Abstract
A core cleavage complex (CCC) consisting of CPSF73, CPSF100, and Symplekin is required for cotranscriptional 3′ end processing of all metazoan pre-mRNAs, yet little is known about the in vivo molecular interactions within this complex. The CCC is a component of two distinct complexes, the cleavage/polyadenylation complex and the complex that processes nonpolyadenylated histone pre-mRNAs. RNAi-depletion of CCC factors in Drosophila culture cells causes reduction of CCC processing activity on histone mRNAs, resulting in read through transcription. In contrast, RNAi-depletion of factors only required for histone mRNA processing allows use of downstream cryptic polyadenylation signals to produce polyadenylated histone mRNAs. We used Dmel-2 tissue culture cells stably expressing tagged CCC components to determine that amino acids 272–1080 of Symplekin and the C-terminal approximately 200 amino acids of both CPSF73 and CPSF100 are required for efficient CCC formation in vivo. Additional experiments reveal that the C-terminal 241 amino acids of CPSF100 are sufficient for histone mRNA processing indicating that the first 524 amino acids of CPSF100 are dispensable for both CCC formation and histone mRNA 3′ end processing. CCCs containing deletions of Symplekin lacking the first 271 amino acids resulted in dramatic increased use of downstream polyadenylation sites for histone mRNA 3′ end processing similar to RNAi-depletion of histone-specific 3′ end processing factors FLASH, SLBP, and U7 snRNA. We propose a model in which CCC formation is mediated by CPSF73, CPSF100, and Symplekin C-termini, and the N-terminal region of Symplekin facilitates cotranscriptional 3′ end processing of histone mRNAs.
Keywords: Symplekin, CPSF73, CPSF100, cotranscriptional, Drosophila, mRNA 3′ end processing
INTRODUCTION
Metazoan pre-mRNAs require extensive processing before becoming mature mRNAs. One important maturation step is 3′ end processing. Canonical polyadenylated [poly(A)] pre-mRNAs are cleaved between a highly conserved AAUAAA and a less conserved G- or G/U-rich downstream element followed by polyadenylation of the upstream mRNA. Maturation of histone pre-mRNAs only requires a single cleavage event between an upstream stem–loop (SL) analogous to the AAUAAA in canonical poly(A) mRNAs, and the histone downstream element (HDE) (Dominski et al. 1999). Histone mRNAs are not polyadenylated, ending in the SL. These different cis elements recruit either histone or canonical poly(A) mRNA specific processing factors. The SL is bound by the stem–loop binding protein (SLBP) while the HDE interacts with the U7 RNA component of the U7 snRNP (Marzluff et al. 2008). The U7snRNP recruits processing factors via an interaction between Lsm11 and FLASH (Burch et al. 2011; Sabath et al. 2013). The highly conserved AAUAAA characteristic of canonical poly(A) mRNAs binds CPSF30 and WDR33 (Chan et al. 2014; Schönemann et al. 2014) and CstF64 contacts the less conserved G- or G/U-rich element (Takagaki and Manley 1997). While both types of mRNAs utilize unique proteins, cleavage and polyadenylation specificity factor (CPSF)73, CPSF100, and Symplekin are essential for proper processing of all metazoan mRNAs (Sullivan et al. 2009). CPSF73 is the endonuclease responsible for catalyzing the cleavage reaction (Ryan et al. 2004; Dominski et al. 2005a; Mandel et al. 2006). CPSF100 forms a heterodimer with CPSF73 and Symplekin acts as a scaffolding protein onto which other cleavage and polyadenylation proteins bind (Takagaki and Manley 2000; Dominski et al. 2005b).
Because the CCC plays an integral role in 3′ end processing of all metazoan pre-mRNAs, determining biologically relevant binding interactions in this complex is important. Details describing CPSF73, CPSF100, and Symplekin interactions are not available as no structural data corresponding to the CCC have been published and X-ray crystal structures of individual components are limited. The three-dimensional structure of human CPSF73 reveals that the N-terminal 460 amino acids contain β-metallo lactamase and β-CASP domains (Fig. 1A; Mandel et al. 2006). The interface of these two domains form the nuclease active site (Mandel et al. 2006). The crystal structure of yeast CPSF100 closely resembles the domain architecture of CPSF73 (Mandel et al. 2006). CPSF100 has both a metallo-β-lactamase-like domain and a β-CASP domain, but CPSF100 active site residues are mutated such that CPSF100 does not function as a nuclease (Fig. 1A; Mandel et al. 2006). Previous experiments led to hypotheses that C-terminal CPSF73/CPSF100 residues may be important for heterodimerization (Jenny et al. 1996). Structures of the N-terminal 271 amino acids of Symplekin reveal seven pairs of α-helices comprising a HEAT domain (Fig. 1A; Kennedy et al. 2009; Xiang et al. 2010). This N-terminal HEAT domain in human Symplekin mediates protein–protein interactions and interacts directly with Ssu72, a RNA polymerase II carboxy-terminal domain (RNAPII CTD) Ser5 phosphatase that is required for mRNA 3′ end processing in yeast (He et al. 2003; Krishnamurthy et al. 2004; Xiang et al. 2010). The C-terminal 85 amino acids of the Symplekin yeast homolog, Pta1, interact with Ysh1 (the yeast CPSF73 homolog) in a directed yeast-two hybrid assay and full-length Pta1 interacts with yeast CPSF100 in an in vitro pull-down assay (Ghazy et al. 2009). Additionally, full-length Pta1 interacts directly with the C-terminus of Ysh1 (Zhelkovsky et al. 2006). While the structure function relationship has been investigated for regions of individual CCC components, molecular details for the complex are sparse.
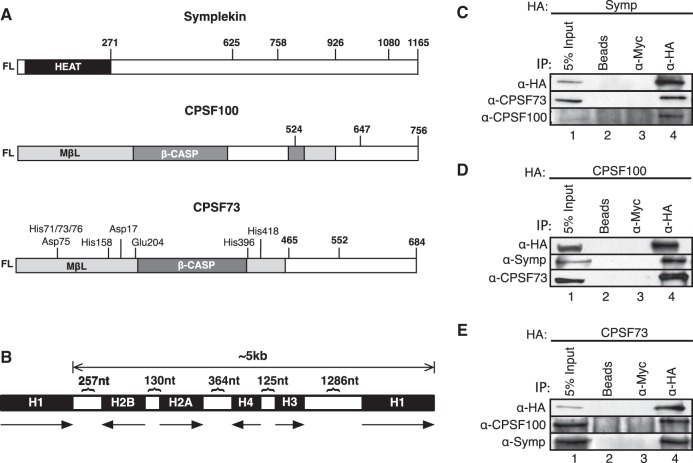
FIGURE 1.
Stably expressed, HA-tagged full-length CCC components interact with endogenous binding partners to form mixed CCCs. (A) Schematic representations of Symplekin, CPSF100, and CPSF73 are shown. HEAT, metallo-β-lactomase (MβL), and β-CASP shaded boxes indicate the location of distinct structural domains. Numbers listed within and above boxes indicate amino acid positions. Those numbers with three letter amino acid abbreviations correspond to catalytic residues. (B) Schematic of one repeat (∼5 kb) within the Drosophila histone gene locus. The direction of transcription is shown directly below each histone gene (H1, H2A, H2B, H3, and H4). The number of nucleotides between each histone gene is shown above. (C–E) Full-length HA-tagged Symplekin (C), CPSF100 (D), and CPSF73 (E) were immunoprecipitated (IP) from whole-cell lysates with an anti-HA antibody. IPs were separated with SDS-PAGE, transferred to a membrane, and probed with anti-HA, anti-Symplekin, anti-CPSF73, and anti-CPSF100 antibodies. Lane 1 is 5% input, lanes 2,3 are controls where IPs were performed in the absence of antibody (beads) or with a nonspecific antibody (anti-Myc), and lane 4 shows the experimental IP. The top panel of each WB set is the IP of HA-tagged protein while the lower panels represent the co-IPs of CCC-binding partners.
We define CPSF73, CPSF100, and Symplekin as the core cleavage complex (CCC). These three proteins form a tight complex in vivo as evidenced by co-immunoprecipitation (co-IP) in stringent buffer conditions and codepletion of the other two CCC factors when one CCC component is RNAi-depleted (Sullivan et al. 2009). Previous investigations in Drosophila tissue culture (Dmel-2) cells of CCC function on endogenous histone mRNAs reveal that this complex is required for proper histone 2A (H2A) 3′ end processing. If a CCC component is RNAi-depleted, cleavage at the site between the SL and the HDE is less efficient resulting in a read through (RT) H2A mRNA with an extended 3′ untranslated region (UTR). (Wagner et al. 2007; Sullivan et al. 2009). The RT product is the dominant misprocessed H2A product in these cells. RNAi-depletion of histone-specific factors U7 snRNA, SLBP, and FLASH also result in less-efficient use of the proper H2A processing site. But, in contrast to RNAi-depletion of CCC components, these cells can use cryptic poly(A) signals downstream from the proper cleavage site to process these mRNAs resulting in poly(A) histone mRNAs (Lanzotti et al. 2002; Sullivan et al. 2009; Tatomer et al. 2014).
mRNA 3′ end processing by the CCC occurs cotranscriptionally and coupling of RNA synthesis and processing is coordinated by modifications in the repetitive heptad of the RNAPII CTD (Greenleaf 1993; Hsin and Manley 2012). Direct interactions between the RNAPII CTD, CPSF and CstF have been observed and removal of the RNAPII CTD causes inefficient mRNA 3′ end processing (McCracken et al. 1997). 3′ end processing factor CPSF73 occupies both 5′ and 3′ ends of candidate mammalian genes while polyadenylation factors CstF50 and CstF77 predominately occupy the 3′ ends (Glover-Cutter et al. 2008; Ni et al. 2008; Sullivan et al. 2009). Additional data show that the CPSF complex is recruited to the 5′ end by TFIID and remains associated with RNAPII throughout transcription elongation (Dantonel et al. 1997). Recruitment of mRNA 3′ end processing factors to the RNAPII CTD correlates with specific phosphorylation of CTD heptad repeat positions Ser5 and Ser2. In Drosophila tissue culture cells, impairment of Ser2 phosphorylation results in less-efficient recruitment of mRNA 3′ end processing factors to the 3′ ends of genes (Ni et al. 2008). Finally, the N-terminal 271 amino acids of human Symplekin interact with Ssu72, a RNAPII CTD Ser5 phosphatase that is required for mRNA 3′ end processing in yeast (He et al. 2003; Krishnamurthy et al. 2004; Xiang et al. 2010). Taken together, these data support a model in which mRNA 3′ end processing is cotranscriptional.
Previous work indicates that histone pre-mRNA 3′ end processing is also cotranscriptional. In Drosophila, the histone gene locus consists of 100 tandem repeats of a 5-kb region containing a single copy of each replication-dependent histone gene separated by <500 nt (Fig. 1B). Each alternating gene is transcribed from the opposite strand, requiring coordinated transcription termination and 3′ end processing to prevent transcription into neighboring genes (Sullivan et al. 2009). Evidence supporting this model is observed in histone mRNA 3′ end sequences obtained from the modENCODE project (Celniker et al. 2009). Sequencing reads are not observed downstream from the histone 3′ SL and cleavage site. Additionally, RNAPII pause sites downstream from the histone mRNA 3′ end cleavage site have been identified that affect histone mRNA processing (Adamson and Price 2003) and CCC components Symplekin and CPSF73 ChIP to histone 3′ ends (Sullivan et al. 2009). Finally, phosphorylation of RNAPII CTD heptad residue Thr4 is specifically required for histone mRNA 3′ end processing. SLBP and CPSF100 are not recruited to histone genes if Thr4 is not phosphorylated (Hsin et al. 2011). Collectively, these data support a model in which pre-mRNA 3′ end processing, transcription and termination are tightly coordinated.
Recent X-ray crystallographic investigations of CPSF73, CPSF100, and Symplekin have focused on individual components and have resulted in only partial structures (Mandel et al. 2006; Kennedy et al. 2009; Xiang et al. 2010). Additionally, previous analyses of CCC component binding interactions are incomplete concentrating on only two of the three factors or were performed in vitro or using bioinformatics (Jenny et al. 1996; Kyburz et al. 2003; Dominski et al. 2005b; Zhelkovsky et al. 2006). To further define CPSF73, CPSF100, and Symplekin binding interactions, we developed a novel Drosophila cell culture system in which tagged, mutant components were individually expressed. Interaction of these exogenous factors with endogenous-binding partners identified important binding regions of CPSF73, CPSF100, and Symplekin in vivo. Furthermore, the activity of mutant component containing CCCs was assessed on an endogenous histone mRNA to demonstrate domains important for cotranscriptional histone mRNA 3′ end processing.
RESULTS
Development and validation of a Dmel-2 expression system to study in vivo protein interactions in the CCC
Our goal was to understand CCC interactions in vivo. Therefore, we needed to develop a system to express tagged CCC components that could both integrate into complexes with endogenous-binding partners (form mixed CCCs) and be immunoprecipitated (IPed). To accomplish this goal, full-length, wild-type CPSF73, CPSF100, and Symplekin sequences were first cloned downstream from a constitutive promoter in plasmids conferring either an N- or C-terminal HA tag. These plasmids were then transfected into Dmel-2 cells and maintained under selection to create polyclonal Dmel-2 cells stably expressing CCC factors. Stable expression of CCC factors did not affect the viability of Dmel-2 cells. Extracts used for IP were prepared from these cells and anti-HA Western blot (WB) confirmed expression of full-length HA-tagged CCC proteins (Fig. 1C–E, lane 1 or Fig. 2B). To validate that stably expressed CCC factors could be IPed and form mixed CCCs, each CCC component was individually IPed from Dmel-2 stable cell line whole-cell lysates followed by WB for co-IP of endogenous-binding partners. IP of HA-tagged Symplekin successfully co-IPed endogenous CPSF73 and CPSF100 (Fig. 1C). IP of HA-tagged CPSF100 co-IPed endogenous Symplekin and CPSF73 (Fig. 1D). IP of HA-tagged CPSF73 co-IPed endogenous Symplekin and CPSF100 (Fig. 1E). In each case, significant amounts of endogenous-binding partners were observed (Fig. 3B,D,F). No IP of exogenous factors or co-IP of endogenous proteins was observed in no (Beads) or nonspecific (α-Myc) antibody controls (Fig. 1C–E, lanes 2,3). These data indicate that HA-tagged CCC components can be stably expressed in Dmel-2 cells without affecting cell viability. Also, the HA fusion does not prevent CCC factor expression or CCC formation with endogenous-binding partners. RNAi-depletion of endogenous CCC factors is not required to assess interactions within the CCC.
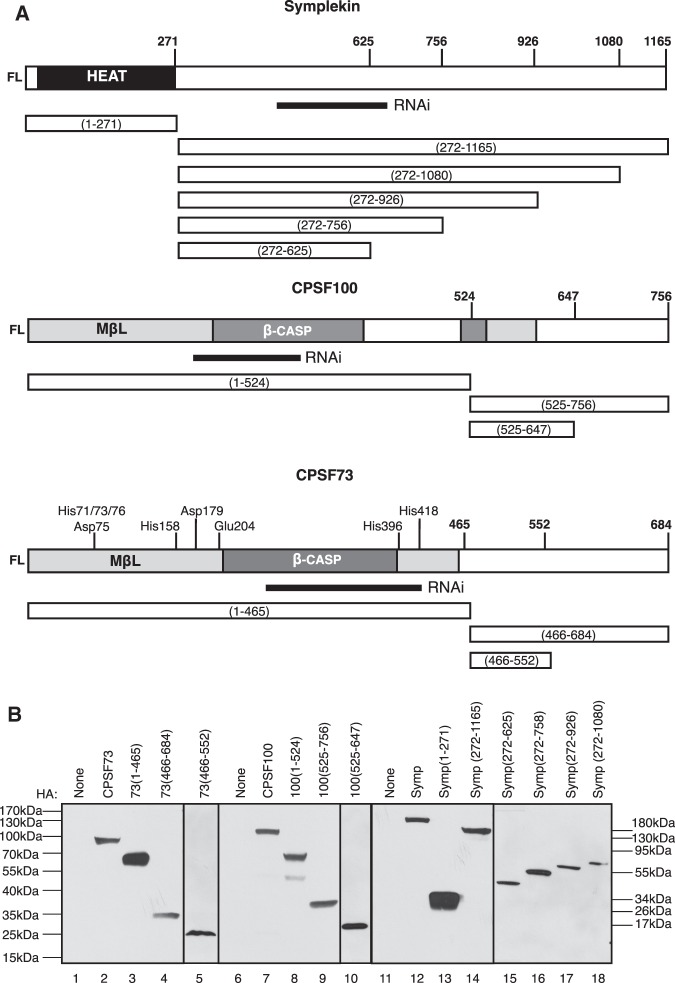
FIGURE 2.
HA-tagged deletion mutants of Symplekin, CPSF100, and CPSF73 can be stably expressed in Dmel-2 cells. (A) Schematics of Symplekin (top), CPSF100 (middle), and CPSF73 (bottom) deletion mutants used for in vivo binding assays are shown below each full-length protein. Each mutant is defined by the amino acids contained within the deletion, for example (1–271). The C-terminal amino acid of each mutant is written above the full-length CCC factor. The nucleotides targeted by dsRNA are defined by a solid rectangle. (B) HA-tagged full length (lanes 2,7,12) and N- and C-terminally truncated CPSF73 (lanes 3–5), CPSF100 (lanes 8–10), and Symplekin (lanes 13–18) deletion mutants were stably expressed in Dmel-2 cells. Whole-cell lysates from these strains were separated with SDS-PAGE, transferred to a membrane, and probed with an anti-HA antibody to confirm expression. Amino acids comprising the deletion mutant are shown following the protein name above the WB. Molecular weight (MW) markers for lanes 1–14 are on the left. MW markers for lanes 15–18 are on the right.
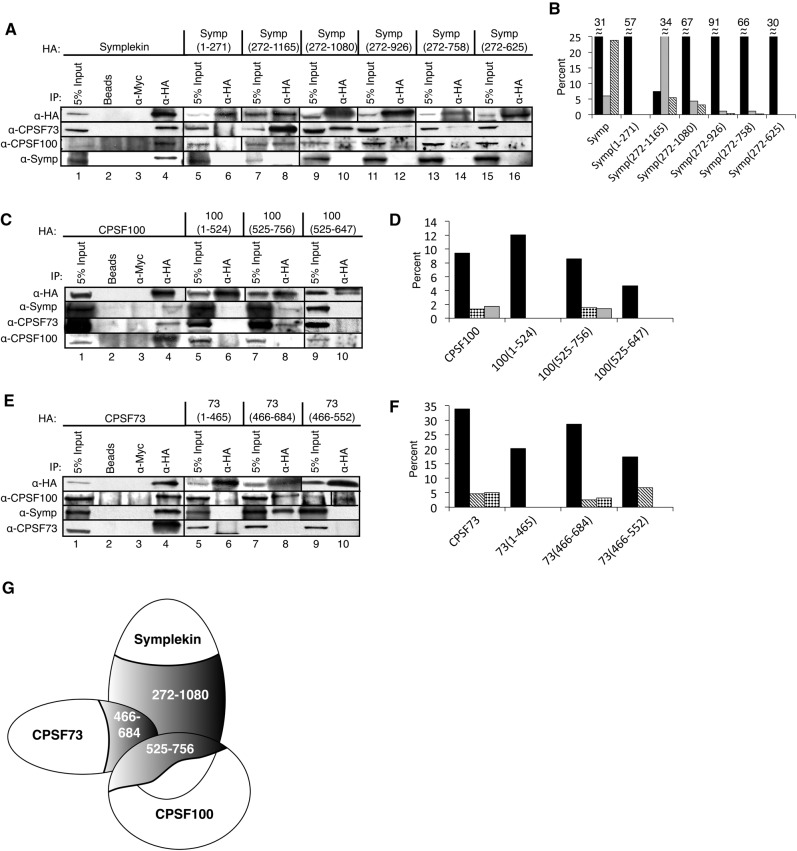
FIGURE 3.
Symplekin amino acids 272–1080, CPSF100 amino acids 525–756, and CPSF73 amino acids 465–684 are sufficient for CCC formation. (A,C,E) HA-tagged Symplekin mutants (A), HA-tagged CPSF100 mutants (C), and HA-tagged CPSF73 mutants (E) were IPed, separated, and probed with HA- and CCC-specific antibodies. Numbers under the protein name correspond to the amino acid residues comprising each mutant. IP of full-length proteins and controls (lanes 1–4) are included for reference. (B,D,F) Percentage of IP or co-IPed proteins corresponding to WBs in A, C, and E are plotted. Black bars are percent HA IP, gray bars are percent CPSF73 IP, bars with diagonal lines are percent CPSF100 IP, and bars with black diamonds are percent Symplekin IP. In (B), the numbers above the bars correspond to percent IP for that sample. (G) A CCC-binding model is shown. The C-termini of Symplekin, CPSF73, and CPSF100 are sufficient for CCC formation.
Symplekin amino acids 927–1080, CPSF100 amino acids 648–756, and CPSF73 amino acids 552–684 are required for CCC formation
IP of mixed CCCs from Dmel-2 whole-cell lysates confirms that CCCs having both exogenous and endogenous components can be formed and isolated. Our next goal was to determine which regions of CPSF73, CPSF100, and Symplekin are required for CCC formation. To address this, we constructed N- and C-terminal CPSF73, CPSF100, and Symplekin mutants. Deletion positions were determined using Phyre2 prediction software (Kelley and Sternberg 2009) and were only made in disordered regions (Fig. 2A). Symplekin mutants were amino acids 1–271 (N-terminal), 272–1165 (C-terminal), 272–1080, 272–926, 272–756, and 272–625. CPSF100 mutants were amino acids 1–524 (N-terminal), 525–756 (C-terminal), and 525–647. CPSF73 mutants were amino acids 1–465 (N-terminal), 466–684 (C-terminal), and 466–552. These mutants were constructed, transfected, and stably expressed as their full-length counterparts in Dmel-2 cells. Expression was confirmed in whole-cell lysates with an anti-HA WB (Fig. 2B). All mutants IP with an α-HA antibody were detected by WB with an α-HA antibody (Fig. 3A,C,E, top WB).
The large C-terminal HA-tagged Symplekin mutant (Symp(272–1165)) binds both CPSF73 and CPSF100, while the N-terminal mutant (Symp(1–271)) binds neither protein (Fig. 3A, lanes 6,8, 3B). Symp(272–1165) IPs more CPSF73 than full-length Symplekin (Fig. 3B). Increasing deletion of C-terminal residues from Symp(272–1165) results in progressively less-efficient CCC formation (Fig. 3B). Symp(272–1080) co-IPs CPSF73 comparable with full-length Symplekin (Fig. 3B). Symp(272–926) and Symp(272–758) also bind CPSF73 and CPSF100, but to a lesser extent than full-length Symplekin (Fig. 3B). Symp(272–625) does not co-IP CPSF73 or CPSF100 (Fig. 3B). Two additional Symplekin mutants, Symp(272–525) and Symp(272–395), having even fewer C-terminal residues also did not co-IP endogenous CPSF73 or CPSF100 (data not shown). These data indicate that the first 271 amino acids of Symplekin are not required for CCC formation. Symplekin amino acids 272–1080 are sufficient for CCC formation while amino acids 927–1080 are essential for interaction with CPSF73/CPSF100.
IP of HA-tagged C-terminal CPSF100 (100(525–756)) also resulted in co-IP of endogenous CCC-binding partners (Fig. 3C,D); 100(525–756) binds CPSF73 and Symplekin comparable with full-length CPSF100 (Fig. 3D), while N-terminal 100(1–524) does not co-IP CCC-binding partners (Fig. 3D). Deletion of an additional 109 amino acids from the C-terminus of the CPSF100 C-terminal mutant (100(525–647)) abolishes co-IP of CPSF73 and Symplekin (Fig. 3D). These data indicate that amino acids 525–756 of CPSF100 are sufficient for CCC formation and amino acids 1–524 are dispensable for interaction with CPSF73 and Symplekin. CPSF100 amino acids 648–756 are required for CCC formation.
IP of HA-tagged C-terminal CPSF73 (73(466–684)) co-IPed Symplekin and CPSF100 comparable with full-length CPSF73 (Fig. 3E,F), while the N-terminal CPSF73 mutant (73(1–465)) cannot participate in CCC formation (Fig. 3F). Deletion of an additional 133 amino acids from the C-terminus of the C-terminal CPSF73 mutant (73(466–552)) prevents Symplekin binding, but interaction with CPSF100 is maintained (Fig. 3F). These data indicate that CPSF73 amino acids 466–684 are sufficient for interaction with CPSF100 and Symplekin while amino acids 1–465 cannot facilitate CCC formation. Amino acids 553–684 are essential for binding Symplekin while CPSF100 may interact with CPSF73 amino acids 466–552.
To ensure that phenotypes observed for mutant CCCs are attributable to the mutant CCC component, we investigated inclusion of corresponding full-length endogenous CCC components in CCCs having HA-tagged full-length and mutant factors. CCCs including full-length HA-tagged proteins do not also contain endogenous CCC factors (Fig. 3A,C,E, bottom WB, lanes 1–4). While CCC antibodies react with both endogenous and exogenous CCC factors, the full-length HA-tagged protein in each IP is slightly larger than the corresponding band in the input indicating that the HA-tagged protein dominates in the IP (Fig. 3A,C,E, bottom WB, cf. lanes 1,4). The HA-tagged protein is expressed at less than endogenous levels and therefore is not visible in the input (Fig. 3A,C,E, bottom WB, lane 1). All HA-tagged deletion mutants differ significantly in size from their full-length counterparts allowing detection of full-length endogenous CCC components in CCC IPs. None of the mixed complexes contain the corresponding endogenous-binding partners (Fig. 3A,C,E, bottom WB).
Together, these data support a model in which one Symplekin, one CPSF73, and one CPSF100 interact to form the CCC. Generally, the C-termini of each protein mediate CCC formation. Specifically, amino acids 272–1080 of Symplekin, amino acids 466–684 of CPSF73, and amino acids 525–756 of CPSF100 are sufficient for binding, while Symplekin amino acids 927–1080, CPSF100 amino acids 648–756 and CPSF73 amino acids 552–684 are required for CCC formation (Fig. 3G). CPSF73 amino acids 466–552 may contact CPSF100 while CPSF73 C-terminal residues 553–684 are essential for binding Symplekin.
HA RNAi-resistant CCC factors express simultaneously with RNAi-depletion of endogenous proteins and behave as wild-type CCC components
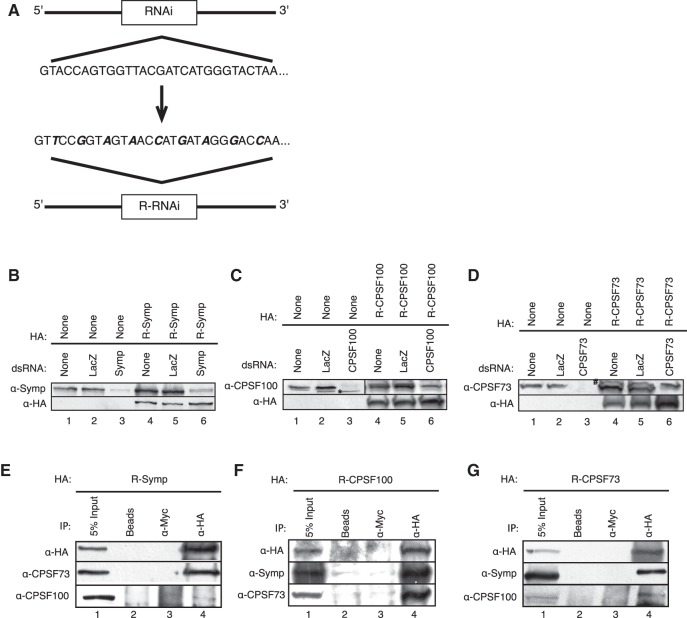
After elucidating binding interactions in the CCC, we wanted to investigate in vivo activity of mutant CCCs. While previous experiments with HA-tagged CPSF73, CPSF100, or Symplekin containing CCCs were performed in the presence of endogenous complexes, functional wild-type CCCs mask in vivo activity of mutant CCCs. To focus on only mixed CCCs, we RNAi-depleted endogenous CCC components while expressing RNAi-resistant (RNAi-R) CCC factors. While this powerful technique has been utilized before (Ruepp et al. 2011; Chen et al. 2013), it is a novel way to study CCC function. RNAi-R mRNAs were constructed by introducing silent mutations at the wobble base of each codon in the RNAi-targeted region (Fig. 4A; Schulz et al. 2009).
FIGURE 4.
RNAi-resistant (RNAi-R) CCC factors are expressed during simultaneous RNAi-depletion of corresponding endogenous proteins and form complexes with endogenous-binding partners. (A) RNAi-R CCC components were constructed by changing the third (wobble) base of every codon within the region targeted by exogenously introduced dsRNA. (B–D) HA-tagged R-Symplekin (B), R-CPSF100 (C), and R-CPSF73 (D) were expressed in Dmel-2 cells simultaneously RNAi-depleted of the corresponding endogenous protein. The proteins were IPed, separated, and probed as described in Figure 1. Lane 3 of each top panel shows RNAi-depletion of endogenous CCC components. Simultaneous RNAi-depletion of endogenous CCC factors and exogenous expression of the corresponding protein is shown in lane 6. The “*” denotes a slightly faster migrating cross-reacting band. The “#” identifies HA-CPSF73. (E–G) RNAi-R full-length CCCs were IPed as described in Figure 1. R-Symplekin (E), R-CPSF100 (F), and R-CPSF73 (G) IP were detected with anti-HA antibody (lane 4, top panels). co-IP was assessed with anti-CCC factor antibodies. Beads only (lane 2) and α-Myc (lane 3) are nonspecific controls.
Stable lines expressing HA-tagged full-length R-Symplekin, R-CPSF73, and R-CPSF100 were RNAi-depleted of the corresponding endogenous proteins. Symplekin, CPSF73, and CPSF100 are efficiently RNAi-depleted when compared with either no double-stranded or nonspecific RNA (LacZ) controls (Fig. 4B–D, top panels, lanes 1–3). WBs with an anti-HA antibody show that RNAi-R CCC components are successfully expressed in the presence of RNAi targeting the corresponding CCC factors (Fig. 4B–D, bottom panels, lane 6). Concurrent expression of HA-tagged proteins and RNAi-depletion is best observed for CPSF73; endogenous CPSF73 is clearly RNAi-depleted while the slightly larger HA-tagged, R-CPSF73 is expressed (Fig. 4D, lanes 4–6). This phenomenon is also observed for Symplekin, although HA-tagged Symplekin is less of the total Symplekin pool as assessed by comparing the amount of HA-tagged and endogenous Symplekin in lane 6 (Fig. 4B, lanes 4–6). HA-tagged and endogenous CPSF100 are not resolved, but reduced CPSF100 levels indicate RNAi-depletion of endogenous protein with concurrent expression of HA-tagged CPSF100 (Fig. 4C, lanes 4–6). These experiments were repeated for each HA-tagged CCC factor mutant to ensure that RNAi-R proteins were successfully expressed simultaneously with RNAi-depletion of endogenous CCC components (Supplemental Fig. 1). Finally, RNAi-R CCC factors IP endogenous-binding partners much like their wild-type counterparts as compared with no and nonspecific antibody controls (Fig. 4E–G).
The C-terminal 232 amino acids of CPSF100 are sufficient for formation of functional CCCs
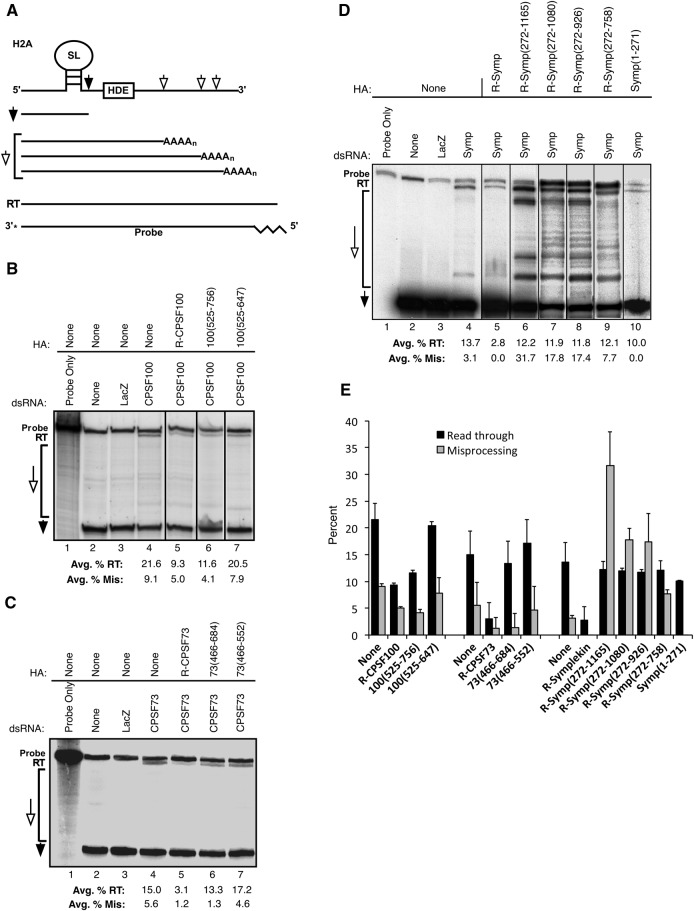
To investigate in vivo activity of mutant CCC complexes, we visualized histone H2A 3′ ends using an S1 nuclease assay. In Drosophila, the 3′ ends of replication-dependent histone mRNAs are properly processed with a single cleavage between a conserved stem–loop (SL) and the histone downstream element (HDE) (Fig. 5A, filled arrow). Misprocessing of H2A occurs when downstream cryptic polyadenylation signals recruit 3′ end processing factors resulting in polyadenylated H2A mRNAs (Fig. 5A, open arrows). When CPSF73, CPSF100, or Symplekin are RNAi-depleted, neither the SL/HDE nor the cryptic polyadenylation signals can be utilized for histone mRNA processing, resulting in an extended 3′ UTR or read through (RT) product (Fig. 5A). These processed H2A mRNAs are clearly observed following hybridization with a radioactively labeled probe and S1 nuclease digestion (Fig. 5A, probe). This assay has been used previously to investigate H2A 3′ end processing (Lanzotti et al. 2002; Sullivan et al. 2009). We performed each S1 nuclease assay in duplicate followed by quantitation of the amount of properly processed, 3′ end misprocessed and RT H2A mRNAs. Averages of percent nonproperly processed H2A mRNAs are included.
FIGURE 5.
CCCs with Symplekin C-terminal residues can use downstream poly(A) sites for H2A mRNA 3′ end processing. (A) A schematic of the histone 2A (H2A) mRNA 3′ end is shown. Cis elements required for proper processing are the stem–loop (SL) and the histone downstream element (HDE). The appropriate cleavage site and the properly processed H2A 3′ end are marked with filled arrows. Downstream cryptic polyadenylation signals and misprocessed products resulting from use of these signals are labeled with open arrows. If neither the proper cleavage site nor cryptic polyadenylation signals can be used for processing, an extended 3′ UTR product results (RT). The 3′ end labeled DNA S1 nuclease assay probe will hybridize to all of these products. The 5′ 20 nt of the probe (zig-zag line) cannot anneal to the H2A 3′ end products. (B) H2A 3′ ends in Dmel-2 cells containing wild-type and mutant CPSF100 CCCs and control cells were visualized using a S1 nuclease assay. Addition of dsRNA is indicated just above the gel while expression of either full-length or mutant CCC components is described above the solid line. Gel positions of potential products are marked on the left with the same labeling as in (A). (C) H2A 3′ ends in Dmel-2 cells containing wild-type and mutant CPSF73 CCCs and control cells were visualized using a S1 nuclease assay. The gel is labeled as in (B). (D) An S1 nuclease assay was performed to visualize endogenous H2A mRNA 3′ ends in Dmel-2 cells with Symplekin mutant CCCs. The gel is labeled as in (B). (E) The percentages of RT, misprocessed, and properly processed H2A mRNAs were calculated for two independent experiments. The averages of RT and misprocessed products for each experiment were plotted with the corresponding standard deviation and the values are indicated below each gel. The black bars are the percent of RT product while the gray bars represent percent misprocessed H2A mRNA.
RNAi-depletion of CPSF100 causes H2A misprocessing when compared with negative controls (Fig. 5B, cf. lanes 2,3 to lane 4). RT mRNA is 21.6% of total H2A mRNA in this sample and is the primary misprocessed product (Fig. 5E). Previous experiments are consistent with this result (Sullivan et al. 2009). When R-CPSF100 is expressed simultaneously with RNAi-depletion of endogenous CPSF100, the amount of RT is 9.3% (Fig. 5B,E). Expression of R-CPSF100 partially rescues the RNAi-depletion defect showing that mixed CCC are functional. Expression of CPSF100(525–756) results in 11.6% RT H2A misprocessing product indicating this mutant also rescues the effect of CPSF100 RNAi-depletion (Fig. 5B,E). Expression of CPSF100(525–647) does not rescue CPSF100 RNAi-depletion as indicated by RT levels (20.5%) comparable with the CPSF100 RNAi sample (Fig. 5B,E). Double-stranded RNA for RNAi targets the N-terminal region of full-length CPSF100 (Fig. 2A), therefore, RNA-R forms of CPSF100(525–756) and CPSF100(525–647) did not have to be constructed. Collectively, these data indicate that CCCs containing the minimal region of CPSF100 required for CCC formation are functional and that the first 524 amino acids of CPSF100 are dispensable for proper H2A processing.
CCCs with the C-terminal 218 amino acids of CPSF73 misprocess histone mRNAs
To investigate the in vivo activity of CCC containing CPSF73 deletion mutants, a similar approach was used. An S1 nuclease protection assay using a probe to H2A (Fig. 5A) shows properly processed H2A mRNA in wild-type Dmel-2 cells and controls treated with nontargeting dsRNA (Fig. 5C, lanes 2,3). When CPSF73 is RNAi-depleted, RT mRNA is 15.0% of the total H2A product (Fig. 5C,E). Few products resulting from use of downstream cryptic poly(A) signals were observed (5.6%). These data are consistent with previously published findings (Sullivan et al. 2009). When full-length R-CPSF73 is expressed simultaneously with RNAi-depletion of endogenous CPSF73, the H2A 3′ end processing defect is partially rescued as only 3.0% of RT H2A is present in this sample (Fig. 5C,E). Therefore, mixed CCC with full-length R-CPSF73 are functional. When CPSF73(466–684) is expressed, 13.3% RT H2A misprocessing is observed (Fig. 5C,E). Although this mutant forms CCCs (Fig. 3E), CPSF73(466–684) does not rescue H2A misprocessing, presumably because it does not contain the CPSF73 active site (Fig. 2A). RT H2A misprocessed 3′ ends also dominate in samples where CPSF73(466–552) is expressed (17.2% of H2A) (Fig. 5C,E), indicating that this mutant does not rescue H2A misprocessing. Neither CPSF73(466–684) nor CPSF73(466–552) act dominantly as cells concurrently expressing a mutant and wild-type full-length endogenous CPSF73 do not result in H2A misprocessing (Supplemental Fig. 2B). As the dsRNA used for RNAi-depletion of CPSF73 targets the N-terminal region (Fig. 2A), RNAi-R forms of CPSF73(466–684) and CPSF73(466–552) did not need to be constructed. Collectively, these data indicate that although CPSF73(466–684) can form CCCs, these CCCs are nonfunctional.
CCCs containing Symplekin lacking the first 271 amino acids use downstream cryptic poly(A) sites for H2A 3′ end processing
We also investigated the activity of mutant Symplekin CCCs by visualizing the 3′ ends of H2A mRNAs in Dmel-2 cells containing these complexes (Fig. 5D). RNAi-depletion of Symplekin causes H2A misprocessing when compared with negative controls (Fig. 5D, cf. lane 4 to lanes 2,3). RT H2A mRNA is the primary misprocessed H2A product (13.7% of total H2A mRNA) as has been observed previously (Sullivan et al. 2009). When R-Symplekin is expressed simultaneously with RNAi-depletion of endogenous Symplekin, RT H2A mRNA is 2.8% of total H2A (Fig. 5D,E) indicating that exogenous Symplekin can partially rescue the defect caused by RNAi-depletion of endogenous Symplekin. As with CCCs including R-CPSF73 and R-CPSF100, R-Symplekin containing CCCs are functional. When Symp(272–1165) is expressed in cells lacking endogenous Symplekin, misprocessed H2A dramatically increases to 31.7% and use of cryptic poly(A) signals is abundant (Fig. 5D,E). We continue to observe this pattern as residues are progressively removed from the Symplekin C-terminus, although the total amount of misprocessed H2A decreases; 17.8% of H2A mRNAs are misprocessed by Symp(272–1080) containing CCCs, 17.4% of H2A mRNAs are misprocessed by Symp(272–926) containing CCCs, and 7.7% of H2A mRNAs are misprocessed by Symp(272–758) (Fig. 5D,E). When Symp(1–271) is expressed in cells RNAi-depleted of endogenous Symplekin, RT H2A mRNA is again the primary misprocessing product (10.0% of total H2A mRNAs) (Fig. 5D,E). None of the Symplekin mutants act dominantly as cells concurrently expressing a mutant and wild-type full-length endogenous Symplekin do not result in H2A misprocessing (Supplemental Fig. 2A).
These data indicate that 3′ end misprocessed H2A mRNAs are the dominant misprocessed product in samples having CCCs with only Symplekin C-terminal amino acids. These complexes can use downstream poly(A) signals for 3′ end processing of H2A mRNAs. This is a novel result as only read through transcription has been observed when full-length CCC components are RNAi-depleted or with nonfunctional CCCs.
DISCUSSION
Pre-mRNA 3′ end processing is an essential step in mRNA maturation. The 3′ ends of all metazoan mRNAs are cleaved by the core cleavage complex (CCC) consisting of CPSF73, CPSF100, and Symplekin (Sullivan et al. 2009). Interactions in this important complex have not been investigated. Furthermore, relationships between protein domains in the CCC and their function in cotranscriptional histone mRNA 3′ end processing have not been characterized. Here, we use a Dmel-2 tissue culture system to determine that the C-terminal ∼200 amino acids of CPSF73 and CPSF100 and amino acids 272–1080 of Symplekin are sufficient for CCC formation. Additionally, in vivo assessment of mutant CCC activity reveals that the N-terminal 524 amino acids of CPSF100 are not required for formation of CCCs that properly process histone mRNAs. Lastly, CCCs containing Symplekin lacking the first 271 amino acids can use poly(A) signals downstream from the proper cleavage site for 3′ end processing. This is a phenotype also observed when histone-specific processing factors are RNAi-depleted but in contrast to the read through product normally produced when CCC components are knocked down. These data support a model in which the first 271 amino acids of Symplekin facilitate cotranscriptional 3′ end processing of histone mRNAs.
A novel in vivo approach to study CCC interactions and activity
We took a novel in vivo approach to investigate both CCC interactions and mutant CCC histone mRNA 3′ end processing. To identify important CCC component binding regions, tagged mutant CCC components were first stably expressed in Dmel-2 cells. These proteins were then immunoprecipitated and co-IP of endogenous CCC-binding partners was identified. This approach was validated using full-length, wild-type tagged CCC factors for IP and ensuring co-IP of endogenous CCC components (Fig. 1C–E). Unlike previous investigations of interactions within multiprotein complexes involved in RNA processing, mixed CCCs having both exogenous and endogenous components did not require RNAi-depletion of endogenous factors (Chen et al. 2013; Lyons et al. 2014).
Analysis of mutant CCC activity in this system necessitated an additional modification. Previous data indicate that misprocessed histone mRNAs are a small percentage of total histone mRNA in Dmel-2 cells RNAi-depleted of CCC components (Sullivan et al. 2009). Therefore, we hypothesized that misprocessed histone mRNA resulting from mutant CCC activity in our system would not be detected in the presence of functional wild-type CCCs. To alleviate this problem, we constructed and stably expressed RNAi-resistant CCC mutants simultaneously with RNAi-depletion of the corresponding endogenous CCC component (Fig. 4). This approach ensured we were only observing activity of CCCs containing mutant factors. The activity of these complexes was then assessed on H2A 3′ ends. Expression of tagged full-length, wild-type CCC components rescued histone mRNA 3′ end processing when the corresponding endogenous component was RNAi-depleted indicating that mixed CCCs are also functional (Fig. 5). This novel Dmel-2 cell culture system could be used to further investigate interactions in the CCC using CPSF73, CPSF100, and Symplekin point mutants. Other factors required for cotranscriptional histone mRNA 3′ end processing could also be studied using this approach.
The C-termini of CPSF73, CPSF100, and Symplekin are sufficient for CCC formation
Previous to this study, little was known about CCC interactions in vivo. The three-dimensional structures of the first ∼400 amino acids of human CPSF73 and yeast CPSF100 (Mandel et al. 2006) and the N-terminal ∼300 amino acids of Drosophila and human Symplekin were individually determined by X-ray crystallography (Kennedy et al. 2009; Xiang et al. 2010, 2012). These structures include important residues comprising the CPSF73 active site (Fig. 2) and a N-terminal Symplekin HEAT domain interacting with the RNAPII CTD phosphatase Ssu72, but do not incorporate C-terminal CPSF73, CPSF100, and Symplekin regions important for CCC formation. Additionally, previous bioinformatics, yeast-two hybrid and pull-down analyses have led to hypotheses that the C-termini of CPSF100, CPSF73, and Symplekin interact (Jenny et al. 1996; Kyburz et al. 2003; Dominski et al. 2005b; Zhelkovsky et al. 2006; Mandel et al. 2008; Ghazy et al. 2009), but these studies never included all three CCC components, instead focusing on only two factors. The experiments described here are the first systematic, in vivo approach to defining simultaneous CPSF73, CPSF100, and Symplekin binding interactions in the CCC. We provide evidence that amino acids 272–1080 of Symplekin, amino acids 466–684 of CPSF73 and amino acids 525–756 of CPSF100 are sufficient for CCC formation.
While our data are generally consistent with previous hypotheses and observations, our experiments reveal the following novel insights. Symplekin amino acids 272–1165 co-IP more CPSF73 than full-length Symplekin, yet removing 85 additional C-terminal amino acids (Symp(272–1080)) results in CPSF73 binding comparable with full-length Symplekin (Fig. 3A,B). These data indicate that while the N-terminal 271 amino acids of Symplekin are not required for CCC formation, these residues inhibit CPSF73 binding to the C-terminal end of Symplekin. The C-terminal 85 amino acids of Symplekin enhance CPSF73 binding. Also, CPSF73(466–552) maintains binding to CPSF100 but not Symplekin, while CPSF73(466–684) IPs both CPSF100 and Symplekin (Fig. 3E,F). These data support a model in which CPSF73 amino acids 466–552 interact with CPSF100 while CPSF73 amino acids 466–684 are required for binding Symplekin.
The in vivo binding data presented here, considered with previous X-ray crystal structure and bioinformatic analyses, indicate dramatic regional separation of functions within CPSF73, CPSF100, and Symplekin. Our N-terminal CPSF73(1–465) mutant encompasses the entire active site (Fig. 2A; Mandel et al. 2006), but these amino acids are not required for CCC formation (Fig. 3E,F). The region of CPSF73 responsible for endonuclease activity is completely distinct from the residues required for CCC formation. We hypothesize that use of an independent domain for CCC formation allows substrate RNA access to the CPSF73 active site. Additionally, CPSF100 amino acids 648–756 are required for CCC formation (Fig. 3C,D). None of the residues comprising the β-CASP or the mettalo-β-lactamase domain are contained within this region of CPSF100. The first 271 amino acids of Symplekin comprise a HEAT domain that can interact with and stimulate activity of RNAPII CTD phosphatase Ssu72 (Xiang et al. 2010). This function of Symplekin is confined to the N-terminal region while C-terminal residues are required for CCC formation. Data presented here support a model in which each CPSF73, CPSF100, and Symplekin domain has a discreet function related to either mRNA 3′ end processing or CCC formation.
Poly(A) sites downstream from the H2A 3′ end processing site are used inefficiently when the number of functional CCCs is low
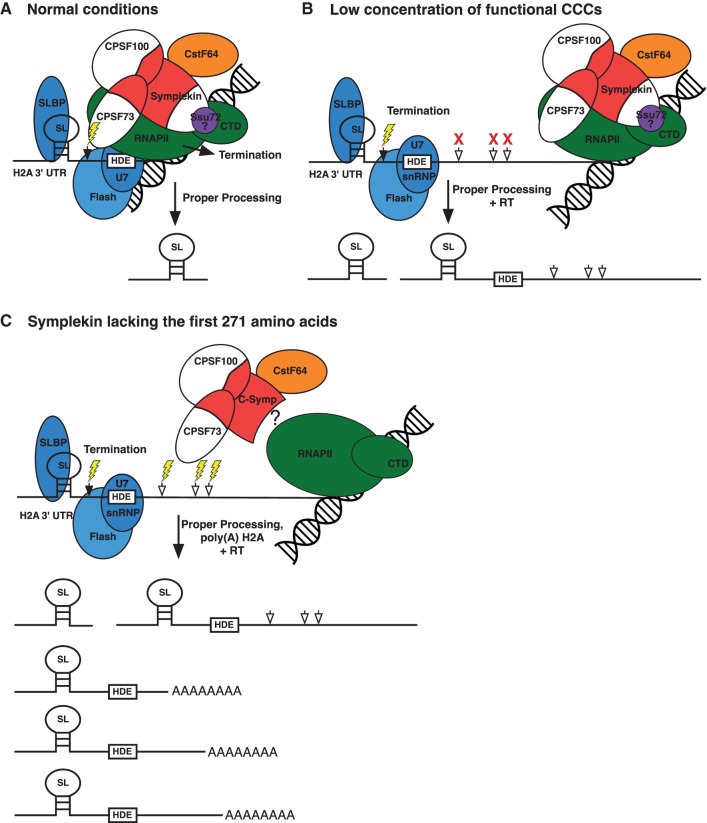
In Drosophila, histone mRNAs are cotranscriptionally processed by the CCC followed by RNAPII termination (Fig. 6A; Sullivan et al. 2009). These events require SLBP binding to the SL, U7 snRNP interaction with the HDE and are coordinated by FLASH (Sabath et al. 2013). Interaction of the CCC with the RNAPII CTD may not be direct, but instead mediated by an additional protein(s). For instance, Ssu72 links Symplekin to the RNAPII CTD in yeast and humans (Xiang et al. 2010). Under normal conditions, RNAPII pauses ∼35 nt downstream from the histone mRNA 3′ end processing site (Adamson and Price 2003). Presumably, this pause facilitates recruitment of all trans factors and allows for correct alignment of the RNAPII associated CCC to the cleavage site. Processing is followed by transcription termination. These events result in a proper 3′ processing of histone mRNAs (Fig. 6A).
FIGURE 6.
Histone mRNA 3′ end processing models. (A) A model for cotranscriptional histone mRNA 3′ end processing under normal conditions is shown. Following transcription of the cleavage site (black arrow) and HDE, RNAPII pauses allowing recruitment and proper orientation of the CCC, SLBP, U7 snRNP, and FLASH to the cleavage site. This results in H2A mRNA processing at the most proximal cleavage site (lightning bolt) and transcription termination. (B) A model for cotranscriptional histone mRNA 3′ end processing under low CCC concentration conditions is shown. If a CCC can be cotranscriptionally recruited to the cleavage site (black arrow), the H2A mRNA 3′ end will be properly processed (lightning bolt). When no functional CCC can be recruited, RNAPII continues transcribing and use of downstream poly(A) sites (white arrows) is inefficient resulting in read through (RT) H2A mRNAs. (C) A model for H2A mRNA 3′ end processing by CCCs containing Symplekin lacking the first 271 amino acids is shown. A high concentration of functional CCCs containing N-terminally truncated Symplekin can use downstream poly(A) sites (white arrows) for H2A mRNA 3′ end processing. We hypothesize that the first 271 amino acids of Symplekin mediate cotranscriptional histone mRNA 3′ end formation.
Previous data show that when CCC components CPSF73, CPSF100, or Symplekin are RNAi-depleted, all three factors are knocked down, significantly reducing the total number of CCCs and causing H2A 3′ end misprocessing (Wagner et al. 2007; Sullivan et al. 2009). A low concentration of CCCs results in primarily read through misprocessing as cryptic poly(A) signals downstream from the proper H2A mRNA cleavage site are used less often (Sullivan et al. 2009). This phenotype is also observed when CCCs do not properly assemble as with CPSF100(525–647) and CPSF73(466–552) (Fig. 3C–F). These mutants cannot increase the concentration of functional CCCs, therefore, read through H2A mRNAs dominate (Fig. 5B,C). When the concentration of CCCs is comparable with wild type (Fig. 3E,F), but the CCCs do not contain functional CPSF73, read through H2A misprocessing product is also prevalent (Fig. 5C,E). These data are in stark contrast to the rescue of H2A misprocessing seen with CCCs containing the C-terminal 231 amino acids of CPSF100 (Fig. 5B,E). The first 524 amino acids of CPSF100 are dispensable for cotranscriptional H2A 3′ end processing.
These data support a model in which reducing the number of functional CCCs prevents proper 3′ end processing and transcription termination resulting in H2A mRNAs with an extended 3′ UTR (Fig. 6B). If a functional CCC can be recruited to the appropriate cleavage site, proper H2A 3′ ends are formed as all other factors required for processing are present. If a functional CCC is not available, cleavage and termination do not occur and RNAPII keeps transcribing. Downstream cryptic poly(A) signals are not efficiently used for H2A mRNA 3′ end processing when the concentration of functional CCCs is low although poly(A)-specific processing factors are recruited to these sites (Sullivan et al. 2009). The mechanistic basis for this observation remains an active area of investigation.
CCCs lacking the first 271 amino acids of Symplekin use downstream poly(A) sites for H2A mRNA 3′ end processing
Amino acids 272–1165 of Symplekin are sufficient to form functional CCCs (Fig. 3A,B). As these CCCs contain full-length endogenous CPSF73, we expected H2A mRNAs in Dmel cells expressing this Symplekin mutant to be properly processed. Surprisingly, almost 32% of the H2A mRNAs in these cells are misprocessed (Fig. 5D,E). While read through H2A mRNA is the dominant misprocessed product upon CCC factor RNAi-depletion, CCCs containing N-terminally truncated Symplekin can efficiently use downstream poly(A) sites for H2A mRNA 3′ end processing (Fig. 5D,E). The amount of read through product in samples containing these mutants is approximately the same (Fig. 5D,E). These data indicate that the ability to utilize downstream poly(A) sites for H2A mRNA 3′ end processing is specific to the N-terminal 271 residues of Symplekin. The efficiency of CCC formation (Fig. 3A,B) for each N-terminal Symplekin deletion mutant is reflected in the total amount of H2A 3′ end misprocessing. CPSF73 is recruited more efficiently to Symp(272–1165) than to full-length Symplekin and we hypothesize that a higher concentration of active CCCs causes increased use of downstream poly(A) signals. As residues are deleted from the Symplekin C-terminus and less functional CCC are formed, total H2A 3′ end misprocessing decreases and the read through H2A mRNAs dominate (Fig. 5D,E).
Our evidence supports a model in which cotranscriptional histone mRNA 3′ end processing is mediated by the N-terminal 271 amino acids of Symplekin (Fig. 6C). When the number of functional CCCs is high, but Symplekin amino acids 1–271 are absent, cryptic downstream poly(A) signals can be efficiently utilized for H2A mRNA 3′ end processing. This is in contrast to read through products observed when the concentration of functional CCCs is low but full-length Symplekin is present (Fig. 6B). Previous experiments specify a role for the N-terminal region of Symplekin in cotranscriptional polyadenylation. Amino acids 30–340 of human Symplekin were cocrystallized with RNAPII CTD phosphatase Ssu72 and a RNAPII CTD peptide supporting strong interactions between these proteins (Xiang et al. 2010). The human Symplekin N-terminal domain inhibits transcription coupled polyadenylation in vitro (Xiang et al. 2010). Additionally, the use of downstream poly(A) sites for H2A mRNA 3′ end processing observed for N-terminally truncated Symplekin containing CCCs is similar to the processing pattern of H2A mRNAs when histone-specific processing factors SLBP, U7 snRNP, and FLASH are RNAi-depleted (Lanzotti et al. 2002; Tatomer et al. 2014). SLBP, U7 snRNP, and FLASH are hypothesized to be required for cotranscriptional 3′ end processing of histone mRNAs. Taken together, these data best support two hypotheses. Either the first 271 amino acids of Symplekin are required for interaction with histone-specific 3′ end processing factors or they mediate interaction indirectly or directly with RNAPII. These ideas are being tested in vivo using our Drosophila tissue culture system.
MATERIALS AND METHODS
Gene cloning and plasmid construction
Full-length CPSF73, CPSF100, and Symplekin were PCR amplified from Drosophila gene collection (DGC) clones (Open Biosystems) using specific primers. The amplified genes were directionally cloned into pENTR D-TOPO (Invitrogen) to create CPSF73, CPSF100, and Symplekin::pENTRD-TOPO. Proper gene insertion was confirmed via restriction digest and Sanger sequencing. CPSF73, CPSF100, and Symplekin::pENTRD-TOPO were each recombined with pAHW and pAWH destination vectors (Drosophila Gateway vector collection, Carnegie Institution for Science) using Clonase II (Life Technologies). Proper gene recombination was confirmed via restriction digest and Sanger sequencing. Deletion mutants for each of the three CCC proteins were cloned and recombined as described for their FL counterparts.
RNAi-resistant (RNAi-R) Symplekin, CPSF73, and CPSF100 were designed using an online tool described in Schulz et al. (2009). This tool changes wobble bases within a targeted area while also being sensitive to codon bias. A DNA fragment corresponding to the ∼500 bp target region was synthesized as a GeneArt Strings DNA Fragment by Life Technologies. Each RNAi-R GeneArt String and corresponding Symplekin, CPSF73, or CPSF100::pENTRD-TOPO were digested with restriction enzymes and the RNAi-R fragment was substituted for the wild-type piece to create RNAi-R Symplekin, CPSF73, and CPSF100::pENTRD-TOPO. Ligation was confirmed via restriction digest and Sanger sequencing. RNAi-R Symplekin, CPSF73, and CPSF100::pENTRD-TOPO were each recombined with pAHW and pAWH destination vectors as described above. Proper recombination was again confirmed via restriction digest and Sanger sequencing.
Creation of stable Drosophila Dmel-2 tissue culture lines
Full-length/mutant CCC factor::pAHW or full-length/mutant CCC factor::pAWH were transfected into Drosophila Dmel-2 cells with Effectene Transfection Reagent (Qiagen) according to the manufacturer's protocol. The pCoBlast vector (Invitrogen) containing a Blasticidin resistance gene was cotransfected to enable selection of successfully transfected cells. Cells were grown in SF-900 II SFM (Gibco) and maintained at 27°C under normal atmospheric conditions. Forty-eight hours post-transfection, Blasticidin (25 μg/mL) was added to the media to select for stably transfected cells. Cells were split and passaged into fresh selective media every 5 d to a concentration of 1 × 106/mL. Stable transfection was confirmed via Western blot with anti-HA antibody (Covance).
RNAi, whole-cell lysate preparation, nuclear extract preparation and RNA isolation
RNAi was performed as described (Sullivan et al. 2009). On Day 5, the cells were counted with a Nexcelom cell counter and harvested for analysis. To make whole-cell lysates, 5 × 106 RNAi-depleted or control cells were lysed with RIPA buffer (50 mM Tris–HCl (pH 8), 150 mM NaCl, 1% Nonidet P-40, 0.25% Na deoxycholate) plus 5 μL 100× Halt Protease inhibitor (Thermo Scientific), and rotated at 4°C for 10 min and centrifuged for 15 min at 14,000 rpm to remove cellular debris. The supernatant was transferred to a new tube and filtered through a 1 mL syringe with a 26 Gauge needle to shear the chromatin. NEs were prepared as described (Sullivan et al. 2009) from 1 × 108 cells and then flash frozen in liquid N2 and stored at −80°C. Whole-cell lysates and NEs were used for immunoprecipitations (IPs) and protein expression analysis. Total RNA was extracted from RNAi-depleted or control cells with TRIzol Reagent (Sigma) as described previously for use in S1 assays (Sullivan et al. 2009).
Antibodies
Monoclonal and polyclonal HA antibodies (Cat #s MMS-101R and PRB-101C, respectively, Covance) were used for both IP (3 μL) and WB (1:1000). Anti-CPSF73, anti-Symplekin, and anti-CPSF100 antibodies were described previously (Sullivan et al. 2009; Yang et al. 2009). For WB, antibodies were used at 1:1000.
Immunoprecipitation and S1 nuclease protection assay
IP of HA-tagged proteins were performed as described (Sullivan et al. 2009). S1 nuclease protection assay was also performed as described (Sullivan et al. 2009).
IP quantitation
Blots were scanned at 600 PPI and analyzed using ImageJ software. Intensity of protein bands representing IP or co-IP were determined and compared with the intensity of the corresponding input band (representing 5% of the sample). The ratio of IP or co-IP intensity to input intensity was multiplied by 5 to determine the percentage of total protein immunoprecipitated from the sample.
S1 quantification
Two independent experiments were quantitated for each S1 experiment. Dried 6% Urea/TBE sample gels were placed on phosphoscreens overnight. Phosphoscreens were scanned using a STORM Scanner and images were saved as TIFF files. The TIFF files were imported into ImageJ software and subsequently analyzed. Peak areas were determined for bands corresponding to RT, misprocessed and properly processed H2A mRNAs. The sum total of all peaks in each lane was then determined and the area for each peak (band) was divided by the sum total of peaks in that lane to give a percentage. The percentages were averaged and the standard deviation calculated for all products in each sample.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Office of Extramural Research, National Institutes of Health (NIH) grant R15GM107931 to M.S., and UMSL start-up funds to M.S. The CPSF100 antibody was a generous gift of Dr. Zbigniew Dominski (UNC-Chapel Hill). We thank Ryan Kosovich, Nathan Ponzer, and Andrew Harrington for technical assistance and Dr. William F. Marzluff, Dr. Ambrose R. Kidd III, Dr. Joseph Russo, Dr. Wendy M. Olivas, and Dr. Bethany K. Zolman for critical review of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.049551.115.
REFERENCES
- Adamson TE, Price DH. 2003. Cotranscriptional processing of Drosophila histone mRNAs. Mol Cell Biol 23: 4046–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17: 1132–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Waltenspiel B, Warren WD, Wagner EJ. 2013. Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J Biol Chem 288: 4867–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Zheng LX, Sanchez R, Marzluff WF. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol 19: 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X-C, Marzluff WF. 2005a. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang X-C, Purdy M, Wagner EJ, Marzluff WF. 2005b. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25: 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy MA, He X, Singh BN, Hampsey M, Moore C. 2009. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol Cell Biol 29: 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf AL. 1993. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci 18: 117–119. [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J-P, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26: 2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J-P, Sheth A, Manley JL. 2011. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334: 683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514–1517. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kennedy SA, Frazier ML, Steiniger M, Mast AM, Marzluff WF, Redinbo MR. 2009. Crystal structure of the HEAT domain from the pre-mRNA processing factor Symplekin. J Mol Biol 392: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394. [DOI] [PubMed] [Google Scholar]
- Kyburz A, Sadowski M, Dichtl B, Keller W. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res 31: 3936–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. 2002. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol 22: 2267–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Ricciardi AS, Guo AY, Kambach C, Marzluff WF. 2014. The C-terminal extension of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA 20: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez J-R, Webb WW, Lis JT. 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol 28: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp M-D, Schweingruber C, Kleinschmidt N, Schümperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: implications on localization and function. Mol Biol Cell 22: 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath I, Skrajna A, Yang X-C, Dadlez M, Marzluff WF, Dominski Z. 2013. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 19: 1726–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L, Kühn U, Martin G, Schäfer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JG, David G, Hassan BA. 2009. A novel method for tissue-specific RNAi rescue in Drosophila. Nucleic Acids Res 37: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 1997. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 17: 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer DC, Rizzardi LF, Curry KP, Witkowski AM, Marzluff WF, Duronio RJ. 2014. Drosophila Symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus 5: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28: 692–699. [DOI] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. 2010. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467: 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Manley JL, Tong L. 2012. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev 26: 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-C, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A, Tacahashi Y, Nasser T, He X, Sterzer U, Jensen TH, Domdey H, Moore C. 2006. The role of the Brr5/Ysh1 C-terminal domain and its homolog Syc1 in mRNA 3′-end processing in Saccharomyces cerevisiae. RNA 12: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.