Abstract
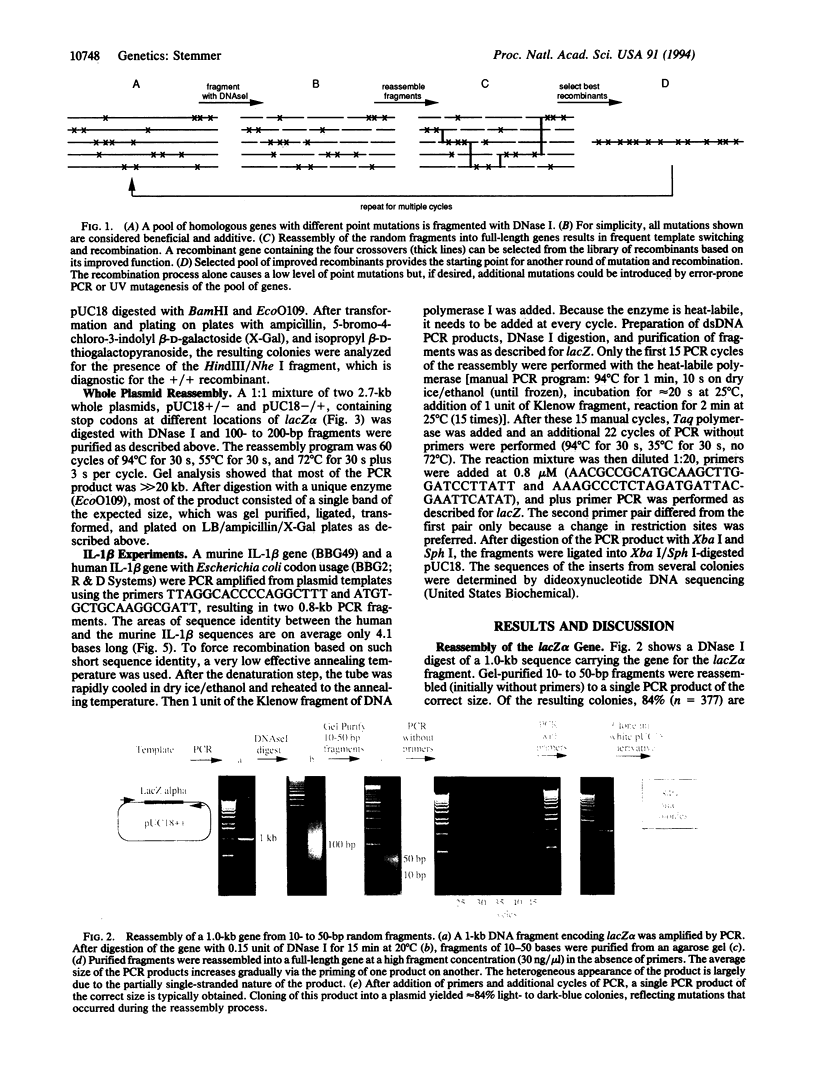
Computer simulations of the evolution of linear sequences have demonstrated the importance of recombination of blocks of sequence rather than point mutagenesis alone. Repeated cycles of point mutagenesis, recombination, and selection should allow in vitro molecular evolution of complex sequences, such as proteins. A method for the reassembly of genes from their random DNA fragments, resulting in in vitro recombination is reported. A 1-kb gene, after DNase I digestion and purification of 10- to 50-bp random fragments, was reassembled to its original size and function. Similarly, a 2.7-kb plasmid could be efficiently reassembled. Complete recombination was obtained between two markers separated by 75 bp; each marker was located on a separate gene. Oligonucleotides with 3' and 5' ends that are homologous to the gene can be added to the fragment mixture and incorporated into the reassembled gene. Thus, mixtures of synthetic oligonucleotides and PCR fragments can be mixed into a gene at defined positions based on homology. As an example, a library of chimeras of the human and murine genes for interleukin 1 beta has been prepared. Shuffling can also be used for the in vitro equivalent of some standard genetic manipulations, such as a backcross with parental DNA. The advantages of recombination over existing mutagenesis methods are likely to increase with the numbers of cycles of molecular evolution.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkin A. P., Youvan D. C. An algorithm for protein engineering: simulations of recursive ensemble mutagenesis. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7811–7815. doi: 10.1073/pnas.89.16.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., Szostak J. W. Isolation of new ribozymes from a large pool of random sequences [see comment]. Science. 1993 Sep 10;261(5127):1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Bock L. C., Griffin L. C., Latham J. A., Vermaas E. H., Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992 Feb 6;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Cadwell R. C., Joyce G. F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992 Aug;2(1):28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cano R. J., Poinar H. N., Pieniazek N. J., Acra A., Poinar G. O., Jr Amplification and sequencing of DNA from a 120-135-million-year-old weevil. Nature. 1993 Jun 10;363(6429):536–538. doi: 10.1038/363536a0. [DOI] [PubMed] [Google Scholar]
- Cull M. G., Miller J. F., Schatz P. J. Screening for receptor ligands using large libraries of peptides linked to the C terminus of the lac repressor. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1865–1869. doi: 10.1073/pnas.89.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwirla S. E., Peters E. A., Barrett R. W., Dower W. J. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Donato A., de Nigris M., Russo N., Di Biase S., D'Alessio G. A method for synthesizing genes and cDNAs by the polymerase chain reaction. Anal Biochem. 1993 Jul;212(1):291–293. doi: 10.1006/abio.1993.1328. [DOI] [PubMed] [Google Scholar]
- Forrest S. Genetic algorithms: principles of natural selection applied to computation. Science. 1993 Aug 13;261(5123):872–878. doi: 10.1126/science.8346439. [DOI] [PubMed] [Google Scholar]
- Gram H., Marconi L. A., Barbas C. F., 3rd, Collet T. A., Lerner R. A., Kang A. S. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3576–3580. doi: 10.1073/pnas.89.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R. E., Russell S. J., Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J Mol Biol. 1992 Aug 5;226(3):889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- Hermes J. D., Blacklow S. C., Knowles J. R. Searching sequence space by definably random mutagenesis: improving the catalytic potency of an enzyme. Proc Natl Acad Sci U S A. 1990 Jan;87(2):696–700. doi: 10.1073/pnas.87.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G. F. Directed molecular evolution. Sci Am. 1992 Dec;267(6):90–97. doi: 10.1038/scientificamerican1292-90. [DOI] [PubMed] [Google Scholar]
- Kauffman S. A. Applied molecular evolution. J Theor Biol. 1992 Jul 7;157(1):1–7. doi: 10.1016/s0022-5193(05)80753-2. [DOI] [PubMed] [Google Scholar]
- Mattheakis L. C., Bhatt R. R., Dower W. J. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J., Griffiths A. D., Winter G., Chiswell D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990 Dec 6;348(6301):552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J. P., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990 Apr 11;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. Searching for peptide ligands with an epitope library. Science. 1990 Jul 27;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Stemmer W. P., Morris S. K., Wilson B. S. Selection of an active single chain Fv antibody from a protein linker library prepared by enzymatic inverse PCR. Biotechniques. 1993 Feb;14(2):256–265. [PubMed] [Google Scholar]
- Stemmer W. P. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994 Aug 4;370(6488):389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Waterhouse P., Griffiths A. D., Johnson K. S., Winter G. Combinatorial infection and in vivo recombination: a strategy for making large phage antibody repertoires. Nucleic Acids Res. 1993 May 11;21(9):2265–2266. doi: 10.1093/nar/21.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]