Abstract
Background
Inflammatory bowel disease (IBD) involves dysregulation of mucosal immunity in response to environmental factors such as the gut microbiota. The bacterial microbiota is often altered in IBD, but the connection to disease is not fully clarified, and gut fungi have recently been suggested to play a role as well. In this study, we compared microbes from all three domains of life–bacteria, archaea, and eukaryota–in pediatric patients with IBD and healthy controls.
Methods
A stool sample was collected from patients with IBD (n=34) or health control subjects (n=90), and bacterial, archaeal, and fungal communities were characterized by deep sequencing of rRNA gene segments specific to each domain.
Results
IBD patients (Crohn’s disease or ulcerative colitis) had lower bacterial diversity and distinctive fungal communities. Two lineages annotating as Candida were significantly more abundant in IBD patients (p = 0.0034 and p=0.00038, respectively) while a lineage annotating as Cladosporium was more abundant in healthy subjects (p=0.0025). There were no statistically significant differences in archaea, which were rare in pediatric samples compared to those from adults.
Conclusions
Pediatric IBD is associated with reduced diversity in both fungal and bacterial gut microbiota. Specific Candida taxa were increased in abundance in the IBD samples. These data emphasize the potential importance of fungal microbiota signatures as biomarkers of pediatric IBD, supporting their possible role in disease pathogenesis.
Keywords: Microbiota, IBD, Fungus, Bacterial, DNA
Introduction
The three domains of life–bacteria, archaea, and eukaryote–are all represented in human gut communities (1–5). Numerous disorders, both intestinal and systemic, have been associated with alterations in microbial community structure, termed “dysbiosis” (6–8). The dysbiotic bacterial microbiota has been extensively characterized for the inflammatory bowel diseases (IBD) and Clostridium difficile infection, although the composition of the archaea and eukaryota domains and their interaction with bacteria has not been studied in detail. The pathogenesis of IBD is associated with an inappropriate and persistent inflammatory response to the commensal gut microbiota in genetically susceptible individuals. In animal studies, dysbiosis has been associated with the development of intestinal inflammation (reviewed in (6, 8, 9)). Many of the genetic risk alleles for IBD involve regulation of innate immune responses that protect the host from bacterial invasion or that regulate the adaptive immune system (reviewed in (10, 11)). In Crohn’s disease (CD), the phylum Firmicutes is commonly reduced in proportional abundance (12–17), notably Faecailbacterium prausnitzii (18–20) and members of the Proteobacteria phylum such as Enterobacteriaceae (21, 22), including E. coli (7, 17, 23) are commonly increased.
The importance of fungi in the human gut to health or disease is not fully understood (2, 24). Fungi, such as Aspergillus, Histoplasma, and Cryptococcus are known to cause pathogenic intestinal infections. Fungi are also part of the commensal gut microbiota. Increases in gut Candida species are hypothesized to cause systemic yeast infections in immunocompromised patients via translocation through the intestinal barrier (25). Fungal-bacterial interactions are just beginning to be described (1, 5, 6, 26–28), and this may be important in IBD (2).
Several lines of evidence link fungi and IBD. Innate immune receptor activation by fungi may augment the development of colitis. The main pattern recognition receptors for fungi include Dectin-1, Dectin-2, DC-SIGN, mannose receptor, and mannose-binding lectin (29). Dectin-1 is a C-type lectin receptor, which recognizes β-glucans in the fungal cell wall. A recent study (30) demonstrated that mice lacking Dectin-1 had increased susceptibility to chemically induced colitis due to their altered responses to indigenous fungi. A polymorphism in the gene encoding Dectin-1 (CLEC7A) was subsequently associated with a severe form of ulcerative colitis (UC) in humans (30). There may also be a relationship between high dietary concentrations of yeast and increased disease activity in patients with CD (31). Treatment with fluconazole, an antifungal, may reduce inflammation in animal models of colitis and in patients with IBD (32), and elevated levels of Saccharomyces cerevisiae antibodies are a biomarker for CD (33). Finally, preliminary studies using denaturing gradient gel electrophoresis (DGGE) suggest that there may be alterations in gut fungal populations associated with IBD (25, 34).
Alterations in the archaeal microbiota may also be relevent to IBD. Methane production from archaea has been shown to be altered in patients with IBD (35). Additionally, the syntrophic interaction between archaea and fermentative bacteria leading to the increase in short chain fatty acid (SCFA) production (36) may have an impact on mucosal immunity via to the impact of SCFA on T regulatory cells and immune tolerance (37). Studies of a modest number of subjects are beginning to suggest that fungi may be dysbiotic in IBD (34) as well as bacteria.
In this study, we use deep sequencing of rRNA gene tags to characterize the composition of the bacterial, fungal, and archaeal microbiota in pediatric patients with IBD (n=32) and healthy controls (n=90), revealing a distinctive signature in the mycobiome.
Methods
Subjects
All subjects studied were from the Philadelphia area and <21 years of age (Supplemental Table 1). Subjects provided written consent or, when appropriate, assent. Subjects were excluded if they had been treated with antibiotics or probiotics within the two weeks prior to sample collection, if there was presence of an ostomy, or if they had been treated with supplemental nutrition (TPN, enteral nutritional therapy, or other nutritional supplements) accounting for more than 50% of the total caloric intake for the one week prior to sample collection. Pediatric Crohn’s Disease Activity Index (PCDAI) (38) or Pediatric Ulcerative Colitis Activity Index (PUCAI) (39) were calculated as described. Stool samples were collected between February and September 2012 and kept frozen at −80° C until they were processed for DNA extraction. The sequence samples obtained from the healthy pediatric and adult subjects and used as controls are described in (5, 40). Briefly, healthy volunteers were screened to be free from any chronic disease, to have a normal bowel movement frequency, and to have a body mass index (BMI) between 18.5 and 35 kg/m2. One stool sample was provided per subject and kept frozen at −80° C until processed for DNA extraction.
DNA Isolation
DNA was isolated from approximately 200 mg of stool using the PSP Spin Stool DNA Plus Kit (Cat #10381102, Invitek, Berlin, Germany) as per the manufacturer’s instructions.
16S rDNA Gene and ITS1 Region Gene Sequencing
Isolated DNA was quantified using the Picogreen system and 50 ng of DNA was amplified in each PCR reaction. Primers were barcoded to label each sample as described previously (41, 42). PCR reactions were carried out in triplicate using Accuprime (Invitrogen, Carlsbad, CA, USA). Each reaction contained 50 nanograms of DNA and 10 pM of each primer. Primers annealing to the V1V2 region of the 16S bacterial gene were used for amplification. The PCR protocol for 16S amplicons was described previously (43). The development of the archaeal specific 16S rDNA primers and ITS1 fungal primers and PCR cycling conditions are described in [42]. Amplified 16S rDNA and ITS1 fragments were purified using a 1:1 volume of Agencourt AmPure XP beads (Beckman-Colter, Brea, CA, USA). The purified products from the stool samples were pooled in equal amounts and analyzed by pyrosequencing using the Roche/454 Genome Sequencer Junior. DNA pools were separated by amplicon type. DNA-free water was subjected to the same amplification and purification procedure, and no DNA product was observed in agarose gels. Sequences of oligonucleotides used in this study are in Supplemental Table 2.
Sequence Analysis
Sequence data was processed using QIIME (44), augmented by the R package QIIMER (http://cran.r-project.org/web/packages/qiimer). Taxonomy was assigned to the sequences using Ribosomal Database Project (RDP) for 16S (45) and the UNITE database for ITS. Further information on methods can be found in the Supplemental Information.
We fit a Random Forest model to predict health versus disease (IBD) using 16S bacterial OTUs, ITS fungal OTUs, or a combination of the two. One thousand bootstrapped iterations were performed to obtain an estimate of the misclassification rate. The classification error rate was compared to guessing, e.g. if the dataset consisted of 32 subjects with IBD and 68 healthy subjects, a sample would be randomly classified as a sample from a subject with IBD 32% of the time and as a sample from a healthy subject 68% of the time.
Bacterial OTUs with greater than 100 sequences across all IBD samples (128 OTUs) were correlated with ITS fungal OTUs with greater than 100 sequences across all IBD samples (27 OTUs). P-values were calculated using a two-sided Pearson correlation test. Bacterial OTUs with at least one correlation to a fungal OTU producing a Bonferroni-corrected p-value less than 0.05 were displayed on the final heatmap.
Results
Patient cohort studied
The bacterial component of the microbiome has been studied extensively in IBD, and is known to be dysbiotic (6, 8, 9, 46). Thus, in this study, we sought to assess effects on the fungal and archaeal components of the microbiome. A cross-sectional analysis was performed whereby a single stool sample was obtained from pediatric patients (3 to 21 years of age) with CD, UC, or indeterminate colitis (IC) (n=32). The diagnosis of IBD was based on endoscopic, radiologic, laboratory, and clinical findings. Clinical phenotypes of all patients and their therapies are described in Supplemental Table 1. Microbiome sequencing results for IBD patients were compared to results for healthy pediatric and adult subjects studied previously (n=90) (40).
Reduced richness in the intestinal bacterial microbiota of patients with IBD
We first characterized the bacterial composition of the microbiota by purifying and amplifying DNA using PCR primers recognizing the V1V2 region of the16S rRNA gene, and pyrosequencing an average of 2,864 reads per sample. We analyzed fecal samples from two IC, four UC, and 26 CD patients (Supplemental Table 1). Reads were clustered into operational taxonomic units (OTUs) at 97% sequence identity, and representative sequences were aligned to databases for taxonomic attribution. Comparison of data among the disease classes revealed few differences among groups (Supplemental Figure 1). Most samples were dominated with Bacteroides. A few had Clostridia as the dominant taxa.
We compared a dataset from healthy adults and children to the IBD samples (40). OTU composition was compared in a pairwise fashion between samples using UniFrac, and healthy and IBD sample sets were compared to each other using PERMANOVA. We observed a clear separation between the IBD and healthy sample sets (p<0.0001) (Supplemental Figure 2).
We also investigated the diversity of the IBD samples and healthy controls, since previously several studies have reported lower diversity in IBD (15). We assessed diversity by calculating the Shannon Index for each sample, and found that the IBD samples had significantly lower diversity compared to either healthy adult or pediatric control subjects (p-value <0.05) (Supplemental Figure 3).
Comparison of archaea in patients with IBD and healthy controls
We compared the archaeal microbiota composition in the 32 IBD samples with 90 healthy controls by amplification with PCR primers selective for archaea, then pyrosequencing and alignment to databases. Only three IBD samples contained detectable Methanobrevibacter (Supplemental Figure 4), the major archaeal lineage in healthy adults (5). Six samples had a few reads (<5 reads each) mostly matching “Unclassified Archaea”. Querying those reads against NCBI’s nucleotide database did not provide clearer taxonomic resolution. We validated these findings by quantitative PCR targeting Archaea, which confirmed that only three IBD samples, Subject004_CD, Subject006_CD, and Subject037_CD, had detectable archaea. There was a lower recovery of archaeal sequences from pediatric IBD samples compared to healthy adults (p<1×10−6), but pediatric IBD patients were not significantly different from healthy children (p=0.128) (Supplemental Figure 5). This indicates that children have lower archaeal colonization than do adults, and that alterations in the archaeal colonization of the gut are not related to IBD.
A distinctive fungal signature in the microbiota of pediatric patients with IBD
The fungal representation in the fecal samples was characterized by amplification of stool DNA with PCR primers targeting the fungal rRNA ITS region, followed by pyrosequencing using the 454/Roche platform, formation into OTUs at 95% sequence identity, and alignment to databases. The ITS amplicon has been reported to have biases in recovery of certain lineages (47), but it does successfully capture a broad range of taxa (5, 26, 28). Thirty-two IBD subjects were compared to 90 healthy adults and children (40). IBD samples had significantly lower fungal diversity than healthy children (p-value <0.05) (Figure 1).
Figure 1.
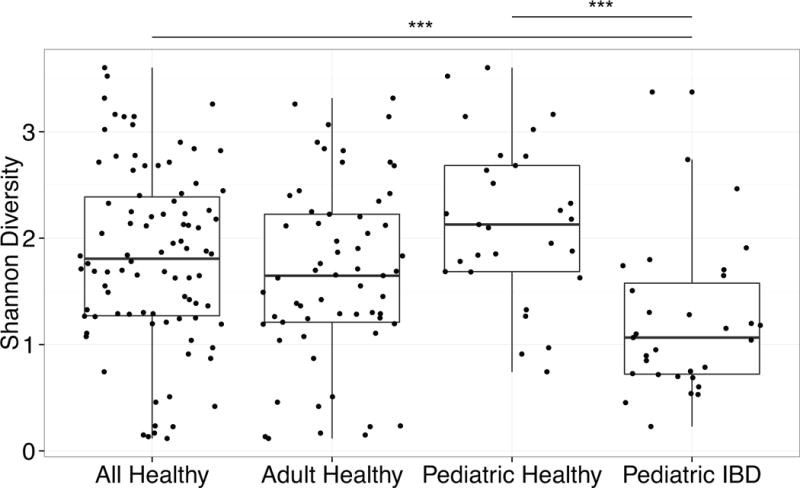
Diversity of fungal communities is decreased in IBD patients compared to healthy subjects.
The Shannon diversity index was calculated based on the OTU-level classification tables. The boxplots show the distribution of diversity values for: (1) all healthy subjects, (2) only the adult healthy subjects, (3) only the pediatric healthy subjects, and (4) the pediatric IBD subjects. Each black dot represents a different subject. *** p<0.0005 on Wilcox test.
Ordination analysis based on the presence or absence of fungal species revealed a separation between healthy and IBD pediatric subjects (p=0.004) (Figure 2). Samples from healthy adults overlapped with both pediatric groups (data not shown). Ordination based on the abundance (rather than presence/absence) of each fungal species also showed separation between healthy and IBD pediatric subjects (p=0.044) (Supplemental Figure 6). A focused analysis of CD only, in which the IC and UC patients were excluded, also revealed significant separation between healthy and IBD pediatric subjects (Supplemental Figure 7 p< .0001).
Figure 2.
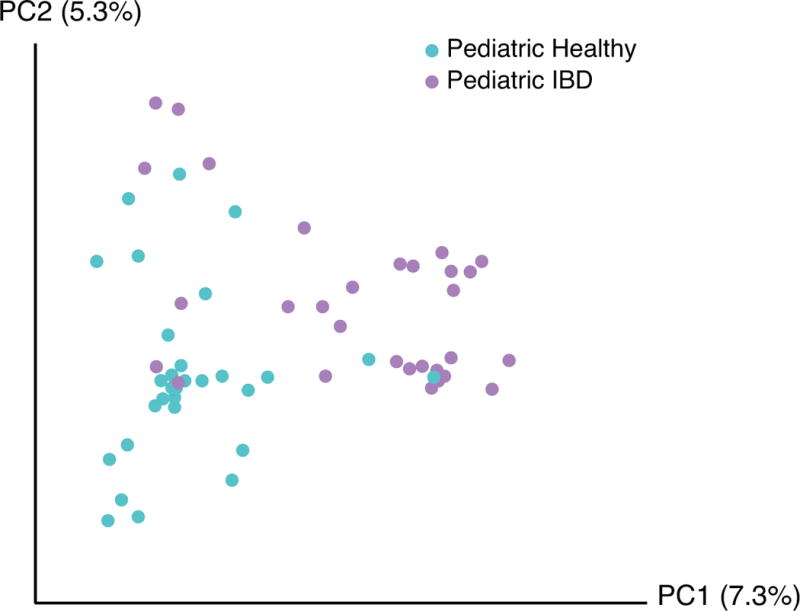
Comparison of pediatric healthy and pediatric IBD subjects’ fungal community composition using principal coordinate ordination.
Principal coordinate analysis was used to depict the relatedness of fungal communities based on presence or absence. The axes represent the two most discriminating axes using the binary Jaccard index distance metric. Pediatric healthy subjects are depicted in cyan and pediatric IBD subjects are depicted in lavender. The two groups clustered separately (p=0.004).
Next, we investigated whether fungal representation differed in IBD and healthy control samples (Figure 3). The most commonly observed order was Saccharomyetales, which contains the genus Candida. IBD samples averaged 2,675 Candida sequences (proportionally 72.9%) while healthy controls averaged 1,320 reads (proportionally 32.9%) (p=0.0107 based on read counts). Healthy adults had slightly higher, non-significant average proportions of Candida than did pediatric healthy subjects (1428 reads (37%) vs. 1055 reads (23%); p=0.535 based on read counts).
Figure 3.

Taxonomic heatmap of fungal community members in healthy and IBD subjects.
Proportions of fungal OTUs in adult healthy, pediatric healthy, and IBD subjects. The color bar on the top indicates the health status of each subject. Each column indicates a different subject. The color bar on the right side indicates the average relative abundances of these genera in each subject.
The Candida reads were categorized in 97 different OTUs with the majority belonging to one OTU (genbank ID: KP132001) (Figure 4A). This OTU annotates as single species, which has previously been given multiple different names, including Pichia jadinii, Candida utilis, Cyberlindnera jadinii, Torula yeast, and Candida guilliermondii var. nitratophila. This OTU was significantly enriched in pediatric IBD patients (p-value<0.01 using raw reads or proportion for both pediatric IBD versus all healthy (adult and pediatric) and pediatric IBD versus pediatric healthy). A second Candida OTU (genbank ID: EF197997), annotating as Candida parapsilosis, was also more common in the pediatric IBD samples (Figure 4B, p-value <0.01 based on number of reads). A third OTU (genbank ID: KJ596320), annotating as Cladosporium cladosporioides, was more common in healthy subjects (pediatric or adult healthy versus pediatric IBD, p-value <0.01 based on number of reads; Figure 4C).
Figure 4.

Abundance of selected fungal OTUs
Barcharts showing the number of reads from three selected fungal OTUs: (A) Candida (accession KP132001), (B) Candida (accession EF197997) and (C) Cladosporium (accession KJ596320). Adult healthy subjects, pediatric healthy subjects, and pediatric IBD subjects are shown in red, blue, and purple respectively. A negative control sample (NC) that was processed alongside the samples is shown at the right in green. No reads matching any of the three lineages mentioned above were found in the negative control sample.
A microbial signature for pediatric IBD
Random Forest, a supervised machine-learning algorithm, was used to generate a classifier capable of sorting IBD and healthy controls based on microbial community composition. Classifiers were developed comparing pediatric IBD to pediatric healthy (Figure 5A), and pediatric IBD versus pooled adult and pediatric healthy controls (Figure 5B). Classifiers were compared that used bacterial 16S rRNA gene data only, fungal ITS sequence data only, or both.
Figure 5.
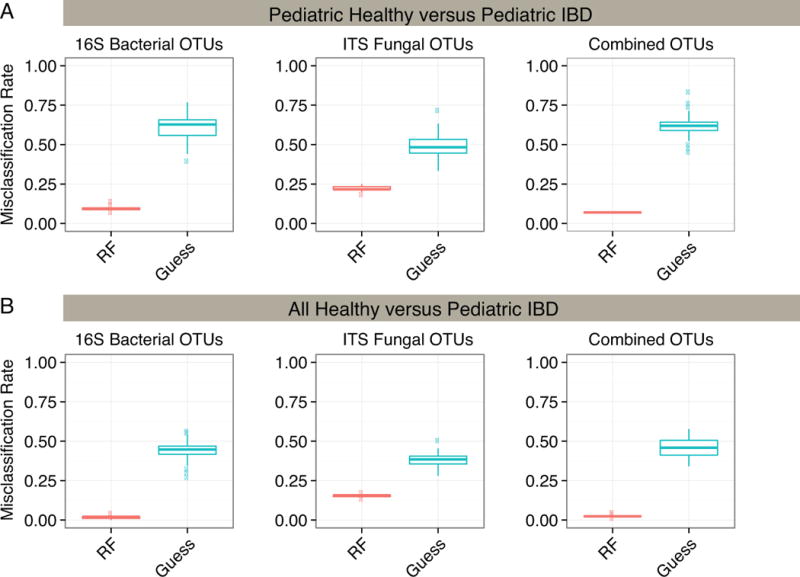
Random Forest Classification Accuracy for Healthy and IBD subjects.
A random forest classifier was used to group samples into either healthy or IBD categories. Random forest accuracy (red) was compared to random guessing (blue) with misclassification rate indicated on the y-axis. The classifier was run using 16S bacterial OTUs, ITS fungal OTUs and the combination of both bacterial and fungal OTUs. Results were also compared using (A) pediatric healthy and pediatric IBD subjects or (B) both adult and pediatric healthy subjects and IBD subjects.
The models successfully partitioned the pediatric samples by disease status (Figure 5A) and partitioned all healthy controls (children and adults) from IBD children (Figure 5B). For pediatric samples, the16S or ITS OTUs used alone showed a median accuracy of 90% and 83% respectively, while the combination of OTUs showed greater than 93% accuracy. The bacterial OTUs that could best distinguish IBD patients from healthy controls annotated as Bacteroides, Parabacteroides, and Faecalibacterium prausnitzii. For fungi, the classifier was dominated by OTUs annotating as Pichia jadinii/Candida utilis. The full list of distinguishing taxa is presented in Supplemental Tables 3–6.
Bacterial and fungal correlations in IBD
Two-sided, Pearson tests were used to measure correlation between bacterial and fungal OTUs across IBD samples. Of the 128 bacterial OTUs tested, 33 showed FDR-corrected, significant (p-value <0.05) correlation with at least one fungal OTU. (Figure 6). The most commonly found fungal species in IBD subjects, Candida OTU KP132001, did not correlate strongly with any bacterial species. Many more significant correlations between fungal and bacterial OTUs were detected in healthy subjects (Supplemental Figure 8). Evidently fungal and bacterial species co-vary, but this was not prominently associated with the lineages implicated as important in IBD in these subjects.
Figure 6.
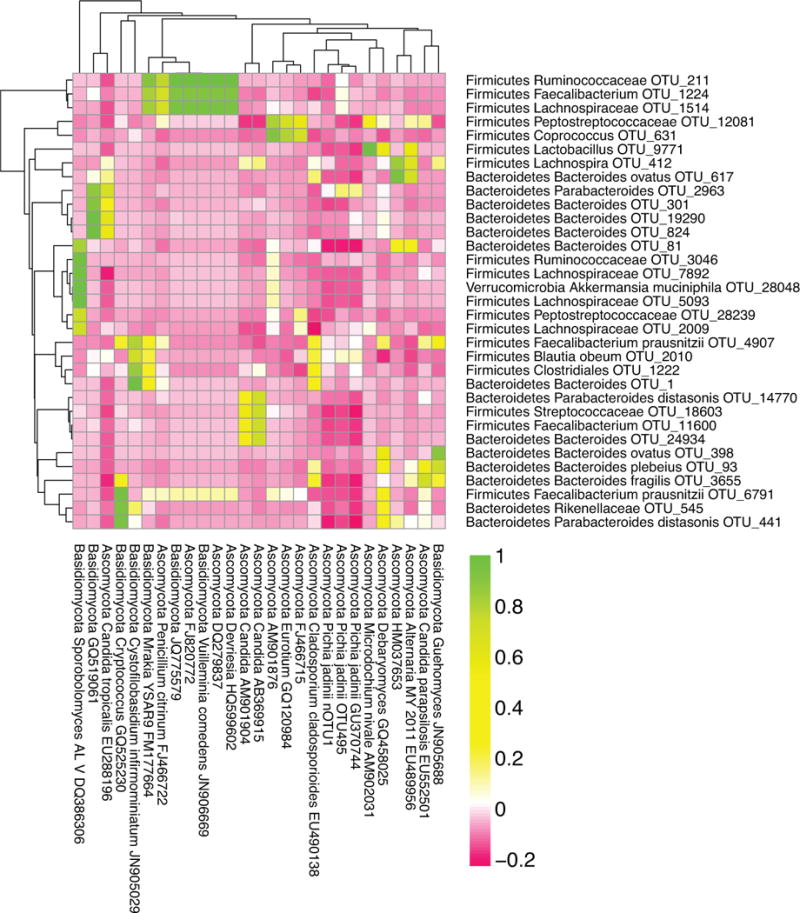
Correlations between Bacterial and Fungal OTUs in pediatric IBD.
The Pearson correlation coefficient between fungal and bacterial OTUs in pediatric IBD patients was calculated. OTUs were included in this heatmap if they had greater than 100 sequences in pediatric IBD patients. OTUs were included if they significantly correlated with at least one other OTU (two-sided correlation, where the p-value exceeded 0.05 after Bonferroni correction).
Discussion
Here, we analyzed the composition and diversity of the fungal, bacterial and archaeal microbiota in pediatric IBD patients compared to healthy controls. In contrast to the many studies of the bacterial component of the gut microbiota, there is relatively little known about the fungal and archaeal microbiota and their roles in IBD. Fungi and archaea are known to be normal components of the gut microbiota (1, 4, 5, 48). In this study, we recovered abundant fungal reads from both IBD patients and healthy controls. In contrast, archaea were rare in the pediatric samples. The potential importance of fungi in IBD is well recognized. Antibodies to baker’s yeast (Saccharomyces cerevisiae), termed ASCA, are detected more frequently in patients with CD than in healthy controls or in patients with UC (29–69% of patients with CD (33) (49–51)). The fungal antigen recognized is thought to be phosphopeptidomannan of the Saccharomyces cerevisiae cell wall (52). ASCA positivity may precede the development of IBD (53), and ASCA are found more commonly in healthy relatives of patients with CD (54). The difference in ASCA prevalence raises the possibility of a differential fungal microbiota in patients with CD versus healthy individuals, and our data documents such a difference.
Although the ASCA epitope is associated with S. cerevisiae, it is also associated with Candida albicans (55) and potentially other related yeasts. Candida colonization has been associated with multiple diseases of the gastrointestinal tract including CD, UC, esophagitis, oral mucositis, and even gastric ulcers (56). Candida strains colonize gastric ulcers, particularly when the ulcers are large or perforated (56) and candida esophagitis responds to antifungal therapy demonstrating the role of Candida colonization in the pathogenesis of inflammatory processes. Increased colonization with Candida in IBD might be a consequence of mucosal or immune system alterations as well as the use of antibiotics(28). The health consequences are unknown but may be adverse.
We found three fungi with differential abundance in IBD patients versus healthy controls. Fungal phylogeny is in a state of transition, so species level attributions are not fully secure. For example, until recently, sexual forms (teleomorphs) were commonly placed in separate genera from asexual forms (anamorphs) of the same fungal organisms (57). We found one Candida OTU (genbank ID: KP132001) that was more abundant in patients with IBD, which has been associated with five named species, including Pichia jadinii, Candida utilis, Cyberlindnera jadinii, Torulopsis utilis, and Torula utilis (58), illustrating the challenges of current fungal taxonomy. A common name for this group is Torula yeast. In its inactive form, Torula yeast is widely used as a flavoring which imparts a savory taste to soups, sauces, and snack products (59). Possibly the pediatric subjects with IBD were eating diets more enriched in Torula yeasts, though there is no universal diet for patients with IBD, so this seems unlikely. Patients with IBD frequently identify dietary components that cause increased symptoms and so follow restricted diets when the disease is active, but most subjects studied here had relatively low disease activity scores and so were not expected to be on specialized diets. Also, a review of dietary records kept by the patients in our study did not reveal an obvious bias towards the consumption of foods rich in Torula yeast (data not shown). The second explanation is that there is truly increased Torula yeast colonization in patients with IBD, which at present seems more likely. Members of the fungal genus Pichia can be pathogens in humans, especially in immunocompromised hosts (60), Pichia species may also lead to enteritis in animals (61, 62), and pathogenesis by Candida species is well described. Further study is needed to clarify whether colonization is related to disease pathogenesis or whether it is a consequence of gut inflammation or immune suppressive therapy.
We found a second Candida OTU (genbank ID: EF197997) that was more abundant in patients with IBD, and a Cladosporium cladosporioides OTU (genbank ID: KJ596320) that was more abundant in healthy controls. All of the IBD patients in our cohort were undergoing various therapies, so it is not clear whether these taxonomic changes were associated with the IBD itself or were a consequence of treatment, for example immunosuppression (63). Other Candida yeasts, in contrast, are used as probiotics (64), raising the possibility that the yeast lineages present contribute a mixture of positive and negative influences.
Studies in animal models suggest a functional role for fungi in intestinal inflammation. Mice lacking the Dectin-1 receptor, a pattern recognition receptor known to identify fungi, were more susceptible to colonic inflammation in a dextran sodium sulfate (DSS) colitis model (30). Administration of Candida tropicalis resulted in a more severe DSS-induced colitis in this model, and treatment with fluconazole, an anti-fungal agent, reduced disease(30). Another PRR, SIGNR3, was recently found to be involved in recognizing fungal commensals. Mice lacking SIGNR3 also exhibit increased intestinal inflammation upon exposure to DSS (65). Additional investigation is needed in humans to determine the mechanisms responsible for the observed alterations in the fungal microbiota in patients with IBD and the role they may play in disease pathogenesis via pattern recognition receptors.
To probe distinctive lineages in health and disease, and to develop noninvasive biomarkers, we developed a Random Forest classifier to sort patients with IBD versus healthy controls. Using a combination of bacterial and fungal OTUs allowed greater than 93% accuracy in distinguishing samples. The currently-available biomarkers focus on the evaluation of patients already diagnosed with IBD such as differentiating CD from UC, differentiating quiescent from active disease, and predicting disease course (66). Drug levels and anti-drug antibodies are also used as biomarkers (66). Unfortunately, the biomarkers currently used to evaluate patients with symptoms of IBD, such as C – reactive protein, erythrocyte sedimentation rate, and fecal calprotectin, are not specific to IBD. Additionally, methods are needed to establish early diagnosis of at-risk individuals, such as asymptomatic family members of patients with IBD. It will be useful to evaluate the performance of the assay targeting both bacteria and fungi described here in these settings.
In a recently described analysis of the archaeal microbiome of healthy human subjects, about 45% of subjects were found to have archaea present in the stool (5). The vast majority of sequences annotated as Methanobrevibacter (5). In our study, we found only 3 pediatric patients with IBD with detectable Methanobrevibacter. The previous literature has been inconsistent, with some studies reporting increased prevalence of methanogens or decreased methane production in patients with IBD (67–70) and other studies demonstrating increased methane production in patients with IBD (71). The low number of pediatric IBD subjects found to have archaeal species from the stool in our study is consistent with a report of methane production increase with aging (72). Thus our data suggest that archaeal colonization may be more characteristic of the adult gut microbiota.
In conclusion, we report that pediatric IBD was associated with decreased fungal diversity and also altered taxonomic composition. We found an increased proportion of two Candida OTUs in IBD patients. The fungal microbiota may be important in IBD given the increased prevalence of ASCA and the more recent work suggesting a relationship between fungal pattern recognition receptors and IBD in mice. We also report a compositional signature elucidated in the Random Forrest analysis that may be useful in diagnosing IBD patients. Thus, these findings 1) clarify the existence and nature of fungal dysbiosis in pediatric IBD, 2) pose the question of whether specific yeast lineages may be more pro-inflammatory or colonize associated with inflammation, 3) motivate the investigation of fungal blooms in response to treatment of IBD, and 4) provide a signature of dysbiosis in pediatric patients potentially useful in individualized molecular diagnosis.
Supplementary Material
Acknowledgments
This work was supported by Project UH2DK083981; R01 GM103591; the Penn Center for Molecular Studies in Digestive and Liver Disease including the Molecular Biology/Gene Expression Core (P30 DK050306); The Joint Penn-CHOP Center for Digestive, Liver, and Pancreatic Medicine; S10RR024525; UL1RR024134, and K24-DK078228; and the University of Pennsylvania Center for AIDS Research (CFAR) P30 AI 045008. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources, National Institutes of Health, or Pennsylvania Department of Health. Sequences were submitted to the Sequence Read Archive with accession numbers SRP050217.
Footnotes
Disclosures: None of the authors have disclosures related to this manuscript.
References
- 1.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richard ML, Lamas B, Liguori G, et al. Gut Fungal Microbiota: The Yin and Yang of Inflammatory Bowel Disease. Inflamm Bowel Dis. 2014 doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 3.Hansen EE, Lozupone CA, Rey FE, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuel BS, Hansen EE, Manchester JK, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartor RB. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc Natl Acad Sci U S A. 2008;105:16413–16414. doi: 10.1073/pnas.0809363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS letters. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 10.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ek WE, D’Amato M, Halfvarson J. The history of genetics in inflammatory bowel disease. Annals of gastroenterology : quarterly publication of the Hellenic Society of Gastroenterology. 2014;27:294–303. [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderploeg R, Panaccione R, Ghosh S, et al. Influences of intestinal bacteria in human inflammatory bowel disease. Infect Dis Clin North Am. 2010;24:977–993. ix. doi: 10.1016/j.idc.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Van de Merwe JP, Schroder AM, Wensinck F, et al. The obligate anaerobic faecal flora of patients with Crohn’s disease and their first-degree relatives. Scand J Gastroenterol. 1988;23:1125–1131. doi: 10.3109/00365528809090179. [DOI] [PubMed] [Google Scholar]
- 14.Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gophna U, Sommerfeld K, Gophna S, et al. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, et al. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 19.Prescott NJ, Fisher SA, Franke A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 20.Swidsinski A, Loening-Baucke V, Vaneechoutte M, et al. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 21.Seksik P, Rigottier-Gois L, Gramet G, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 23.Mangin I, Bonnet R, Seksik P, et al. Molecular inventory of faecal microflora in patients with Crohn’s disease. FEMS Microbiol Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Kohler JR, Casadevall A, Perfect J. The Spectrum of Fungi That Infects Humans. Cold Spring Harbor perspectives in medicine. 2014:5. doi: 10.1101/cshperspect.a019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott SJ, Kuhbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 26.Dollive S, Peterfreund GL, Sherrill-Mix S, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome biology. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bittinger K, Charlson ES, Loy E, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome biology. 2014;15:487. doi: 10.1186/s13059-014-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dollive S, Chen YY, Grunberg S, et al. Fungi of the Murine Gut: Episodic Variation and Proliferation during Antibiotic Treatment. PLoS One. 2013;8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZK, Yang YS, Stefka AT, et al. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther. 2014;39:751–766. doi: 10.1111/apt.12665. [DOI] [PubMed] [Google Scholar]
- 30.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barclay GR, McKenzie H, Pennington J, et al. The effect of dietary yeast on the activity of stable chronic Crohn’s disease. Scand J Gastroenterol. 1992;27:196–200. doi: 10.3109/00365529208999948. [DOI] [PubMed] [Google Scholar]
- 32.Zwolinska-Wcislo M, Brzozowski T, Budak A, et al. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa. J Physiol Pharmacol. 2009;60:107–118. [PubMed] [Google Scholar]
- 33.Prideaux L, De Cruz P, Ng SC, et al. Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm Bowel Dis. 2012;18:1340–1355. doi: 10.1002/ibd.21903. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Wang C, Tang C, et al. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. Journal of clinical gastroenterology. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nature reviews Gastroenterology & hepatology. 2012;9:504–518. doi: 10.1038/nrgastro.2012.85. [DOI] [PubMed] [Google Scholar]
- 36.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013 doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 38.Hyams JS, Ferry GD, Mandel FS, et al. Development and validation of a pediatric Crohn’s disease activity index. Journal of pediatric gastroenterology and nutrition. 1991;12:439–447. [PubMed] [Google Scholar]
- 39.Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423–432. doi: 10.1053/j.gastro.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 40.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna P, Hoffmann C, Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS pathogens. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann C, Minkah N, Leipzig J, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 49.Ruemmele FM, Seidman EG. Cytokine–intestinal epithelial cell interactions: implications for immune mediated bowel disorders. Zhonghua Min Guo Xiao Er Ke Yi Xue Hui Za Zhi. 1998;39:1–8. [PubMed] [Google Scholar]
- 50.Quinton JF, Sendid B, Reumaux D, et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791. doi: 10.1136/gut.42.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peeters M, Joossens S, Vermeire S, et al. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. Am J Gastroenterol. 2001;96:730–734. doi: 10.1111/j.1572-0241.2001.03613.x. [DOI] [PubMed] [Google Scholar]
- 52.Peyrin-Biroulet L, Standaert-Vitse A, Branche J, et al. IBD serological panels: facts and perspectives. Inflamm Bowel Dis. 2007;13:1561–1566. doi: 10.1002/ibd.20226. [DOI] [PubMed] [Google Scholar]
- 53.Israeli E, Grotto I, Gilburd B, et al. Anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic antibodies as predictors of inflammatory bowel disease. Gut. 2005;54:1232–1236. doi: 10.1136/gut.2004.060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Standaert-Vitse A, Sendid B, Joossens M, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol. 2009;104:1745–1753. doi: 10.1038/ajg.2009.225. [DOI] [PubMed] [Google Scholar]
- 55.Standaert-Vitse A, Jouault T, Vandewalle P, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14:386–391. doi: 10.1016/j.mib.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawksworth DL. A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA fungus. 2011;2:155–162. doi: 10.5598/imafungus.2011.02.02.06. Available at: http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=4903. Accessed 11/3/2014, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Festring D, Hofmann T. Discovery of N(2)-(1-carboxyethyl)guanosine 5′-monophosphate as an umami-enhancing maillard-modified nucleotide in yeast extracts. J Agric Food Chem. 2010;58:10614–10622. doi: 10.1021/jf102899j. [DOI] [PubMed] [Google Scholar]
- 60.Moses A, Maayan S, Shvil Y, et al. Hansenula anomala infections in children: from asymptomatic colonization to tissue invasion. Pediatr Infect Dis J. 1991;10:400–402. doi: 10.1097/00006454-199105000-00010. [DOI] [PubMed] [Google Scholar]
- 61.Suchodolski JS, Morris EK, Allenspach K, et al. Prevalence and identification of fungal DNA in the small intestine of healthy dogs and dogs with chronic enteropathies. Vet Microbiol. 2008;132:379–388. doi: 10.1016/j.vetmic.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Pettoello-Mantovani M, Nocerino A, Polonelli L, et al. Hansenula anomala killer toxin induces secretion and severe acute injury in the rat intestine. Gastroenterology. 1995;109:1900–1906. doi: 10.1016/0016-5085(95)90757-2. [DOI] [PubMed] [Google Scholar]
- 63.Dave M, Purohit T, Razonable R, et al. Opportunistic infections due to inflammatory bowel disease therapy. Inflamm Bowel Dis. 2014;20:196–212. doi: 10.1097/MIB.0b013e3182a827d2. [DOI] [PubMed] [Google Scholar]
- 64.Sanders ME, Guarner F, Guerrant R, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eriksson M, Johannssen T, von Smolinski D, et al. The C-Type Lectin Receptor SIGNR3 Binds to Fungi Present in Commensal Microbiota and Influences Immune Regulation in Experimental Colitis. Front Immunol. 2013;4:196. doi: 10.3389/fimmu.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826 e1812. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levine J, Ellis CJ, Furne JK, et al. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93:83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- 68.Pimentel M, Mayer AG, Park S, et al. Methane production during lactulose breath test is associated with gastrointestinal disease presentation. Digestive diseases and sciences. 2003;48:86–92. doi: 10.1023/a:1021738515885. [DOI] [PubMed] [Google Scholar]
- 69.Rana SV, Sharma S, Malik A, et al. Small intestinal bacterial overgrowth and orocecal transit time in patients of inflammatory bowel disease. Digestive diseases and sciences. 2013;58:2594–2598. doi: 10.1007/s10620-013-2694-x. [DOI] [PubMed] [Google Scholar]
- 70.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eadala P, Matthews SB, Waud JP, et al. Association of lactose sensitivity with inflammatory bowel disease–demonstrated by analysis of genetic polymorphism, breath gases and symptoms. Aliment Pharmacol Ther. 2011;34:735–746. doi: 10.1111/j.1365-2036.2011.04799.x. [DOI] [PubMed] [Google Scholar]
- 72.Peled Y, Gilat T, Liberman E, et al. The development of methane production in childhood and adolescence. Journal of pediatric gastroenterology and nutrition. 1985;4:575–579. doi: 10.1097/00005176-198508000-00013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


