cytoplasmic proteins and organelles under conditions of nutrient deprivation18. Autophagy is characterized by autophagosome formation and generation of the proteolyzed form of autophagosome markers LC3A and LC3B24. IFN-γ induced comparable generation of the proteolyzed isoforms LC3A-II and LC3B-II as did rapamycin (Fig. 3d). These results further support that IFN-γ inhibits mTORC1 activity and suggest that one functional outcome of such inhibition is increased autophagy, which promotes microbial killing and antigen presentation24. Inhibition of mTORC1 resulted in increased inflammatory cytokine production (Supplementary Fig. 3b), supporting the notion that attenuation of mTORC1 activity by IFN-γ contributes to its activating functions. mTORC1 activation occurs on lysosomal membranes and requires recruitment of mTORC1 to late endosomes and lysosomes by the Ragulator-Rag complex18, 25. We used immunofluorescence microscopy to test whether IFN-γ affected localization of mTOR to lysosomes. Control macrophages exhibited prominent colocalization of staining for mTOR and the lysosome and late endosome marker LAMP1 (Fig. 3e, upper panels). In contrast, IFN-γ-treated macrophages exhibited a clear dissociation of mTOR and LAMP1 staining (Fig. 3e, lower panels; p = 0.0001). Collectively, reduced 4E-BP1 and p70S6K phosphorylation, enhanced autophagy, and disruption of mTOR lysosomal localization converge to the same conclusion that IFN-γ suppresses mTORC1 activity in human macrophages.
The results that IFN-γ suppressed mTORC1 activity and in parallel suppressed HES1 translation suggested that translation and expression of HES1 protein might depend on mTORC1 signaling. We tested this prediction using rapamycin to inhibit mTORC1 and measured HES1 by immunoblot. Rapamycin inhibited TLR2-induced expression of HES1 protein in a dose dependent manner (Fig. 3f) but had no effect on HES1 mRNA (Supplementary Fig. 3c). Thus, mTORC1 activity is required for translation of HES1 mRNA into protein. These results link the inhibition of mTORC1 activity by IFN-γ with IFN-γ-mediated suppression of HES1 translation and provide an additional functional outcome of suppression of mTORC1 by IFN-γ.
IFN-γ suppresses localization of mTORC1 to lysosomes
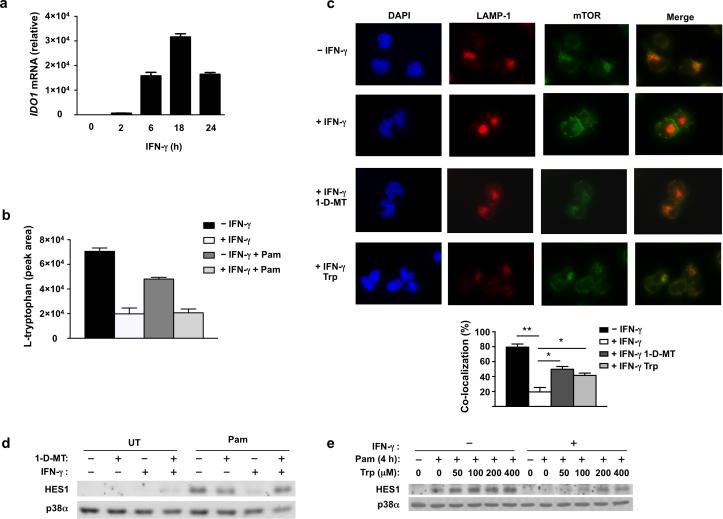
Next we sought to identify mechanisms by which IFN-γ inhibits mTORC1 in human macrophages. mTORC1 activation requires dual inputs, one from the extracellular environment, typically mediated by growth factor-PI3K-Akt-TSC1/2-Rheb signaling, and a second from intracellular nutrient availability, typically conveyed by amino acid-mediated activation of the Ragulator-Rag complex that recruits mTORC1 to lysosomal membranes, where mTOR is activated by GTP-binding protein Rheb (Supplementary Fig. 3d). We wished to elucidate how IFN-γ suppressed the localization of mTORC1 to lysosomes, as was observed above. We thus investigated whether IFN-γ regulates the amino acid pathway of mTORC1 activation. The most potent amino acid activator of mTORC1 recruitment is leucine26, but mTORC1 activation is also partially dependent on tryptophan and certain additional amino acids27. Measurement of intracellular amino acid concentrations using ninhydrin reaction-based colorimetric detection analysis showed that intracellular concentrations of most amino acids, including leucine, were not notably diminished, although this technique does not detect tryptophan in our system (data not shown). IFN-γ is a strong inducer of indoleamine 2,3-dioxygenase (IDO), which catalyzes tryptophan degradation and has been shown to deplete extracellular tryptophan and thus suppress lymphocyte proliferation28, 29. We reasoned that IFN-γ might induce sufficient IDO expression in human macrophages to deplete intracellular tryptophan and thereby suppress mTORC1 activity. We observed that IFN-γ strongly induces IDO expression with sustained kinetics in our system (Fig. 4a). Direct measurement of intracellular tryptophan and its catabolites by HPLC followed by mass spectrometry revealed striking depletion of tryptophan (70% decrease after IFN-γ priming) with parallel accumulation of its catabolites in the IDO-mediated degradation pathway (Fig. 4b and Supplementary Fig. 3e-f). To test whether this IDO-mediated depletion of intracellular tryptophan contributes to reduced mTORC1 activity we treated cells with IDO inhibitor 1-D-MT and performing immunofluorescence microscopy (Fig. 4c). 1-D-MT restored the colocalization of mTOR and LAMP1 in IFN-γ-treated macrophages (Fig. 4c, lower panels; p = 0.0008). Furthermore, addition of exogenous tryptophan to IFN-γ-treated cells also partially recovered the lysosomal distribution of mTOR (Fig. 4c), supporting that IDO-mediated depletion of tryptophan was responsible for IFN-γ-induced mTOR subcellular redistribution, although it is possible that tryptophan metabolites could also contribute to modulation of mTORC1 activity. To further establish the role of IDO in regulating mTORC1 activity, we used mTORC1-dependent HES1 protein expression as a downstream readout of mTORC1 signaling. Consistent with its reversal of IFN-γ-mediated suppression of mTORC1 lysosomal localization (Fig. 4c), 1-D-MT reversed IFN-γ-mediated suppression of HES1 protein expression (Fig. 4d lanes 5-8, p = 0.0003). Consistent with this and with restoration of lysosomal localization of mTOR (Fig. 4c), supplementation of tryptophan partially restored HES1 expression in IFN-γ-treated macrophages in a dose-dependent manner (Fig. 4e). Taken together, these data implicate depletion of intracellular tryptophan by IDO in IFN-γ-mediated suppression of mTORC1 localization to lysosomes and thus suppression of mTORC1 activity.
Figure 4. IDO-mediated tryptophan depletion suppresses mTOR lysosomal localization and HES1 protein expression.
(a) qPCR analysis of IDO1 mRNA in human primary macrophages treated with IFN-γ (100 U/ml) for 0-24 h (error bars, s.d.). Data are shown as means + SD of triplicate determinants and are normalized relative to GAPDH mRNA. (b) HPLC-MS measurement of intracellular L-tryptophan concentration in control or IFN-γ-primed human primary macrophages treated with or without Pam3CSK4 (10 ng/ml) for 4h (error bars, s.d.). Data are shown as means + SD of triplicate determinants. (c) Upper panels: immunofluorescence images of LAMP1 (red) and mTOR (green) co-staining in control macrophages (row 1), IFN-γ-primed macrophages (row 2), IFN-γ-primed macrophages pretreated for 30min with IDO inhibitor 1-D-MT (200 μM) (row 3), and IFN-γ-primed macrophages supplemented with tryptophan (Trp) (800 μM) (row 4). All cells were stimulated with Pam3CSK4 (10 ng/ml) for 4 h; nuclei were counter-stained with DAPI (blue). Lower panel: quantitation of co-localization between LAMP1 and mTOR (error bars, s.e.m.). Data are presented as mean + SEM of the percentage of co-localized cells from 800 cells counted in two independent experiments; overall p = 0.0008 by one way ANOVA followed by Bonferroni's multiple comparison post-test; *p<0.05, ** p<0.0001. (d, e) Immunoblot analysis of HES1 in control or IFN-γ-primed macrophages treated for 30min with 1-MT (200 μM) (d) or the indicated concentrations of tryptophan (Trp) (e), and then stimulated with Pam3CSK4 (10 ng/ml) for 4h; p38α serves as loading control. Data are representative of at least three (a, d, e) or two (b, c) independent experiments.
IFN-γ suppresses mTORC1 activation by extracellular cues
Inhibition of lysosomal localization of mTOR by IFN-γ was not complete (Fig. 3e) and was only partially reversed by 1-D-MT or Trp (Fig. 4), suggesting IFN-γ also inhibits another pathway required for mTORC1 activity. Thus we considered the possibility that IFN-γ also suppresses PI3K-Akt-TSC signaling (Supplementary Fig. 3d). PI3K-Akt-TSC signaling is a major regulator of mTOR activity in response to extracellular factors, such as growth factors, and is also activated by inflammatory stimuli such as TLR ligands30. We found that TLR2-induced Akt phosphorylation was attenuated by IFN-γ (Fig. 5a). The PI3K inhibitor LY294002 abrogated Akt phosphorylation, confirming this inducible phosphorylation was due to PI3K activation (Fig. 5a). To corroborate suppression of Akt signaling by IFN-γ, we measured downstream induction of β-catenin, which is dependent on Akt-GSK3 signaling. The accumulation of β-catenin induced by TLR2 signaling was effectively inhibited by IFN-γ, further supporting that IFN-γ inhibits Akt signaling (Fig. 5b). Akt activates mTORC1 by phosphorylating and thereby deactivating the negative regulator TSC2. Consistent with suppression of Akt signaling, IFN-γ also suppressed TSC2 phosphorylation (Fig. 5c). These results indicate that IFN-γ attenuates TLR2-activated signals that lead to Rheb-mediated activation of mTORC1 (Supplementary Fig. 3d). We also tested whether IFN-γ could inhibit signaling by growth factors that maintain basal mTORC1 activity in macrophages by stimulating Akt activity. First, we confirmed that serum components and M-CSF (macrophage colony stimulating factor) were key factors for maintaining basal mTORC1 activity in human macrophages (Supplementary Fig. 4a, b). M-CSF promotes Akt signaling; although basal Akt phosphorylation was difficult to detect secondary to limited sensitivity of immunoblotting, we found that IFN-γ effectively blocked M-CSF-induced Akt phosphorylation in macrophages that had been serum- and M-CSF-starved for 4 hours (Fig. 5d). These results indicate that IFN-γ suppresses activation of mTORC1 by extracellular factors by suppressing the activation of Akt.
Figure 5. IFN-γ inhibits PI3K-Akt-TSC1/2 signaling and M-CSFR expression.
(a-c) Immunoblot analysis of whole cell lysates from control or IFN-γ-primed macrophages that were stimulated with Pam3CSK4 (10 ng/ml) for the indicated times. (d) Immunoblot analysis of phosphorylated (p-)Akt in control or IFN-γ-primed macrophages that were serum- and M-CSF-starved for 4 h, followed by pretreatment with vehicle control DMSO or LY294002 (10 μM) for 30 min, and then stimulated with M-CSF (100 ng/ml) for 0-30 min; Akt serves as loading control. (e) qPCR analysis (left panel) of CSF1R mRNA in control or IFN-γ-primed macrophages (error bars, s.d.). Data are shown as means + SD of triplicate determinants and are normalized relative to GAPDH mRNA. Immunoblot analysis (right panel) of M-CSFR in control or IFN-γ-primed macrophages; p38α serves as loading control. (f) qPCR analysis of MYC mRNA in monocytes cultured with M-CSF (20 ng/ml) with or without IFN-γ for indicated times (error bars, s.d.). Data are shown as means + SD of triplicate determinants and are normalized relative to GAPDH mRNA. (g) Immunoblot analysis of M-CSFR in human primary monocytes treated with vehicle control DMSO or Myc inhibitor 10058-F4 (60 μM) and then cultured with M-CSF (20 ng/ml) for indicated time points; p38α serves as loading control. (h) Immunoblot analysis of p-4E-BP1 in human primary monocytes treated with vehicle control DMSO or Myc inhibitor 10058-F4 (60 μM) for 30 min. (i) Immunoblot analysis of c-Myc in nuclear extracts of control or IFN-γ-primed macrophages that were stimulated with Pam3CSK4 (10 ng/ml) for 0-4h; TBP serves as loading control. Data are representative of at least three independent experiments (a-i).
We investigated mechanisms by which IFN-γ suppresses growth factor-mediated Akt signaling in macrophages. We found that IFN-γ suppressed the expression of M-CSFR (CSF1R) mRNA and protein (Fig. 5e), which corroborated previous results31. Decreased M-CSFR expression could explain diminished Akt activation by M-CSF and had minimal effect on cell survival under IFN-γ-primed conditions (Supplementary Fig. 4c). IFN-γ-mediated suppression of TLR2-induced Akt activation was mediated by an okadaic acid-sensitive phosphatase (Supplementary Fig. 2g) but was not associated with changes in phosphatases SHIP or PTEN that regulate this pathway (data not shown); future work will address how IFN-γ suppresses TLR2-induced Akt phosphorylation. To investigate mechanisms by which IFN-γ suppresses M-CSFR expression, we tested the role of Myc, which has been previously shown to promote mTORC1 activity in lymphocytes, although underlying mechanisms have not been fully clarified. We found that Myc and M-CSFR expression were induced during culture of cells with M-CSF (Fig. 5f and 5g), and inhibition of Myc with the compound 10058-F4 suppressed induction of M-CSFR expression (Fig. 5g), suggesting that Myc is required for M-CSFR expression and mTORC1 activity in macrophages. Consistent with this notion, inhibition of Myc effectively suppressed 4E-BP phosphorylation (Fig. 5h), and also the downstream induction of mTORC1-dependent HES1 protein expression (Supplementary Fig. 4d); similar results were obtained when Myc expression was knocked down using siRNA (Supplementary Fig. 4e). IFN-γ suppressed Myc expression (Fig. 5f and 5i). These results suggest that IFN-γ suppresses expression of the transcription factor Myc to downregulate M-CSFR expression and downstream activity of mTORC1, and identifies regulation of Myc as a link between IFN-γ and cell metabolism. Collectively, the results indicate that IFN-γ suppresses Akt signaling and thereby attenuates signals from extracellular factors that are required for mTORC1 activity (Supplementary Fig. 3d). This attenuation of signaling, working cooperatively with decreased recruitment of mTORC1 to lysosomal membranes (Fig. 3e), can explain strong inhibition mTORC1 activity by IFN-γ.
Genome-wide analysis of translational regulation by IFN-γ
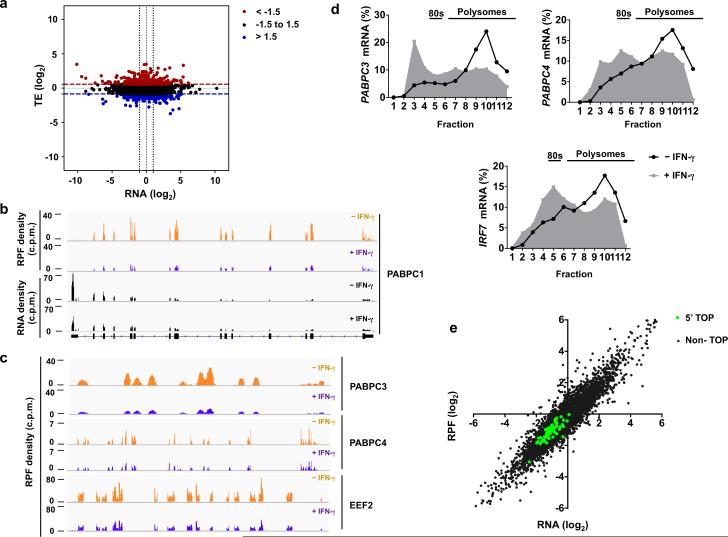
The MNK-eIF4E and 4EBP-eIF4E pathways that were suppressed by IFN-γ selectively regulate translation7, 10, 11, 12, and translational regulation is important for inflammatory responses7. To address the broad functional role of IFN-γ-regulated translation in inflammatory responses in primary human macrophages, we used a genome-wide approach combining high throughput RNA sequencing with ribosome profiling. Ribosome profiling provides a snapshot of the translatome by quantifying ribosome-protected RNA fragments (RPF), the frequency of which on each transcript reflects the translation rate of the corresponding mRNA32. We quantified and compared the abundance of mRNA (counts per million reads (cpm)) and actively translated mRNA (RPF (cpm)) in TLR2-activated macrophages that had been treated with or without IFN-γ in two biological replicates (Supplementary Fig. 5a-b and Online Methods). For each of 9290 genes that passed stringency cut-offs (Online Methods), we calculated the translational efficiency (TE) by dividing the change in RPF reads by the change in cpm; this identifies changes that are solely attributable to changes in translation rate. As expected, IFN-γ treatment altered the abundance of multiple mRNA transcripts (Fig. 6a, x axis, 2976 mRNAs changed by > 2-fold). IFN-γ also induced significant changes in translational efficiency in almost one thousand genes (Fig. 6a, y axis (z-score cut-off ±1.5) and Supplementary Fig. 5c), of which 396 genes were strongly affected (greater than 2-fold changes in TE). In contrast to consistent translational repression of most mRNAs upon complete inhibition of mTOR23, the effects of IFN-γ on translation were bidirectional, and IFN-γ increased and decreased TE of comparable numbers of genes (Fig. 6a and Supplementary Fig. 5c). Thus, ribosome profiling revealed an unappreciated role of IFN-γ in modulating translation of mRNA at a genome-wide level.
Figure 6. Genome-wide ribosome profiling analysis of IFN-γ-mediated translational regulation in macrophages.
(a) Scatter plot comparing the changes in mRNA abundance (x axis) and translational efficiency (TE) (y axis) in response to IFN-γ; RNA fold change (x axis) = log2( RNAIFN-γ/RNAcontrol); TE fold change (y axis) = log2(TEIFN-γ /TEcontrol); mRNAs with suppressed TE (z-score <−1.5) or induced TE (z-score>1.5) are highlighted in blue or red, respectively. (b) Ribosome-protected fragment (RPF) and RNA-seq read density profiles for PABPC1 in control (tracks 1 and 3) and IFN-γ-primed (tracks 2 and 4) macrophages; data are normalized to total mapped reads in each library. (c) Ribosome-protected fragments (RPF) read density profiles for PABPC3, PABPC4 and EEF2 in control (yellow) and IFN-γ-primed (purple) macrophages; data are normalized to total mapped reads in each library. (d) Polysome shift analysis of PABPC3, PABPC4 and IRF7 mRNA. (e) Scatter plot comparing the changes in mRNA abundance and ribosome footprint frequency; RNA (log2) = log2(RNAIFN-γ/RNAcontrol); RPF (log2) = log2(RPFIFN-γ/RPFcontrol); mRNAs with 5’ TOP elements are highlighted in green; Pearson correlation value (R) was calculated by GraphPrism. R2=0.65 for 65 established 5’ TOP genes; R2=0.86 for Non-TOP genes. Data (a, e) was generated from a merged dataset of two biological replicates, or is representative of two (b-c) or three (d) independent experiments.
We wished to confirm our genome wide findings by analyzing individual genes and to corroborate regulation by IFN-γ of transcripts that are known to be targeted at the translational level by mTORC1. We examined the ribosome occupancy pattern on the well-established mTOR target gene PABPC123, 33 (Fig. 6b, gene tracks of normalized RPF and RNA levels (c.p.m.)). Consistent with the depletion of PABPC1 mRNA from polysomes and decreased protein but not mRNA expression in IFN-γ-treated macrophages (Figure 1d, e), the ribosome footprint tag density across the message was suppressed (Fig. 6b, upper 2 tracks) while the RNAseq read profile remained unchanged (Figure 6b, lower 2 tracks). In addition, RPF read tag density was diminished at the mTORC1 target genes PABPC3, PABPC4 and EEF2 (Fig. 6c), although mRNA levels did not change as assessed by RNAseq and qPCR (data not shown). Polysome shift analysis confirmed translational repression of PABPC3 and PABPC4 by IFN-γ (Figure 6d), and also showed translational repression of IRF7 (Fig. 6d), an immunologically important gene previously shown to be dependent on 4E-BPs11. RPF read track density was suppressed in the translated portion of the mRNA for mTORC1 target HES1 (Supplementary Fig. 5d). Canonical mTORC1 target mRNAs possess a hallmark oligopyrimidine (TOP) motif immediately after the 5’cap and mainly encode factors that comprise protein synthesis machinery33. To gain insights into the overall influence of IFN-γ on mTORC1 targets, 65 established TOP mRNAs were analyzed. We analyzed genome-wide the relationship between IFN-γ-induced changes in mRNA and in RPF read density (which measures the summed effects of changes in mRNA and TE). For all genes, the correlation between changes in mRNA and RPF density was strong (Fig. 6e, R2 = 0.86), which is consistent with the results in Fig. 6a that mRNA levels are regulated over a broader dynamic range and a smaller subset of genes is regulated translationally. For 5’TOP mRNAs, both mRNA and RPF read density were suppressed, but the correlation between the two was weaker (Fig. 6e, R2 = 0.65), suggesting that IFN-γ regulates the translation of mTORC1 target mRNAs to a greater extent than non-targets. Taken together, ribosome profiling and polysome shift analysis demonstrated that IFN-γ inhibits the translation of some mTORC1-dependent mRNAs.
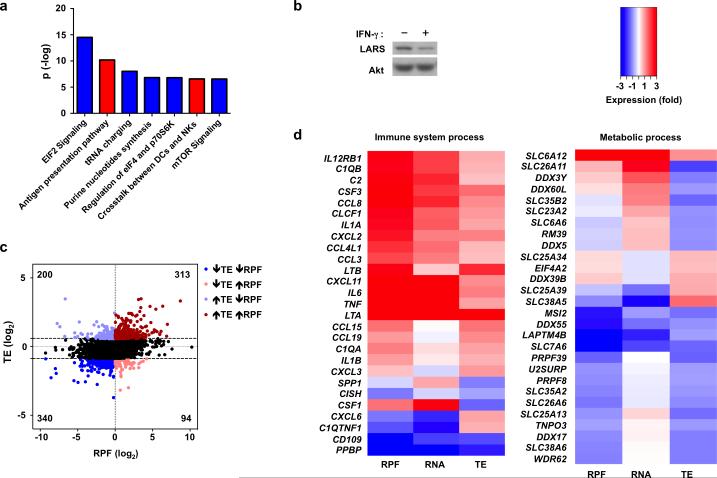
To gain additional insight into the functional role of IFN-γ-mediated translational regulation during inflammatory macrophage activation, we performed pathway and gene ontology (GO) analysis of genes that were regulated at the level of mRNA (c.p.m.), translation (TE), and RPF reads; changes in RPF reads reflect a combination of mRNA and translational regulation and are considered a better predictor of protein abundance32, 34 and thus IFN-γ function. Ingenuity pathway analysis (IPA) of ribosome profiling data (RPF reads) revealed highly statistically significant regulation of well-known IFN-γ-mediated pathways important in immune responses, such as antigen presentation (Fig. 7a, red bars and Table 1). IPA analysis also identified highly statistically significant negative regulation of mTOR signaling, eIF4 signaling and tRNA charging (Fig. 7a, blue bars and Table 1). The patterns of pathway enrichment were consistent among biological replicates and the combined dataset (Supplementary Fig. 6a). These results are in accord with a feed forward loop described in other systems whereby proteins important for translation are themselves regulated at the translational level33. Thus, translational dampening of eIF4 pathway components likely cooperates with signaling attenuation (Figs. 2, 3) to regulate eIF4 function in IFN-γ-treated macrophages. The suppression of tRNA charging was particularly striking as translation of 29 out of 36 detected aminoacyl tRNA synthetases was suppressed (Supplementary Fig. 6b). This included leucyl-tRNA-synthetase (LARS), recently described as a direct amino acid sensor that recruits mTORC1 to the lysosomal surface and mediates its leucine-dependent activation35. Suppression of LARS expression was confirmed using immunoblotting (Fig. 7b, 62% mean suppression in 3 independent experiments, p = 0.013). Decreased LARS expression would contribute to the decreased mTORC1 localization to lysosomal membranes shown in Fig. 4. Collectively, our ribosome profiling results reveal that the cellular translational machinery might represent an unrecognized target for negative regulation by IFN-γ.
Figure 7. Selective translational inhibition of mRNAs involved in metabolic processes and protein synthesis by IFN-γ.
(a) Ingenuity Pathway Analysis (IPA) of canonical pathways most enriched in sets of genes either up- (red) or down- (blue) regulated by IFN-γ at the level of ribosome footprint frequency (RPF); the y-axis indicates the –log10 (p-value) of each enriched pathway. (b) Immunoblot analysis of LARS in control or IFN-γ-primed macrophages; p38α serves as loading control. Data is representative of three experiments. (c) Scatter plot comparing the changes in ribosome footprint frequency and translational efficiency; RPF (log2) = log2(RPFIFN-γ/RPFcontrol); TE (log2) = log2(TEIFN-γ /TEcontrol); mRNAs with decreased TE (log2TE < −0.856) and decreased RPF (log2RPF < 0) are highlighted in dark blue; mRNAs with decreased TE (log2TE < −0.856) and increased RPF (log2RPF > 0) in light red; mRNAs with increased TE (log2TE > 0.578) and increased RPF (log2RPF > 0) in dark red; mRNAs with increased TE (log2TE > 0.578) and decreased RPF (log2RPF < 0) in light blue. (d) Heat map showing changes in RPF, RNA and TE of representative immune and metabolic process genes selected from gene sets identified by Gene Ontology (GO) analysis of translationally-regulated genes. GO analysis and entire enriched gene sets are shown in Supplementary Fig.7 and Supplementary Tables 1 and 2. Data (a, c, d) was generated from merged dataset of two biological replicates.
Table 1.
IPA canonical pathways regulated by IFN-γ
| Canonical pathways | Up-regulated | Down-regulated | Not detected | Total |
|---|---|---|---|---|
| EIF2 Signaling | 9.9% | 79.1% | 11% | 172 |
| Antigen presentation pathway | 70.3% | 13.5% | 16.2% | 37 |
| tRNA charging | 2.6% | 92.1% | 5.3% | 38 |
| Purine nucleotides synthesis | 0% | 90.9% | 9.1% | 11 |
| Regulation of eIF4 and p70S6K | 12.7% | 71.1% | 16.2% | 142 |
| Crosstalk between DCs and NKs | 41.6% | 12.4% | 46.1% | 89 |
| mTOR Signaling | 13.7% | 63.2% | 23.1% | 182 |
Ingenuity Pathway Analysis (IPA) of canonical pathways most enriched at the level of ribosome footprint frequency (RPF); ranked by P-value (indicated in Fig. 7a). The percentages of genes detected in each category (up- or down-regulated by IFN-γ, or not detected) are listed. Data was generated from merged dataset of two biological replicates.
We next addressed the question of whether IFN-γ-induced changes in TE generally reinforce or oppose and modulate the overall IFN-γ response, as measured in an integrated manner by RPF reads. We analyzed the relationship between changes in TE and changes in RPF reads (Fig. 7c). Overall, the correlation between these two values was low (Pearson correlation coefficient R = 0.27, R2 = 0.072), suggesting that at the genome-wide level regulation of translation modulates and fine tunes the transcriptional response to IFN-γ. Consistent with this notion, of the genes with decreased RPF reads and significant changes in TE, 37% had a discordant increase in TE (Fig. 7c, left two quadrants). In contrast, of genes with increased RPF reads, the majority (77%) had a concordant increase in TE (Fig. 7c, right two quadrants, compare red and pink dots). Thus, IFN-γ-mediated increases in TE tend to reinforce and augment upregulation of gene expression. To analyze the function of genes regulated at the level of TE, we selected highly regulated genes (z-score=±1.5 in pooled data set) and stratified them into functional subcategories based on Gene Ontology (Supplementary Figure 7a, 7b). This analysis revealed that genes related to metabolic processes were the most enriched category among translation-regulated genes; immune genes were enriched as well (Supplementary Fig. 7; gene lists are shown in Supplementary Tables 1 and 2 and representative genes are shown in Fig. 7d). Strikingly, inflammatory cytokines and chemokines, such as IL-6, TNF, LTA, LTB, CXCL2 and CXCL3 were included in the immune gene group with increased TE (Fig. 7d, left panel). This suggests that translational regulation contributes to the previously described1 augmentation of TLR-induced inflammatory responses by IFN-γ. Pathway analysis showed that metabolic process genes whose translation was increased were significantly associated with oxidative phosphorylation and mitochondrial pathways, genes whose translation was decreased were associated with stress pathways, and both sets of genes were associated with growth factor pathways (Supplementary Fig. 8a). Transporters of amino acids and other metabolites as well as RNA modification factors were among the genes most strongly downregulated by IFN-γ at the level of TE (Fig. 7d); additional investigation is required to discern the functional effects of translational regulation of these genes. Thus, IFN-γ regulates macrophage metabolism at the translational level. In summary, genome-wide ribosome profiling analysis indicates that IFN-γ changes the translational landscape of human macrophages, likely contributing to the inflammatory response and metabolic reprogramming.
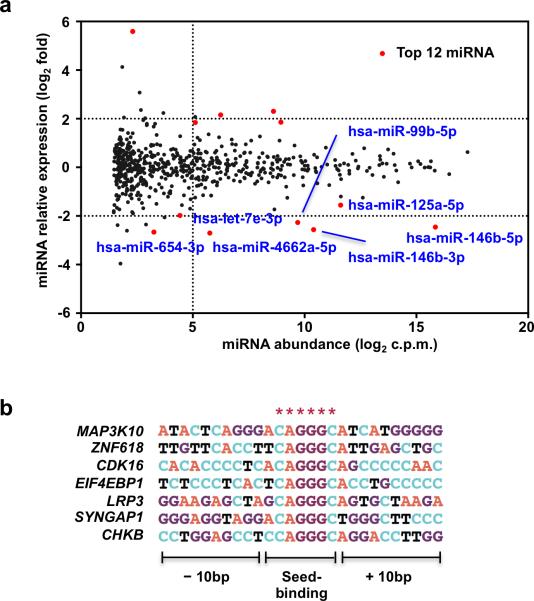
Regulation of MNKs and mTORC1 can explain downregulation of translation by IFN-γ; we wished to investigate distinct mechanisms that could underlie increased translational efficiency of some mRNAs, such as IL-6 and TNF. IFN-γ modestly and transiently decreased phosphorylation of translation factor eIF2α (Supplementary Fig. 8b), which increases translation and could contribute to global maintenance of translation in IFN-γ-treated cells and possibly to enhanced translation of specific transcripts. However, as regulation of eIF2α was modest, we tested the non-mutually exclusive hypothesis that IFN-γ could increase translational efficiency of select mRNAs by downregulating expression of microRNAs that suppress translation of these transcripts. We performed global profiling of miRNA expression in human primary macrophages using microRNAseq. 20 miRNAs were significantly induced by TLR2 stimulation in two independent experiments, including known regulators of inflammatory responses miR-155, miR-146a, miR-9 and miR-132 (Supplementary Table 3). IFN-γ suppressed expression of 54 miRNAs by at least 2-fold in TLR2-stimulated macrophages (Fig. 8a, Supplementary Table 3 and data not shown). IFN-γ suppressed expression of members of the let-7 miRNA family (Fig. 8a and Supplementary Table 4), which suppress inflammation by directly targeting IL-6 mRNA36. This observation supports the hypothesis that IFN-γ augments translation of inflammatory cytokines by suppressing miRNA expression. To analyze whether downregulation of additional miRNAs could contribute to increased TE of select transcripts, we integrated miRNA and ribosome profiling data to identify potential miRNA targets amongst all mRNAs with increased TE (Fig. 6a, z-score cut-off ±1.5) based on complementarity of 3’UTR sequences to miRNA seed sequences. We focused on miRNAs most significantly (p < 1.3E−03, FDR<0.0653) downregulated by IFN-γ in TLR2-stimulated macrophages (Fig. 8a and Table 2). Of these miRNAs, a majority had potential target mRNAs with increased TE; miR-146b-3p, a TLR2-induced miRNA that was suppressed 6 fold by IFN-γ, had several potential target mRNAs whose TE was increased by IFN-γ (Fig. 8b). Taken together, the results suggest that suppression of miRNA expression by IFN-γ may contribute to increased translation of certain mRNAs. Overall the results suggest that IFN-γ regulates human macrophage metabolism and mRNA translation in an integrated manner to specify an activated phenotype (Supplementary Fig. 8c).
Figure 8. IFN-γ downregulates miRNAs that target translationally upregulated genes.
(a) Scatter plot showing normalized miRNA abundance of all conditions analyzed (x axis) and changes in miRNA expression induced by IFN-γ (y axis). Statistical analysis was performed using edgeR; the most significantly regulated genes are highlighted in red. (b) Potential target mRNAs of miR-146b-3p that are translationally upregulated by IFN-γ. * indicates 3’UTR sequence complementary to seed sequence of miR-146b-3p. Data shown was generated from merged dataset of two biological replicates.
Table 2.
miRNAs suppressed by IFN-γ
| miRNA | Expression (log2 fold) | p-value | FDR |
|---|---|---|---|
| hsa-miR-146b-3p | –2.56 | 8.35E–07 | 0.0005 |
| hsa-miR-4662a-5p | –2.70 | 1.71E–06 | 0.0005 |
| hsa-miR-99b-5p | –2.27 | 3.89E–06 | 0.0008 |
| hsa-miR-146b-5p | –2.46 | 3.76E–05 | 0.0050 |
| hsa-miR-654-3p | –2.66 | 8.75E-04 | 0.0559 |
| hsa-miR-125a-5p | –1.56 | 8.96E-04 | 0.0559 |
| hsa-let-7e-3p | –1.98 | 1.30E-03 | 0.0653 |
miRNAs most significantly suppressed by IFN-γ in TLR-stimulated macrophages (p < 1.3E−03, FDR<0.0653). Expression (log2 fold) was calculated as relative miRNA expression by comparing IFN-γ-treated and control condition; Expression (log2 fold)=log2(IFN-γ/control) Statistical analysis was performed using edgeR.
DISCUSSION
The profound activating effects of IFN-γ on macrophages have been predominantly attributed to transcriptional activation of ISGs, and its augmentation of inflammatory responses to signaling crosstalk and chromatin remodeling1, 6. In this study we found that IFN-γ alters macrophage metabolism by suppressing the central metabolic regulator mTORC1 and selectively alters mRNA translation to promote inflammation, further reprogram metabolic pathways, and modulate cellular translational machinery. Translational reprogramming was achieved by targeting mTORC1 and MNK pathways that are upstream of eIF4E, a factor that selectively regulates translation7, 10, 11. Conversely, genome-wide analysis showed that translational regulation modulated expression of genes important in metabolism. Thus, these newly described IFN-γ functions are tightly integrated to specify an activated macrophage phenotype. Inhibition of mTORC1 by pharmacological or genetic means has been previously shown to promote inflammation by suppressing STAT3 activity19 and to augment microbial killing and antigen presentation, in part by increasing autophagy30, 37, 38. Thus, our findings provide insights into mechanisms by which physiological regulation of mTORC1 and translation by IFN-γ can contribute to hallmark IFN-γ functions such as boosting inflammatory responses and anti-microbial mechanisms. These findings also advance our understanding of reprogramming of cell states by IFN-γ to include metabolism and translation.
The importance of cellular metabolism is well established in adaptive immunity and is emerging in innate immunity39, 40, 41. Previous work on innate immune cells has focused TLR responses, which trigger a switch to aerobic glycolysis 42. Full inflammatory activation of macrophages by TLR ligands requires IFN-γ1, 43, and our signaling results and genome-wide analysis of translational regulation suggest that extensive metabolic reprogramming contributes to IFN-γ -mediated polarization of macrophages, which will impact on how macrophages respond to subsequent environmental challenges.
IFN-γ suppressed mTORC1 in a physiological cell culture model using primary human macrophages that models the well known “priming” effects of IFN-γ that enhance responses to inflammatory factors and increase microbial killing1. Physiological regulation of mTORC1 by IFN-γ in human macrophages was partial, but comparable to that achieved by the allosteric mTORC1 inhibitor rapamycin. The level of mTORC1 inhibition achieved by rapamycin, and thus IFN-γ, has potent effects on immune responses and has been shown to augment innate responses in vitro and in vivo in mice and humans19, 20, 44, in part by augmenting inflammatory cytokine production and M1 polarization. Furthermore, partial mTORC1 suppression has been shown to enhance host defense against L. pneumophila38 and pathogenic Shigella, in the latter case by augmenting autophagic clearance mechanisms37. This work supports the biological importance of the newly described regulation of mTORC1 by IFN-γ and helps link our findings with canonical IFN-γ functions. Overall, our results are in accord with the literature suggesting a suppressive role for mTORC1 in innate immunity19, 20, 37, 38. However, as a negative feedback loop induced by constitutive mTORC1 activity in TSC1 knock out mice can augment cytokine production45, and mTOR has been linked to trained innate immunity46, the function of mTORC1 in innate immunity is context dependent and likely determined by the absence or presence of an IFN-γ response and concomitant ISG expression.
Investigation of translational regulation by interferons has predominantly focused on type I IFNs, which globally suppress translation by suppressing eIF2α and by direct effects of ISGs13, 14, 15. Regulation of translation by IFN-γ has been primarily investigated in cell line models of cellular transformation16. Interestingly, in transformed cells IFN-γ, and also type I IFNs, directly but transiently activate Akt-mTORC1 and MNK-eIF4E signaling, resulting in increased ISG mRNA translation and thus augmentation of the early phase of the IFN response16. In contrast, we have not observed the early phase of Akt or MNK activation by IFN-γ in primary human macrophages; instead, IFN-γ suppresses Akt-mTORC1 and MNK-eIF4E at later time points by indirect mechanisms such as inhibition of M-CSFR expression. Thus, the effects of IFN-γ on Akt-mTORC1 and MNK-eIF4E signaling are dependent on cell context and increase with duration of IFN-γ exposure. A striking contrast between translational effects of IFN-γ and type I IFNs is the selective regulation of translation of only a subset of expressed mRNAs by IFN-γ, which may be explained by selective regulation of translation by eIF4E7. In contrast, suppression of eIF2α by type I IFNs has more global effects14. A modest increase in eIF2α activity by IFN-γ may contribute to enhanced translation of specific transcripts, as could downregulation of miRNAs that suppress translation. Overall our work establishes mechanisms by which IFN-γ downregulates translation and provides insights into mechanisms by which IFN-γ could augment translation of select mRNAs.
This study describes the first, to our knowledge, comprehensive genome-wide translational profiling analysis in primary immune cells and in terminally differentiated nonproliferating cells. Analysis of genome-wide data revealed several functions for IFN-γ-mediated translational regulation in macrophages. First, IFN-γ-induced changes in translational efficiency (TE) were superimposed on changes in mRNA abundance to selectively modulate expression of different gene sets. Second, regulation of translational efficiency potentiated inflammatory activation by augmenting production of inflammatory proteins (TNF, IL-6, lymphotoxins) while decreasing production of anti-inflammatory feedback inhibitors (IL-10, HES1). Third, by suppressing expression of components of the translational machinery, IFN-γ could potentiate previously proposed ‘translational skewing’ towards highly expressed transcripts that favors host defense38, although our genome-wide analysis showing selective translational regulation suggests the translational skewing model38, 47 may need to be refined to take into account properties of specific transcripts. Fourth, translational regulation by IFN-γ broadly impacted on proteins in metabolic pathways. Fifth, IFN-γ-mediated translational downregulation of multiple tRNA synthases by may have important consequences for macrophage activation as these proteins have various noncanonical functions independent of their aminoacyl transferase activity48.
In summary, the present study reveals that IFN-γ regulates mTORC1 and mRNA translation in an integrated manner in human macrophages. This regulation and associated changes in intracellular metabolism are linked with key IFN-γ functions such as potentiation of inflammatory activation and autophagy, which is related to microbial killing. These findings extend our understanding of the role of metabolic regulation in innate immune cell activation, identify new functions for translational regulation by IFN-γ, and suggest approaches to targeting metabolic pathways and translation factors to modulate macrophage activation and function.
METHODS
Cell Culture
Primary human CD14+ monocytes were isolated from buffy coats purchased from the New York Blood Center using anti-CD14 magnetic beads (MiltenyiBiotec) as previously described3, using a protocol approved by the Hospital for Special Surgery Institutional Review Board. Monocytes were cultured in RPMI 1640 (Invitrogen) medium supplemented with 10% (vol/vol) FBS (Hyclone) and macrophage colony stimulating factor (M-CSF) (20 ng/ml). IFN-γ (100 U/ml) was added at the initiation of cultures and maintained for at least 24 h prior to subsequent stimulations or harvesting cells.
Reagents and Antibodies
Human IFN-γ and S7 micrococcal nuclease were purchased from Roche; human M-CSF was from Peprotech; Pam3CSK4 was from EMC Microcollections; UltraPure E.Coli LPS was from Invivogen. MG132, CGP57380, Rapamycin and PP242 were purchased from Calbiochem; Cycloheximide, Bafilomycin A1, 1-Methyl-D-tryptophan (1-D-MT), L-Tryptophan and 10058-F4 were from Sigma; Torin 1 was from R&D Systems; LY 294002 was from EMD Millipore. The following antibodies were purchased from Cell Signaling: mTOR (#2983), p-TSC2(Thr1462) (#3617), TSC2 (#4308), p-Akt (Ser473) (#4060), p-p70S6K(Thr421/Ser424) (#9204), 4E-BP1 (#9644), p-4E-BP1(Thr37/46) (#2855), LC3A (#4599), LC3B (#3868), IκB-α (#9242), Mnk1(#2195), p-Mnk1(Thr197/202) (#2111), eIF4E (#9742), p-eIF4E(Ser209) (#9741), p-p38(Thr180/Tyr182) (#9215), p-ERK1/2 (Thr202/Tyr204) (#9101), ERK (#9102), β-catenin (#9562), M-CSFR (#3152). Additionally, p38α (sc-535), HES1 (sc-25392), TBP (sc-204), LAMP1 (sc-20011) and Myc (sc-764) were from Santa Cruz Biotech. PABPC1 (ab6125), β-tubulin (ab11307) and Leucyl tRNA synthetase (ab31534) antibodies were from Abcam. mirVana miRNA isolation kit was purchased from Ambion/Life Technologies. BD™ Cytometric Bead Array (CBA) human inflammatory cytokine kit was purchased from BD Biosciences.
RNA extraction and quantitative PCR
Total RNA was extracted from cells using RNeasy Mini kit (Qiagen), and 500 ng of total RNA was reverse transcribed using the RevertAid First Strand cDNA Synthesis kit (Fermentas). Real-time PCR was performed in triplicate with Fast SYBR Green Master Mix and 7500 Fast Real-time PCR system (Applied Biosystems).
Oligonucleotide primers for human transcripts were as follows:
(Forward primer, reverse primer, listed 5’ > 3’)
GAPDH: ATCAAGAAGGTGGTGAAGCA, GTCGCTGTTGAAGTCAGAGGA.
HES1: CAGGCTGGAGAGGCGGCTAAGGTG, GGAGGTGCCGCTGTTGCTGGTGTA.
ACTB: GGACTTCGAGCAAGAGATGG, AGCACTGTGTTGGCGTACAG.
PABPC1: TTGGAGCTAGGGCAAAAGAA, TCCACAGCTTTCTGTGCATC.
NFKBIA: ACACCTTGCCTGTGAGCAGGGC, GGCAGTCCGGCCATTACAGGG.
IDO1: TTAGAGTCAAATCCCTCAGTCC, TGCAGATGGTAGCTCCTC.
LRS: GAGAGCAAATGGACA, CCAGTTGGCAACCATTTCTT.
CSF1R: GGTACTGCTGTAATGAGCCAA, AGTTTGTGCTTCCTGCTTGGT.
MYC: CGACGCGGGGAGGCTATTCTGC, CGTCGCGGGAGGCTGCTGGTTTT
PABPC3: GACACACGAAGCAGCTGAAA, TCCTTAAGGCGCTCATCATC.
PABPC4: AGCCAAGGAATTCACCAATG, CCTTATTGGCATCCTCGTGT.
IRF7: GAGCCGTACCTGTCACCCT, CTGGGGAATGCTACCTGCTG.
HEY1: TTCTCTTTCGGCTCCTTCCACTTA, TTTCCCCTCCCTCATTCTACATCA.
MKNK1: CGTGTAGCTGGAAGACACCA, TTCCTCCTCCTGTCACCATC.
MKNK2: CCGCTTCTACCTGGTGTTTG, AGAGGATGTTTTCCGGCTTT.
IRF8: TGCGCTCCAAACTCATTCTCGTG, GTCTGGCGGCGGCTCCTC.
DUSP1: CTGCCTTGATCAACGTCTCA, ACCCTTCCTCCAGCATTCTT
DUSP2: TGTGGAGATCTTGCCCTACC, CTCCACCATCTGGTTGTCCT
DUSP4: GGGGAAATGGACCAAGTTTT, ATTCCCAATCTCCAGCCTCT
DUSP8: CTGCTCACACAGGGAGTTCA, GGCATGCCACCTAGAGAGAG
DUSP16: GAAGGAGGTGGGAAAAGAGG, CCTTCGCTTCATAAGCTTGG
Preparation of nuclear extracts
Pefabloc (Roche, 0.5 mg/mL) was added to cells 15 min prior to harvest. Cytoplasmic and nuclear extracts were obtained by incubating cells in Buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, supplemented with protease inhibitor cocktail (Roche, 1X)) for 15 min on ice. The plasma membrane was then solubilized by incubation with NP-40 (0.2%) for 2 min. The nuclear pellet was obtained by centrifugation at 10,000×g for 30 s; supernatants corresponded to cytoplasmic lysates. Nuclear lysates were obtained by directly lysing nuclear pellets in 1× SDS-PAGE loading buffer.
Immunoblotting
All samples for immunoblotting were denatured at 95 °C for at least 10 min. Denatured cell lysates were fractionated on 7.5% or 10% or 12% polyacrylamide gels using SDS-PAGE and transferred to polyvinylidene difluoride membranes for probing. Western Lightning® Plus-ECL, Enhanced Chemiluminescence Substrate (PerkinElmer) was used for detection. Images shown in figure panels were derived from one gel. In Figure 5h intervening lanes were spliced out and a white vertical line indicates lanes that were noncontiguous on the gel.
RNA interference
Immediately after isolation, primary human monocytes were nucleofected with On-Target plus SMARTpool short interfering RNAs (siRNA) purchased from Dharmacon Inc. (Lafayette, Colorado, USA) specific for MNK1, MNK2 or MYC. Non-targeting siRNA#5 was used as control. Human Monocyte Nucleofector buffer (Lonza Cologne, Cologne, Germany) and the AMAXA Nucleofector System program Y001 for human monocytes were used according to the manufacturer's instructions.
Immunofluorescence staining
Primary human macrophages were cultured on poly-D lysine-coated coverslips (BD Biosciences Discovery Labware) for 24 h in the presence or absence of IFN-γ (100 U/ml). Cells were fixed with 2% formaldehyde for 15 min at room temperature and were permeabilized with 0.1% Tween-20 (PBST) at room temperature for 10min. 5% goat serum (Santa Cruz (sc-2043)) was used for blocking at 37 °C for 1 h, and then cells were stained with rabbit antibody to mTOR (Cell Signaling 2983 (7C10)), and mouse antibody to LAMP-1 (Santa Cruz H4A3 sc-20011) simultaneously in the 4°C cold room for at least 12 h. Alexa Fluor 488 goat anti-rabbit (Invitrogen) and Alexa Fluor 594 goat anti-mouse (Invitrogen) secondary antibodies were then used to detect mTOR and LAMP1 primary antibodies, respectively. Coverslips were mounted with Vectashield mounting medium (Vector Laboratories) and images were obtained using a Nikon Eclipse Microscope.
Cytometric Bead Array (CBA) Assay
Culture supernatants of primary human macrophages as indicated in the figure legends were collected and processed immediately or snap frozen on dry ice for storage. Supernatant concentrations of secreted cytokines TNF, IL-6 and IL-10 were measured using BD Cytometric Bead Array (CBA) Human Inflammatory Cytokines Kit according to the manufacturer's instructions. Data were acquired on a BD FACS Canto flow cytometer; results were analyzed using FCAP Array software.
HPLC-MS analysis
For intracellular tryptophan and related metabolites measurement, samples were processed and analyzed by the Donald B. and Catherine C. Marron Cancer Metabolism Center at the Memorial Sloan Kettering Cancer Center. We harvested 10 million cells for each condition and quenched metabolism using 1ml of 80% methanol supplemented with 2μM D5-2HG. Protein was precipitated and removed by centrifugation. The samples were analyzed by Agilent 6230 time-of-flight (TOF) LC-MS instrument.
Ribosome Profiling
To obtain enough material to generate high quality ribosome footprints, human primary macrophage lysates from different blood donors were pooled together (4 donors for replicate #1, 6 donors for replicate #2, Supplementary Fig 5a). Control or IFN-γ-treated human primary macrophages were stimulated for 4 h with Pam3CSK4 (10ng/ml), followed by cycloheximide (100 μg/ml) treatment for 7 min. We used the same pools of lysates for both RPFs (ribosome protected fragments) and RNA libraries construction. We followed a previously published protocol for ribosome profiling49 with the following modifications: S7 micrococcal nuclease (120 U/ml) was used to generate mRNA-associated monosomes, and the monosomes were separated on sucrose density gradient followed by a sucrose cushion purification step to minimize the contamination of other protein-RNA complexes.
A modified reverse transcription primer (/phos/NNNNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGT/idSp/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTTTTTTTTTTTTTTTTTTTVN) was designed to generate cDNA libraries. The first seven degenerate nucleotides were designed to monitor the clonal amplification bias from PCR. Both RPF and RNAseq libraries were sequenced on HiSeq 2000 platform.
Sequence alignment and annotation
The human genome sequence hg19 (Home_Sapiens.GRCh37.75.gtf) was downloaded from ensemble website (http//feb2014.archive.ensembl.org/downloads.html). A gtf file that only contains protein-coding genes was extracted based on the feature defined on the original gtf file. Before alignment, the first seven degenerate nucleotides were trimmed from 5’end; the polyA adapter at 3’end was removed by Clip; low quality reads were filtered out (mean quality score<20). rRNA contamination of ribosome protected fragments (RPF) was removed by Bowtie2 using the default setting. Both RPF and RNAseq reads were aligned to the genome sequence with Tophat2. The BAM files generated from Tophat2 were annotated with Htseq. Here we only analyzed protein-coding genes guided by gtf file described above. Genes that had less than 128 combined reads in two RNA libraries (control plus IFN-γ-primed condition) of each biological replicate were removed for the reasons previously discussed32, 33. Reads that have multiple alignments were filtered out and cpm (counts per million) was calculated as cpm=106(Ci/N), where Ci is the number of reads mapped to exons of gene i, N is the number of mapped reads in the entire library including multiple aligned reads. Data sets from each replicate were analyzed either individually or merged by summing up the cpm for each condition. Translational efficiency (TE) was calculated as ratio of RPF and RNA (TE=RPF/RNA).
microRNA sequencing and data analysis
Control or IFN-γ-primed human primary macrophages from two independent donors were stimulated with or without Pam3CSK4 (10 ng/ml) for 4h. Total RNA was extracted using a mirVana miRNA isolation kit; miRNA libraries were prepared using TruSeq small RNA library prep kits (Illumina). Mirdeep2 was used to align the sequencing reads to human precursor miRNAs (miRBase release 21) as well as to calculate the read counts for each miRNA. Mature miRNAs with non-unique precursors were merged into one entry, and the final expression values were normalized by library size (correspond to counts per million mapped miRNA reads). edgeR was used to merge the two biological replicates and statistical analysis was performed as previously described50. miRNAs of interest (e.g. miR-146-3p) were searched against conserved miRNA binding sites downloaded from TargetScan database (http://www.targetscan.org) to find target genes. Multiple sequence alignment with Clustal W was used to identify seed sequence binding sites with flanking regions in the 3’UTRs of genes showing increased translation efficiency.
Polysome profiling
Control or IFN-γ-treated human primary macrophages were stimulated for 4 h with Pam3CSK4 (10 ng/ml), followed by cycloheximide (100 μg/ml) treatment for 7 min. Cell pellets were lysed in polysome lysis buffer (20 mM Tris (pH 8.0), 140 mM NaCl, 15 mM MgCl2, 100μg /ml cyclohexmide, 0.5% Triton X-100 and protease inhibitor cocktail (Roche, 1X)), followed by 5-6 passages through a 26G needle using a 1ml syringe. Cell lysates were gently laid onto 10%-50% sucrose density gradients to isolate polysome fractions. Gradients were centrifuged in a SW41 ultracentrifuge rotor at 35,000 r.p.m. for 2 h.
Statistics
The student t-test was used to analyze differences in experiments with two conditions; one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison post-test were used for experiments with more than two conditions. Statistical analyses were performed using GraphPad Prism 5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Weill Cornell Medical College Genomics Core for advice about RNA-seq, J. Schulze from UC Davis Proteomics Core for advice about amino acid measurements, B. Zhao and L. Donlin for review of the manuscript, K. Park-Min for providing Myc inhibitor and siRNA oligos, S. Park and Y. Qiao for discussion about bioinformatic analysis. This work was supported by grants from the NIH (L.B.I.) and The Leonard Tow Foundation to the David Z. Rosenseweig Genomics Research Center; this work was supported in part by grants from NIH (C.M.R.) and the Greenberg Medical Research Institute, the Starr Foundation to C.M.R.; and an EU fellowship for Y. Zhong EU PITN-GA-2012-316861.
Footnotes
ACCESSION CODES
GEO: ribosome profiling, RNAseq and miRNAseq data, GSE66810.
AUTHOR CONTRIBUTIONS
X.S. designed and conducted experiments, analyzed data and prepared the manuscript; Y.Y. performed polysome profiling and ribosome profiling experiments and analyzed data; Y.Z. analyzed ribosome profiling, RNAseq and miRNAseq data; E.G.G. analyzed the ribosome profiling and RNAseq data; J.R.C. and H.L. performed LC-MS experiments and analyzed data; X.H., G.R. and C.M.R. provided advice about experiments and data analysis and contributed to manuscript preparation; L.B.I. conceived and supervised experiments and prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31(4):539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24(5):563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29(5):691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1-2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39(3):454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nature immunology. 2014;15(6):503–511. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- 8.Wan Y, Xiao H, Affolter J, Kim TW, Bulek K, Chaudhuri S, et al. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated posttranscriptional control. The Journal of biological chemistry. 2009;284(16):10367–10375. doi: 10.1074/jbc.M807822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nature immunology. 2012;13(7):642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, et al. Translational control of the activation of transcription factor NF-kappaB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nature immunology. 2012;13(6):543–550. doi: 10.1038/ni.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452(7185):323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 12.Larsson O, Perlman DM, Fan D, Reilly CS, Peterson M, Dahlgren C, et al. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic acids research. 2006;34(16):4375–4386. doi: 10.1093/nar/gkl558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature reviews Immunology. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell host & microbe. 2012;12(4):470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroczynska B, Mehrotra S, Arslan AD, Kaur S, Platanias LC. Regulation of interferon-dependent mRNA translation of target genes. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2014;34(4):289–296. doi: 10.1089/jir.2013.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends in biochemical sciences. 2009;34(7):324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29(4):565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Turnquist HR, Cardinal J, Macedo C, Rosborough BR, Sumpter TL, Geller DA, et al. mTOR and GSK-3 shape the CD4+ T-cell stimulatory and differentiation capacity of myeloid DCs after exposure to LPS. Blood. 2010;115(23):4758–4769. doi: 10.1182/blood-2009-10-251488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, et al. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. The Journal of biological chemistry. 1995;270(24):14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature reviews Immunology. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino Acid Sufficiency and mTOR Regulate p70 S6 Kinase and eIF-4E BP1 through a Common Effector Mechanism. Journal of Biological Chemistry. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 28.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1991;5(11):2516–2522. [PubMed] [Google Scholar]
- 29.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nature reviews Immunology. 2004;4(10):762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 30.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annual review of immunology. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji JD, Park-Min KH, Shen Z, Fajardo RJ, Goldring SR, McHugh KP, et al. Inhibition of RANK expression and osteoclastogenesis by TLRs and IFN-gamma in human osteoclast precursors. Journal of immunology. 2009;183(11):7223–7233. doi: 10.4049/jimmunol.0900072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen AR, Gsponer J, Foster LJ. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Molecular systems biology. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 36.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell host & microbe. 2012;11(6):563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Ivanov SS, Roy CR. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nature immunology. 2013;14(12):1219–1228. doi: 10.1038/ni.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493(7432):346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 40.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annual review of immunology. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortese M, Sinclair C, Pulendran B. Translating glycolytic metabolism to innate immunity in dendritic cells. Cell metabolism. 2014;19(5):737–739. doi: 10.1016/j.cmet.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Neill LA. Glycolytic reprogramming by TLRs in dendritic cells. Nature immunology. 2014;15(4):314–315. doi: 10.1038/ni.2852. [DOI] [PubMed] [Google Scholar]
- 43.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercalli A, Calavita I, Dugnani E, Citro A, Cantarelli E, Nano R, et al. Rapamycin unbalances the polarization of human macrophages to M1. Immunology. 2013;140(2):179–190. doi: 10.1111/imm.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, et al. The TSC-mTOR pathway regulates macrophage polarization. Nature communications. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345(6204):1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemaitre B, Girardin SE. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nature reviews Microbiology. 2013;11(6):365–369. doi: 10.1038/nrmicro3029. [DOI] [PubMed] [Google Scholar]
- 48.Lo WS, Gardiner E, Xu Z, Lau CF, Wang F, Zhou JJ, et al. Human tRNA synthetase catalytic nulls with diverse functions. Science. 2014;345(6194):328–332. doi: 10.1126/science.1252943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.