Abstract
TDP-43 is a soluble, nuclear protein that undergoes cytoplasmic redistribution and aggregation in the majority of cases of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. TDP-43 autoregulates the abundance of its own transcript TARDBP by binding to an intron in the 3′ untranslated region, although the mechanisms underlying this activity have been debated. Herein, we provide the most extensive analysis of TARDBP transcript yet undertaken. We detail the existence of a plethora of complex splicing events and alternative poly(A) use and provide data that explain the discrepancies reported to date regarding the autoregulatory capacity of TDP-43. Additionally, although many splice isoforms emanating from the TARDBP locus contain the regulated intron in the 3′ UTR, we find only evidence for autoregulation of the transcript encoding full-length TDP-43. Finally, we use a novel cytoplasmic isoform of TDP to induce disease-like loss of soluble, nuclear TDP-43, which results in aberrant splicing and up-regulation of endogenous TARDBP. These results reveal a previously underappreciated complexity to TDP-43 regulated splicing and suggest that loss of TDP-43 autoregulatory capacity may contribute to the pathogenesis of ALS.
Keywords: TDP-43, ALS, FTLD, autoregulation, splicing
INTRODUCTION
The majority of cases of sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are characterized by the aggregation of ubiquitinated TAR-DNA binding protein of 43 kDa (TDP-43) in the cytoplasm of neurons and glia (Neumann et al. 2006; Brandmeir et al. 2008). TDP-43 is composed of two RNA recognition motifs (RRM) that allow binding to RNA substrates, nuclear localization and export sequences that permit nucleocytoplasmic shuttling, and a glycine-rich C-terminus that binds catalytic protein cofactors (Buratti and Baralle 2001; Ayala et al. 2008; Winton et al. 2008; Ling et al. 2010). By virtue of these properties, TDP-43 binds a wide range of RNA subtypes and mediates their stability, splicing, and biogenesis (Buratti et al. 2001; Strong et al. 2007; Polymenidou et al. 2011; Tollervey et al. 2011; Kawahara and Mieda-Sato 2012; Di Carlo et al. 2013).
In addition to its aggregation in disease, TDP-43 is also hyperphosphorylated and cleaved to produce insoluble C-terminal fragments and normal nuclear expression is lost (Neumann et al. 2006). The shift from a predominantly nuclear to cytoplasmic localization may occur via a process of sequestration in which initially small, insoluble aggregates sequester soluble TDP-43, depleting it from the nucleocytoplasmic shuttling pool (Winton et al. 2008; Nonaka et al. 2009; Che et al. 2011; Furukawa et al. 2011). The resulting loss of TDP-43 nuclear function has been hypothesized to result in dysregulation of TDP-43 mediated RNA metabolism. Silencing of TDP-43 using shRNA in vivo or siRNA in vitro combined with RNA-seq or exon array have demonstrated that loss of TDP-43 induced via these methods results in profound changes in exon splicing and pre-mRNA levels of TDP-43 substrates (Polymenidou et al. 2011; Tollervey et al. 2011). To date, however, it has not been demonstrated that loss of functional TDP-43 by sequestration generates identical splicing abnormalities, nor has it been possible to study processing of TARDBP itself under these TDP-43 loss-of-function conditions.
The TARDBP gene encoding full-length TDP-43 is composed of six exons. Although multiple groups have described possible alternative splicing events across the transcript, this remains poorly characterized. Wang et al. (2004) confirmed the existence of four and three alternative transcripts in mouse brain and human cell lines, respectively. Several groups have reported the existence of an alternative transcript (variously referred to as V2, isoform 3, or TDPs) that is created using a cryptic exon in the 3′ untranslated region (UTR) of TARDBP, with the donor splice site residing in exon 6 (Wang et al. 2002; Ayala et al. 2011; Polymenidou et al. 2011). To date, however, a detailed analysis of the TARDBP transcriptome has not been performed, potentially limiting the understanding of TDP-43 regulation and any relation to disease. Most interestingly, TDP-43 protein autoregulates the reduction of TARDBP transcript levels (Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012; Bembich et al. 2014). Cross linking and immunoprecipitation studies have unambiguously demonstrated that TDP-43 protein binds to TARDBP pre-mRNA in the 3′ UTR (Ayala et al. 2011; Polymenidou et al. 2011). However, the effect of this binding has been contentious. One group demonstrated that TDP-43 enhanced the expression of V2/isoform 3/TDPs (Polymenidou et al. 2011). This process concomitantly excised an intron in the 3′ UTR which resulted in the nonsense-mediated decay of the transcript encoding full-length TDP-43. A second group confirmed the splicing event in the 3′ UTR and additionally demonstrated that this event is coupled to use of an alternative polyadenylation site, but found no evidence for up-regulation of V2/isoform 3/TDPs upon TDP-43 overexpression (Ayala et al. 2011; Avendaño-Vázquez et al. 2012). In subsequent studies, it was demonstrated that the 3′ UTR splicing event is the main determinant of TDP-43 autoregulation, and that it also results in the nuclear retention of TARDBP transcript, preventing translation (Avendaño-Vázquez et al. 2012; Bembich et al. 2014). Given these discrepancies, we sought a more comprehensive understanding of the splicing of Tardbp in order to extend current knowledge regarding TDP-43 autoregulation, function, and pathogenesis.
RESULTS
Tardbp undergoes complex alternative splicing to yield nineteen distinct transcripts
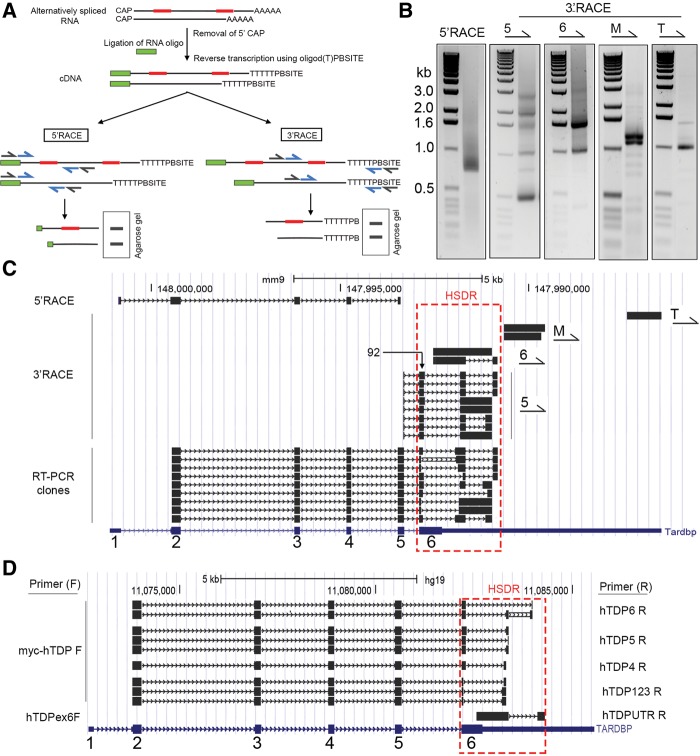
We initially aimed to extend the understanding of Tardbp transcript processing, in particular focusing on the region bound by TDP-43 protein during autoregulation. To determine potential alternative splicing events in this region, we used 3′ and 5′ Rapid Amplification of cDNA Ends (RACE). 3′ RACE is an RT-PCR that uses a reverse primer complementary to the poly(A) tail of all mRNAs, and a transcript-specific forward primer, allowing the amplification of all poly(A) bearing mRNAs in the region of interest (Fig. 1A). Similarly, 5′ RACE uses a forward primer that binds a tag artificially added to the 5′ end of mRNAs and a reverse primer specific to the transcript of interest. As template, we used total RNA isolated from the cortex of a healthy, 2-mo-old naïve female FVB mouse. We reasoned that this would represent typical adult murine expression. Also, a number of our in house TDP-43 transgenic lines are on an FVB background. 5′ RACE using forward primers specific to the artificial 5′ tag and reverse primers specific to Tardbp exon 5 amplified a product of 700 bp (Fig. 1B, left panel). Cloning of this product and sequencing of three individual clones identified this amplicon as exons 1–5 of murine Tardbp (Fig. 1C). Hence, although in silico analysis of Tardbp has previously suggested shorter, potential alternatively spliced transcripts between exons 1 and 5 (Wang et al. 2004), we did not find evidence for their expression in adult mouse cortex. Consequently, if other sequence variants exist, these are likely to be of low abundance.
FIGURE 1.
Complex alternative splicing and polyadenylation result in multiple Tardbp transcripts. (A) Simplified schematic of 3′ and 5′ RACE. Two alternatively spliced transcripts from the same hypothetical gene are shown, differing via the spliced inclusion of two exons (red). The mRNA CAP is first removed and replaced with a synthetic RNA oligo. Reverse transcription using an oligo d(T) primer with a unique 3′ Primer Binding Site (PBSITE) creates cDNA with 5′ and 3′ sequences for primer binding during PCR. Subsequently, two rounds of PCR are performed; an initial round with primers complementary to the artificial sites (black primers), and a second nested PCR using primers inside these initial sites (blue). Products are analyzed on a gel, excised, cloned, and sequenced. (B) 5′ and 3′ rapid amplification of cDNA ends from murine cortex produced multiple PCR products. Primers for 5′ RACE or 3′ RACE (5, 6, M, and T) are shown above each PCR and represent the primer used in reaction. (C) Sequences derived from 5′ RACE, 3′ RACE, and RT-PCR cloning, aligned on murine UCSC Genome Browser. Sequences derived from our studies are in black; blue sequence below represents preexisting Tardbp gene in mouse genome archive. Primers (5, 6, M, and T) used in the 3′ RACE reactions are denoted to the right of aligned sequences. Conventional exon numbering system for Tardbp is shown in blue at bottom. The High Splicing Density Region (HSDR) is designated with a red dash. “92” indicates the first cluster of splice sites, containing eight donor sites. (D) Alignment of sequences from human H4 neuroglioma RT-PCR on human UCSC Genome Browser. The forward and reverse primers used in reactions to generate amplicons are shown on left and right of alignment, respectively.
Prior work had established the existence of V2/isoform 3/TDPs, an alternative transcript that is created using a cryptic exon 7 in the 3′ untranslated region (UTR) of TARDBP, with the donor splice site residing in exon 6 (Wang et al. 2002; Ayala et al. 2011; Polymenidou et al. 2011). We aimed to characterize any additional splice variants in this region and elsewhere in the Tardbp transcript. To maximize our chances of detecting alternative splice variants, we designed four forward 3′ RACE primers to use in separate reactions; these were complementary to exon 5 (primer 5), exon 6 (primer 6), a middle portion of the 3′ UTR (primer M), and a region near the 3′ UTR terminus (primer T). Multiple amplicons were produced by these RACE experiments (Fig. 1B); these were cloned, sequenced, and aligned on the University of Santa Cruz (UCSC) mouse genome browser. As can be seen in Figure 1C, primer T only amplified one major PCR product corresponding to the extreme 3′ end of the gene, with no splicing, and terminating at a polyadenylation site already cataloged in the browser. Primer M amplified two products that corresponded to the use of two different polyadenylation sites (AATAAA) located just 104 bp from one another. Primer 6 in exon 6 of Tardbp amplified two products. The larger RACE product corresponded to the unspliced Tardbp transcript and terminated at a polyadenylation site. The smaller product corresponded to a transcript that splices out a section of the 3′ UTR following the stop codon and terminated at a more 3′ polyadenylation site. Thus, as determined in previous studies (Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012), the transcript encoding full-length Tardbp has two alternative 3′ UTRs. Our work raises the number of known polyadenylation sites in Tardbp to five.
3′ RACE using primers in exon 5 generated multiple products. Sequencing of these identified eight distinct transcripts. As we were interested in the functionality of novel sequence variants, we attempted to clone the full sequence of these transcripts, from exon 1 through to the polyadenylation site. Sequencing of the products of these reactions, however, identified a further nine novel transcript variants (Fig. 1C). Hence, Tardbp is spliced to produce a minimum of 19 transcripts. This variation is generated via the use of 20 splice sites (nine splice donors, 11 splice acceptors) contained in a region of 1938 bp that we have termed the High Splice Density Region (HSDR) (Fig. 1C). Prior to further analysis of these murine transcripts, we wished to confirm the existence of similarly extensive splicing in a human cell line. To do so, we performed RT-PCR using RNA from human H4 neuroglioma cells, using different combinations of reverse and forward primers (Fig. 1D). We identified 10 different possible transcripts generated by alternative splicing, all occurring within a human HSDR.
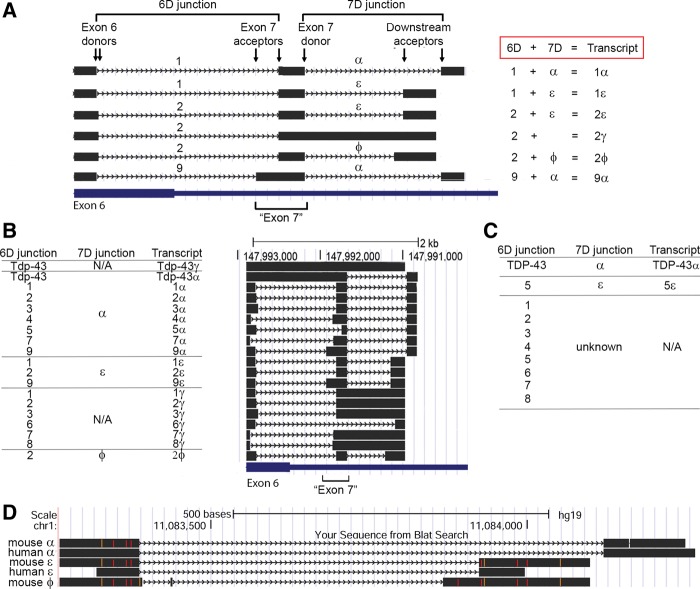
In order to accurately describe the highly complex manner in which the splicing in the HSDR occurs, and to effectively communicate our subsequent experimental observations, it was necessary to generate an entirely new nomenclature as standard nomenclature of introns/exons lacked adequate explanatory power. Splice sites within the HSDR are found in clusters. The first of these is in exon 6, which contains eight distinct donor splice sites within a 92 bp region (labeled “92” in Fig. 1C). With one exception, the downstream splice acceptor sites that pair with these donors to form junctions are contained in a second cluster containing seven splice acceptor sites within a 137 bp region. By using different combinations of these splice donor and acceptor sites, nine different splice junctions are formed, and we differentiate them from one another using numbers 1–9. Examples of this junction formation (junctions 1, 2 and 9) and the nomenclature used are shown in Figure 2A. As it is necessary during our studies to refer to this group of junctions (1–9) collectively, and because they all emanate from exon 6 donor sites, these are collectively referred to as 6D junctions (Fig. 2A).
FIGURE 2.
Defining a new nomenclature for Tardbp splicing. (A) Six transcripts identified in our studies (black) serve as examples for the nomenclature used in this work. Only the murine HSDR is shown. Locations of exon 6 and the region formerly known as exon 7 are indicated at bottom. Splice junctions formed between an exon 6 donor site and downstream exon 7 acceptor are termed 6D junctions and are numbered 1–9; they are exemplified here by junctions 1, 2, and 9. Note how junctions 9 and 1 share the same donor splice site, but differ in their acceptor site; however, junctions 1 and 2 differ in their donor site but share acceptor sites. Splice junctions formed between the donor exon 7 site and downstream acceptors are termed 7D junctions and are designated with the symbols α, ε, and ϕ. Combinations of 6D and 7D form specific transcripts; here these are 1α (composed of the 6D junction 1 and the 7D junction α), 1ε (6D = 1, 7D = ε), 2ε (6D = 2, 7D = ε), 2γ (6D = 2, no 7D splice junction), 2ϕ (6D = 2, 7D = ϕ), and 9α (6D = 9, 7D = α). (B) All murine transcripts identified in our studies, with 6D junction, 7D junction, and total transcript name. Position of the transcript within the HSDR is shown by alignment on the UCSC genome browser to the right of the table. (C) 6D and 7D junctions, and complete transcripts identified by RT-PCR from human neuroglioma cells. Owing to primer position, we only detected one “pairing” of 6D and 7D, hence the 7D junctions that might pair with 6D junctions are majority “unknown.” (D) Conservation between mouse and human 7D splice junctions. Sequences shown are aligned on the human UCSC genome browser. Mismatched base pairs appear as orange or red.
The region of high splice acceptor density described above (seven acceptor sites in 137 bp) has been referred to previously as exon 7. With so many potential acceptor sites, “exon 7” no longer has a definite boundary or meaning. However, it is useful for our studies to maintain this exonic designation to describe this general area. “Exon 7” has a single donor splice site and there are three potential downstream acceptor sites, and therefore three potential junctions. We differentiate the three possible junctions that can be formed with symbols (α, ε, ϕ). These are shown clearly in the examples in Figure 2A. As all these junctions are formed using a donor site in exon 7, we refer collectively to these as 7D junctions.
We found that different Tardbp transcripts are generated in vivo by using a 6D junction, a 7D junction, or both in combination. For transcripts with both, we represent this with the appropriate combination of 6D number and 7D symbol. Five examples (1α, 1ε, 2ε, 2ϕ, 9α) are shown in Figure 2A. For transcripts with only a 6D junction, we use a number followed by γ, which is exemplified in Figure 2A by the transcript 2γ. These various combinations of splicing events (and associated alternative polyadenylation site usage) give rise to 19 alternative transcripts. All possible transcripts we identified are shown in Figure 2B, with their splice site combinations and position in the HSDR (see also sequences submitted to GenBank, accession numbers in Supplemental Table 1). We found only one instance of an alternative transcript generated by 7D splicing alone; this was the transcript encoding full-length Tdp-43 with splicing in the 3′ UTR, the splicing corresponding to an α junction (Tdp-43α). All the splice donor and acceptor sites we refer to were identified from sequenced RACE clones, or clones isolated in subsequent RT-PCR reactions intended to clone the novel transcripts identified from RACE. They were not the result of predictive algorithms or database mining.
We also applied this nomenclature system to human TARDBP splicing which we identified in human H4 neuroglioma cells (Fig. 2C). We identified eight 6D junctions and two 7D junctions. Owing to the position of the primers used in our experiment, we were only able to identify one combination of 6D with 7D junctions (transcript 5ε). Of note, the two 7D junctions α and ε were highly conserved between mouse and human (Fig. 2D). It is important to note that there was insufficient homology in 6D junctions at the nucleotide level to unambiguously identify splice orthologs. As such, murine 6D junction 1 should not be assumed to be the direct ortholog of human junction 1, for example. Interestingly, a large proportion of human and mouse splice junctions uncovered here used noncanonical dinucleotide sequences at the exon–intron boundary. Previous studies have demonstrated that ∼99% of all splice junctions in the human and mouse genome utilize the dinucleotides GU and AG at the 5′ and 3′ end of the excised intron respectively (Parada et al. 2014). Of the total twelve new junctions we identified here in mouse, only eight (67%) were GU-AG sites. The remainder were comprised of AU-AU (n = 1), AU-AG (n = 1) and AU-AC (n = 2). Similarly, of the 10 novel human splice junctions we identified in the HSDR, only six (60%) were GU-AG sites, with the remainder using AU-AC (n = 2) or AU-AG (n = 2).
Tdp-43 autoregulates Tardbp splicing but does not predict whole transcript abundance
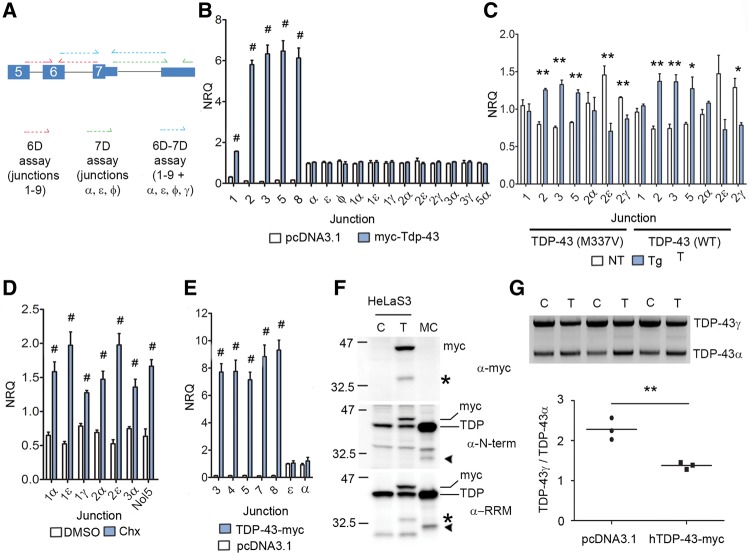
We used quantitative real-time PCR (qPCR) to assess the autoregulatory capacity of TDP-43 on the splice junctions identified in the aforementioned RACE reactions. We designed primers that would amplify specific 6D junctions, 7D junctions, or specific transcripts bearing a combination of 6D-7D (Fig. 3A). Transient expression of N-terminally myc-tagged mouse Tdp-43 (myc-Tdp-43) in a mouse derived neuroblastoma cell line (N2a) resulted in increased expression of all 6D splice junctions tested, relative to vector only controls (Fig. 3B). Although 7D junctions α, ε, and ϕ span the intron bound by Tdp-43, we observed no increase in their levels when myc-Tdp-43 was expressed (Fig. 3B). Importantly, there was no increase in expression of any 6D-7D specific transcripts tested using primers spanning both splice junctions (Fig. 3B). Given in vitro results may not faithfully model in vivo systems, we sought to confirm these findings by assaying the abundance of Tardbp transcripts in the cortex of two transgenic mouse lines that overexpress human wild type or mutant M337V TDP-43 (Cannon et al. 2012; D'Alton et al. 2014). With the exception of junction 1, we observed increased expression of 6D junctions in 2-mo-old transgenic animals relative to nontransgenics (Fig. 3C). However, identical to our results in N2a cells, we did not observe concomitant increases in 6D-7D transcripts when specific combinations of junctions were assayed (Fig. 3C). These 6D-7D transcripts have been shown previously to be degraded by nonsense-mediated decay (NMD) (Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012). We confirmed that 6D-7D transcripts are elevated by conditions of NMD inhibition using treatment of N2a cells with cycloheximide (Chx) (Fig. 3D). This finding also demonstrates that our primers were capable of detecting expression changes in 6D-7D junctions. As splicing mechanisms may differ between mouse and human, we further examined these reactions in a human cell line. We overexpressed C-terminally myc-tagged human TDP-43 (TDP-43-myc) in HeLaS3 cells. Relative to empty vector controls, we detected a significant increase in all 6D junctions assayed in TDP-43-myc transfected cells (Fig. 3E). Identical to that observed in N2a cells, we observed no increase in the abundance of 7D junctions that span the TDP-43 binding region (Fig. 3E). Consequently, we find evidence across species and from in vitro and in vivo systems that TDP-43 universally increases all splicing between the internal exon 6 donor splice site and downstream acceptor sites (6D junctions), but splicing events that surround the intron within which TDP-43 binds remain unchanged (7D junctions). Importantly, despite the increased retention of the 6D splice junctions, we found no evidence that the abundance of total transcripts (6D-7D) bearing them was increased.
FIGURE 3.
TDP-43 mediated splicing does not predict whole transcript abundance. (A) Schematic of primers used in analysis, showing their binding sites on an example transcript. 6D junctions between splice donor in exon 6 and downstream acceptor site in exon 7 assayed using primers in red, 7D junctions between exon 7 and downstream acceptor sites in green. A third set of primers (blue) assayed specific transcripts by incorporating both junctions. (B) qPCR to assay abundance of junctions in neuro2a cells transiently transfected with myc-Tdp-43 or empty plasmid vector for 24 h (N = 5 per group, NRQ = Normalized Relative Quantity). (C) qPCR to assay abundance of splice junctions in cortex of female nontransgenic mice (NT) and female mice overexpressing human wild type or M337V TDP-43 (Tg, N = 3 per genotype). (D) Relative abundance of indicated junctions in either DMSO treated cells or cells exposed to cycloheximide (Chx) as outlined in Materials and Methods; an NMD-sensitive exon in Nol5 was used as positive control (N = 5 per group). (E) Relative abundance of indicated junctions in HeLaS3 cells transiently transfected with human TDP-43-myc or empty plasmid for 24 h (N = 5 per group). (F) Western blotting of SDS lysate from HeLaS3 cells transfected with empty vector control (C) or TDP-43-myc (T), run alongside lysate from murine cortex (MC). Antibodies to myc tag, TDP-43 N-terminus (N-term), or the RRM domains of TDP-43 (RRM) were used. Asterisks denote C-terminal fragment immunopositive for myc and RRM antibodies; arrowheads denote N-terminal and RRM positive alternative isoform. Exogenous full-length TDP-43-myc (myc) and endogenous TDP-43 (TDP) are labeled. (G) 3′ RACE of HeLaS3 cells transfected with empty vector as control (C) or TDP-43-myc (T), using primers specific to full-length TDP-43 encoding transcript. Quantification of the ratio of unspliced TDP-43γ to α-spliced TDP-43α is below. In B, C, D, E, and G: (*) P < 0.05, (**) P < 0.01, (#) P < 0.001, Student unpaired two-tailed t-test, error bars are SEM.
These results run counter to preexisting data which suggested that overexpression of TDP-43 in vitro resulted in increased expression of the 6D-7D junction TDP isoform 3 (using our nomenclature, 1α) at the mRNA level (Polymenidou et al. 2011). Polymendiou et al. also identified a TDP-43 immunoreactive species at a lower molecular weight on western blot, consistent with up-regulation of isoform 3 at the protein level. As we observed increases only in 6D expression and not entire transcripts (6D-7D) at the mRNA level, we investigated the possibility of up-regulation solely at the protein level. Accordingly, we transiently overexpressed TDP-43-myc in HeLaS3 cells and used western blotting using antibodies raised to the N-terminus or RRM domains of TDP-43, or to the myc epitope. In parallel, we also examined lysate from 2-mo-old naïve mouse cortex. Overexpression of TDP-43-myc in vitro generated an ∼35 kDa species, immunopositive for myc and RRM which therefore can only be a cleavage product derived from the exogenous TDP-43 expression (Fig. 3F, asterisk). In murine cortex, we observed an ∼32 kDa species, immunopositive for N-terminal and RRM (Fig. 3F, arrowhead), demonstrating the expression of at least one of these alternative isoforms in vivo. Consequently, although TDP-43 autoregulates 6D junction abundance, we find no evidence that it increases the level of whole transcripts (6D–7D) at either the RNA or protein level.
TDP-43 autoregulates α-splicing of the transcript encoding full-length TDP-43
Previous reports have demonstrated that TDP-43 mediates the splicing of the TARDBP 3′ UTR intron (in our nomenclature, this corresponds to the “α” 7D junction, henceforth termed α-splicing) when fused to reporter constructs (Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012). Avendano-Vazquez et al. also used 3′ RACE to demonstrate that TDP-43 protein catalyzes the endogenous splicing of unspliced TDP-43 (in our nomenclature, TDP-43γ) to spliced TDP-43α (in the publication by Avendano-Vazquez et al., this is referred to as pA1 to pA2 switching), both in vitro and in vivo (Avendaño-Vázquez et al. 2012). We sought to confirm these findings by using 3′ RACE that would specifically amplify transcripts encoding full-length TDP-43 only. In HeLaS3 cells transiently overexpressing TDP-43-myc, we discovered a decrease in the ratio of TDP-43γ:TDP-43α compared with cells transfected with empty vector (Fig. 3G). Therefore, TDP-43 protein does autoregulate α-splicing of endogenous TARDBP transcript encoding full-length TDP-43. However, this autoregulatory capacity does not extend to all transcripts with α-junctions, as our experiments in HeLaS3, N2a, or in vivo in transgenic animals found no evidence of a general effect of TDP-43 protein on α-splicing or any of the transcripts bearing α.
C-terminal sequences dictate subcellular localization of RRM domains
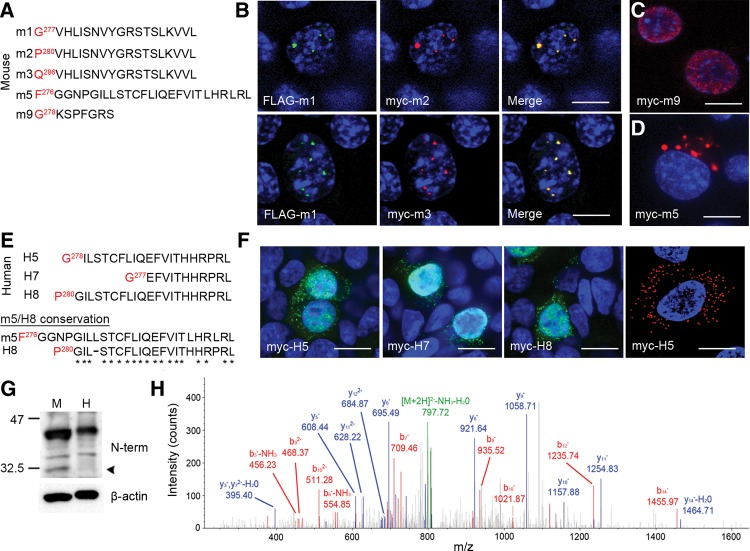
The presence of alternative 6D junctions results in the generation of unique potentially protein coding transcripts. In each case, the stop codon for these novel proteins occurs prior to the 7D splice junction. Therefore, we use the 6D junction number to refer to the protein translated from each transcript. Consequently, in mice, as there are nine 6D junctions there are nine potential protein coding sequences, named m1–m9. Protein m1 is translated from any transcript in which the 6D junction is 1, m2 from any transcript in which the 6D junction is 2, and so on. In humans, there are eight novel protein coding sequences termed H1–H8, and, for example, transcripts with 6D junction 5 translate to H5 protein.
These share the N-terminal sequence of TDP-43 including the RRM domains, but they differ at the C-terminus. One of these transcripts, corresponding to m1 using our nomenclature, has been previously shown to localize to subdomains within the nucleus (Wang et al. 2002). Interestingly, we discovered two additional splice variants (m2 and m3) that shared identical protein sequence with m1 at the C-terminus (VHLISNVYGRSTSLKVVL), but which was preceded by a short stretch of divergent sequence as a result of splice site differences (Fig. 4A). Similar to previous reports, N-terminally FLAG tagged m1 (FLAG-m1) localized to nuclear subdomains when expressed in N2a cells (Fig. 4B). Furthermore, FLAG-m1 colocalized with both N-terminally myc-tagged m2 (myc-m2) and m3 (myc-m3), demonstrating that their shared C-terminal motif dictates their localization, regardless of sequence divergence preceding it (Fig. 4B). In contrast, expression of an isoform that generates a stop codon only 7 amino acids following the 6D splice junction (m9) resulted in diffuse nuclear expression (Fig. 4C). Given these results, we set out to determine the distribution of other isoforms in vitro. Strikingly, expression of m5 with an N-terminal myc tag (myc-m5) yielded entirely cytoplasmic, punctate expression in N2a cells (Fig. 4D). We found no evidence in our human sequence data of any isoform homologous to m1/m2/m3. However, we identified two human isoforms (H5 and H8) containing a C-terminal motif (ILSTCFLIQEFVITHHRPRL) and a third with a partial motif (H7; EFVITHHRPRL) which appear to be orthologs to m5 (Fig. 4E). Similar to m5, myc-H5, myc-H7, and myc-H8 isoforms demonstrated punctate cytoplasmic distribution in addition to diffuse nuclear localization when expressed in human embryonic kidney 293T cells (HEK293T) (Fig. 4F). Furthermore, in human neuroglioma H4 cells, localization was entirely of the cytoplasmic and punctate form (Fig. 4F). These data demonstrate that this novel C-terminal sequence directs localization to the cytoplasm. In sum, these observations suggest that the cellular localization of N-terminal sequences including the RRM domains is modulated by C-terminal sequences.
FIGURE 4.
Alternative isoform expression in vitro and in vivo. (A) Alignment of sequences of murine isoforms cloned and expressed in vitro. Only the sequence after the last amino acid shared with TDP-43 (red) is shown. (B) Coexpression of FLAG-m1 and myc-m2 (top row) and FLAG-m1 and myc-m3 (bottom row) in neuro2a cells; merged images on right. (C) Expression of myc-m9 encoding an isoform with STOP codon only seven amino acids after the 6D splice junction. (D) Expression of myc-m5 in neuro2a cells. (E) Alignment of sequences of human isoforms cloned and expressed in vitro. Conservation between mouse m5 and human H8 sequence shown below. (F) Expression of myc-H5, -H7, and -H8 in HEK293T cells and myc-H5 in H4 neuroglioma cells. In B, C, D, and F, all images counterstained with DAPI; scale bars, 10 µm. (G) Western blotting of 2% SDS lysate from human and murine cortex, probed with antibody to the N-terminus of TDP-43; loading shown with β-actin. Arrowhead denotes expression of alternative isoform in murine cortex. (H) MS/MS spectra of m2 isoform peptide fragment isolated from murine cortex.
As the expression of an additional cytoplasmic isoform in humans would be of potential significance to the pathogenicity of TDP-43 proteinopathies, we searched for evidence of expression in mouse and human cortical lysates. Antibodies raised to the N-terminus of TDP-43 were immunoreactive to an ∼32.5 kDa species in mouse cortex, which was absent in human cortex (Fig. 4G, arrowhead). To verify the nature of this species in mouse cortex, we used mass spectroscopy and identified the peptide fragment FGGNPVHLISNVYGR uniquely corresponding to m2 (Fig. 4H). Consequently, despite the presence of multiple RNA transcripts bearing similar splice patterns—including two that encode proteins with identical functional motifs and predicted localization—only a single isoform is permitted at the protein level. Furthermore, there is a distinct species difference in expression of TARDBP between mouse and human.
Sequestration of TDP-43 is accompanied by TARDBP dysregulation
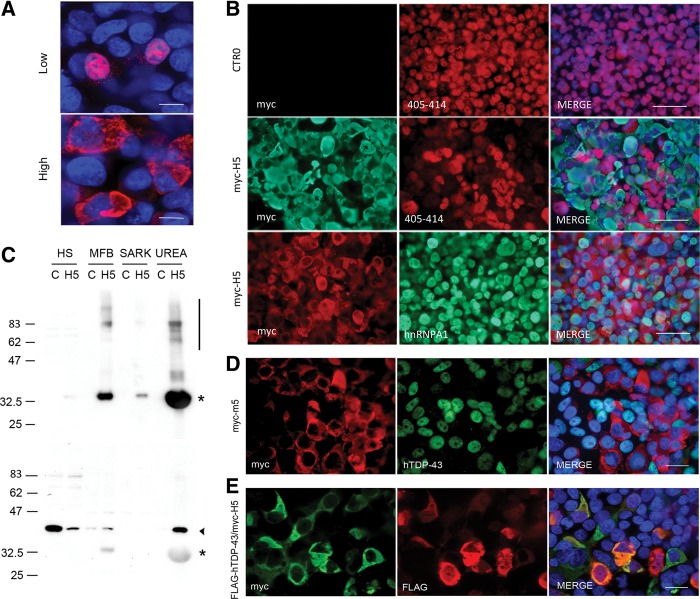
As these cytoplasmic isoforms lack the disease-associated C-terminus of TDP-43, we were curious as to their ability to sequester endogenous TDP-43. Optimizing transfection in HEK293T cells, we observed that high-level expression of myc-H5 resulted in complete cytoplasmic localization and aggregation of myc-H5 (Fig. 5A). Next, we expressed myc-H5 at high levels and used an antibody to amino acids 405–414 to visualize endogenous TDP-43. As isoform H5 lacks the C-terminus, this allowed discrimination between endogenous TDP-43 and H5 by immunofluorescence. HEK293T cells transfected with empty vector demonstrated robust nuclear expression of TDP-43 in all cells (Fig. 5B, top row). Expression of myc-H5 induced widespread loss of endogenous nuclear TDP-43, and in many cells this clearance was complete (Fig. 5B, middle panel). In contrast, another nuclear-cytoplasmic shuttling RNA-binding protein HNRNPA1 remained nuclear in all cells, demonstrating that the effect of H5 was specific to TDP-43 (Fig. 5B, bottom panel). However, loss of nuclear TDP-43 was not accompanied by increased cytoplasmic staining as might be expected in the case of sequestration. Biochemically, sequestered TDP-43 becomes detergent insoluble and urea soluble (Winton et al. 2008). Therefore, we performed biochemical fractionation and Western blotting of HEK293T cells expressing myc-H5. Using an antibody to the N-terminal myc tag, we found that the majority of myc-H5 was insoluble in high salt or detergent and was concentrated in the urea fraction. This fraction also contained a significant amount of high molecular weight species consistent with aggregated material (Fig. 5C, upper panel). Using the antibody to 405–414, we detected vast reduction of TDP-43 in the high salt fraction, which is consistent with the loss of soluble, nuclear TDP-43 observed by immunocytochemistry (Fig. 5C, lower panel). This was accompanied by the appearance of urea-soluble TDP-43, consistent with the shift to insolubility that accompanies sequestration. Given this discrepancy between immunocytochemistry and biochemistry, we hypothesized that the lack of cytoplasmic signal in the former was due to destruction or obstruction of the antigenic epitope upon sequestration. Consequently, we attempted a second means of discrimination between exogenous myc-H5 and TDP-43 by using species specific antibodies. We expressed N-terminally myc-tagged mouse m5 and visualized endogenous TDP-43 using a human-specific antibody. In cells expressing myc-m5, we observed complete loss of endogenous nuclear TDP-43 in many cells, but again without a concomitant increase in cytoplasmic localization (Fig. 5D). To determine if myc-H5 is capable of sequestering TDP-43, we coexpressed myc-H5 with FLAG-hTDP-43. Under these conditions, we observed extensive colocalization in the cytoplasm of myc-H5 and FLAG-TDP-43, demonstrating the ability of myc-H5 to sequester full-length TDP-43 (Fig. 5E). In sum, myc-H5 induces loss of soluble, nuclear TDP-43 via a mechanism that promotes insolubility, and myc-H5 is capable of sequestering TDP-43.
FIGURE 5.
Overexpression of myc-H5 induces insolubility and loss of nuclear TDP-43. (A) Immunofluorescence of HEK293T cells transfected with myc-H5 for low expression and high expression, using myc antibody for detection. Scale bars, 10 µM. (B) Transfection with optimized protocol for high expression of empty CTR0 plasmid (top row) or myc-H5 (middle and bottom rows), followed by immunofluorescence with indicated antibodies. Note the loss of TDP-43 staining in myc-H5 cells compared with empty plasmid. Nuclear staining of another RNA-binding protein HNRNPA1 remains intact. Scale bars, 50 µm. (C) Western blotting of high salt (HS), myelin floatation buffer (MFB), sarkosyl (SARK), and urea fractions from HEK293T cells transfected with empty CTR0 plasmid control (C) or myc-H5 (H5), using myc (top) or antibody to 405–414 C-terminus of TDP-43 (bottom). Asterisk denotes myc-H5, arrowhead TDP-43. Note the high molecular weight smear indicative of protein aggregation (bar in top panel). Also note that the C-terminal antibody nonspecifically binds to myc-H5 under these conditions. (D) HEK293T cells transfected with myc-m5 and immunostained with antibodies to myc and human-specific TDP-43. Merge with DAPI counterstain on right; note cells with m5 expression lack endogenous hTDP-43 staining, both nuclear and cytoplasmic. Scale bar, 20 µm. (E) HEK293T cells cotransfected with myc-H5 and FLAG-hTDP-43, and immunostained using indicated antibodies; merge with DAPI counterstain on right. Note the colocalization of myc and FLAG epitopes in yellow. Scale bar, 20 µm.
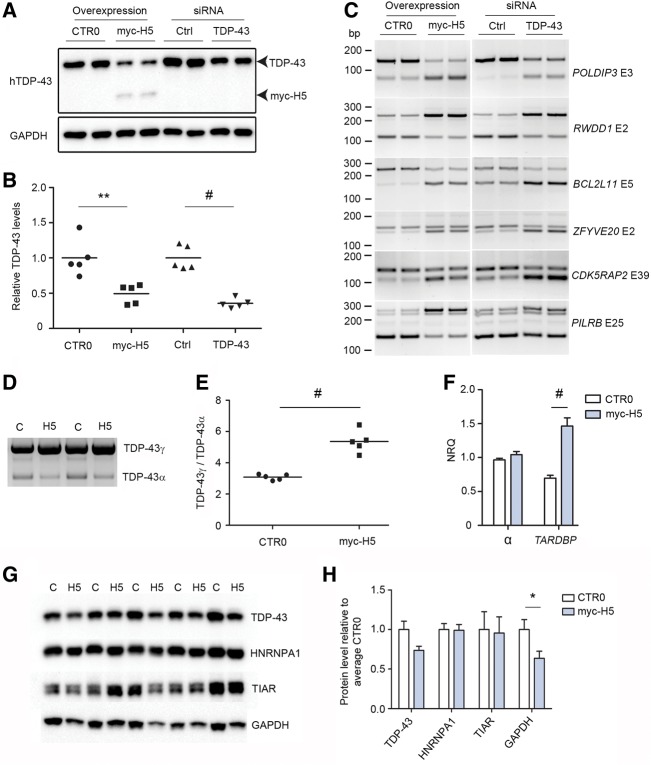
Studies in mice and in neuronal cell lines have demonstrated that loss of TDP-43 by conventional silencing techniques results in profound splicing abnormalities of TDP-43 RNA substrates (Polymenidou et al. 2011; Tollervey et al. 2011). We aimed to determine if the loss of soluble, nuclear TDP-43 observed in our work was accompanied by RNA dysregulation predicted by these previous studies. We expressed empty vector or myc-H5 in HEK293T cells and in parallel-transfected control siRNA or TDP-43 siRNA, and quantified the level of soluble, functional TDP-43 in the high salt fraction (Fig. 6A). TDP-43 levels in both myc-H5 and TDP-43 siRNA-treated cells were reduced relative to their controls (myc-H5 49 ± 0.06% relative to vector only; TDP-43 siRNA 35 ± 0.04% relative to ctrl siRNA, n = 5 per treatment) (Fig. 6B). In parallel, we harvested RNA from identically treated cells to assess splicing of known TDP-43 substrates using semi-quantitative RT-PCR. We chose six targets (POLDIP3, RWDD1, BCL2L11, ZFYVE20, CDK5RAP2, and PILRB) that were previously identified from unbiased searches for TDP-43 RNA substrates in humans and that showed the largest changes in exon inclusion or exclusion upon silencing of TDP-43 in human neuronal cell lines (Tollervey et al. 2011). In TDP-43 siRNA-treated HEK293T cells, we were able to replicate these previous findings, identifying enhanced exon inclusion or exclusion in all six transcripts compared with control siRNA-treated cells (Fig. 6C). Furthermore, we found identical patterns of exon inclusion or exclusion in HEK293T cells transfected with myc-H5 compared with empty vector (Fig. 6C).
FIGURE 6.
Sequestration of nuclear TDP-43 induces dysregulation of RNA metabolism and TARDBP processing. (A) Representative Western blotting of high salt fraction from HEK293T cells transfected with empty CTR0 vector, myc-H5, control (Ctrl) siRNA, or siRNA to TDP-43, GAPDH for loading. (B) Quantification of samples from A; results are shown as levels of TDP-43 in treated conditions (myc-H5 and TDP-43 siRNA) relative to respective controls (empty CTR0 plasmid and ctrl siRNA), N = 5 per group. (C) RT-PCR of HEK293T cells transfected in parallel with the samples from A and B, using primers for specific exons of six TDP-43 binding substrates identified in humans by Tollervey et al. (2011). Gene names are indicated on right in italics, nonitalicized “E” refers to the exon assayed. (D) Representative 3′ RACE of HEK293T cells transfected with empty CTR0 plasmid control (C) or myc-H5 (H5), using primers specific to full-length TDP-43 encoding transcript. (E) Quantification of the ratio of unspliced TDP-43γ to α-spliced TDP-43α from D, N = 5 per group. (F) Quantitative real-time PCR of HEK293T cells as treated in D, assaying total α-splicing, and TARDBP encoding full-length TDP-43 transcript, N = 5 per group. (G) Western blotting analysis of urea-extracted HEK293T cells overexpressing empty vector CTR0 (C) or myc-H5 (H5) using antibodies to the indicated proteins. (H) Quantitation of the blots in G. In B, E, F, and H: (*) P < 0.05, (**) P < 0.01, (#) P < 0.001, two-tailed unpaired t-test, error bars are SEM.
Studies using conventional silencing techniques cannot offer any information on aberrant splicing of TARDBP in response to loss of TDP-43 protein; whereas, the sequestration paradigm above allows for such an analysis. Consequently, we performed 3′ RACE on RNA extracted from HEK293T expressing empty vector or myc-H5 to assess the α-splicing of TARDBP (splicing of pre-mRNA to TDP-43α) encoding full-length TDP-43 under conditions of reduced TDP-43 protein. We observed an increase in the ratio of TDP-43γ:TDP-43α in cells overexpressing H5 relative to vector only controls (Fig. 6D,E). As soluble, nuclear and functional TDP-43 is reduced in cells expressing H5, this finding is consistent with data above demonstrating that overexpression of TDP-43 causes a decrease in this ratio. We also performed qPCR to measure total α-splicing and observed no change (Fig. 6F), further demonstrating that TDP-43 does not regulate α-splicing of all transcripts. As overexpression of TDP-43 in vivo and in vitro leads to decreased TARDBP transcript and TDP-43 protein levels (Ayala et al. 2011; Igaz et al. 2011; Xu et al. 2013; D'Alton et al. 2014), and that increased α-splicing is implicated in this process (Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012), we examined the effect of loss of functional, nuclear TDP-43 and concomitant increase in the TDP-43γ:TDP-43α ratio on TARDBP transcript levels. Using primers that detect all transcripts that encode full-length TDP-43, we observed a doubling of TARDBP transcript in cells overexpressing H5 relative to controls (NRQ empty vector = 0.69 ± 0.04, NRQ myc-H5 = 1.46 ± 0.12, N = 5 per group, P = 0.0003) (Fig. 6F). As increased TDP-43 protein could provide substrate for greater cytoplasmic aggregation and disease propagation, we repeated the above transfection of myc-H5 and empty vector control and harvested cells directly in urea, thus solubilizing all TDP-43 protein in one fraction. Western blotting and raw quantitation of TDP-43 in this fraction revealed a strong trend toward down-regulation of TDP-43 protein in myc-H5 transfected cells relative to empty vector controls (myc-H5 = 0.73 ± 0.05%, empty vector = 1.0 ± 0.11, N = 5 per group, P = 0.06) (Fig. 6G,H). This decrease was not limited to TDP-43, as we also observed a similar decrease in GAPDH (myc-H5 = 0.63 ± 0.09, empty vector = 1.0 ± 0.13, P = 0.048) (Fig. 6G,H). However, this effect was not global in nature as levels of the RNA-binding proteins HNRNPA1 and TIAR were unaffected (Fig. 6G,H).
DISCUSSION
Previous work from several different groups had reported the existence of an alternative isoform of TDP-43 created by splicing of Tardbp or TARDBP from exon 6, and a splicing event in the 3′ UTR necessary for autoregulation (Wang et al. 2002, 2004; Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012). Discovery of these splicing events chiefly relied on searches of preexisting sequence repositories. Herein we have performed the most comprehensive analysis to date of Tardbp using a disease-relevant region as a source of mRNA (cortex) and a technique that allows for the identification of poly(A)-bearing expressed sequences spanning multiple exon junctions (3′ and 5′ RACE). We discovered that the extent and diversity of Tardbp splicing is far more complex than previously anticipated, giving rise to nineteen transcripts, nine potentially protein coding transcripts, and multiple 3′ UTRs and polyadenylation sites. It is likely that this work still underestimates this complexity. For example, of the five poly(A) sites identified, three were located in the middle of the 3′ UTR or at the very distal 3′ end of the transcript. We do not know whether all Tardbp transcripts (including that encoding full-length Tdp-43) or merely a proportion extend to all or some of these sites. Furthermore, the effect of these 3′ UTRs on transcript and TDP-43 protein levels is unknown.
In previous studies, the ability of TDP-43 to autoregulate expression of these isoforms has yielded contradictory results (Ayala et al. 2011; Polymenidou et al. 2011; Avendaño-Vázquez et al. 2012). Herein, we provide data that explain these discrepancies. We demonstrated that TDP-43 nondiscriminately up-regulates the expression of all 6D junctions in vitro and in vivo, a result that is consistent with the observations of Polymenidou et al. (2011), who analyzed the expression of only one transcript. However, we also determined that this up-regulation was limited to single exons (6D) as we found no evidence that whole transcripts (6D–7D) bearing these exons were increased. Although we cannot rule out that we failed to identify a solitary transcript that is up-regulated fully across both 6D and 7D junctions by overexpression of TDP-43, we also found no evidence of up-regulation at the protein level. This exactly matches the findings of Ayala et al. (2011) and Avendaño-Vázquez et al. (2012), who did not detect increases in the expression of full transcripts across both 6D and 7D junctions using RT-PCR. This finding that not all enhanced exon inclusion or exclusion events are reflected in full length, spliced transcripts highlights an important limitation of measuring splicing of single exons. This is particularly important in the study of RNA-binding proteins such as TDP-43, for which current high-throughput sequencing techniques with short read lengths (such as RNA-seq) do not allow for discrimination of total transcripts bearing the changes in the affected exon from those that do not. Indeed, this important consideration is also exemplified by our finding that although TDP-43 protein does autoregulate the expression of α-splicing of transcripts encoding full-length TDP-43, there is no increase in the abundance of α junctions in total. Consequently, regulatory activity of this nature may be missed by current high-throughput sequencing methods. Although TDP-43 protein only regulated α-splicing of transcript encoding full-length TDP-43 in our studies, we cannot definitively say that this is exclusive and rule out that there are no other transcripts regulated in this fashion. However, it does appear that TDP-43 activity regulates a subset of transcripts. Further studies will be needed to determine how this is achieved, but possible scenarios include binding to only a subset of pre-mRNAs or the recruitment of cofactors to a subset of transcripts. Most importantly, these findings are likely to extend to other TDP-43 substrates, and future work will be required to determine if disturbance in the splicing of single exons due to TDP-43 dysfunction is reflected in the final translated or functional RNA.
Interestingly, we determined that many of the alternative transcripts we identified encode unique C-terminal sequences that influence the subcellular distribution of the RRM domains N-terminal to them. Originally, one of these isoforms was found localized in discrete nuclear bodies (Wang et al. 2002). Here, we found that this isoform is one of three in mice that harbor a consensus motif (VHLISNVYGRSTSLKVVL) that is responsible for this localization. Furthermore, we identified a unique consensus sequence in three human and one murine isoform (EFVITHHRPRL) that renders localization cytoplasmic. However, in vivo in murine cortex, we only found evidence for expression of m2 (transcripts containing the 6D junction 2) as protein, and we found no evidence of expression in human cortex. It is possible that other isoforms are translated but are below the detection threshold of our methodology. However, it is interesting to speculate that there is a mechanism in place to distinguish and translate only the transcript encoding m2 from all other, highly related alternative transcripts that are produced, including two that encode highly similar proteins with identical predicted localization. In either case, it is clear that regulation of Tardbp is multifaceted, complex and is controlled at multiple levels. Furthermore, the lack of any expressed protein human ortholog to m2 raises the question of divergent regulation of mouse and human transcripts, which may be a consideration in the study of TDP-43 in mice.
The lack of expression of these isoforms in human cortex means a role in disease is improbable. However, we determined that the H5 isoform forms highly insoluble aggregates in the cytoplasm, and is functionally capable of sequestering exogenous TDP-43. This aggregated H5 was also capable of driving loss of endogenous, soluble, nuclear TDP-43. Although we were unable to visualize cytoplasmic colocalization with aggregated H5, the shift to an insoluble form no longer in the nucleus (i.e., in the cytoplasm) favors sequestration as the underlying mechanism of these effects. As previous studies investigating the effect of loss of soluble, nuclear TDP-43 on RNA metabolism have chiefly used knockdown techniques, we examined what effect sequestration of TDP-43 would have on RNA metabolism. We were able to confirm findings reported previously that knockdown of TDP-43 using siRNA leads to aberrant splicing of the six transcripts tested and we demonstrated that sequestration results in identical changes in the splicing of these known TDP-43 substrates. Furthermore, we were able to examine the effect of loss of soluble, functional TDP-43 protein on TARDBP regulation itself, an analysis that is not possible using conventional silencing paradigms. We determined that loss of endogenous TDP-43 protein is accompanied by an increase in the ratio of TDP-43γ:TDP-43 α (i.e., decreased α-splicing), and an increase in TARDBP transcript encoding full-length TDP-43. These two observations are compatible with data from numerous models in vivo and in vitro (Ayala et al. 2011; Igaz et al. 2011; Avendaño-Vázquez et al. 2012; Xu et al. 2013; D'Alton et al. 2014), which show reduction of TARDBP/Tardbp transcript levels in response to exogenous TDP-43 expression. In our studies, the increase in TARDBP transcript did not result in increased levels of TDP-43 protein; in fact, cells overexpressing H5 strongly trended toward reduced TDP-43 protein. However, in our opinion, this does not rule out increases in TDP-43 due to aberrant processing of TARDBP in disease. Although the paradigm we have used here faithfully models loss of TDP-43 function in the nucleus, and thus RNA misprocessing due to loss of TDP-43, it is unlikely to model other features that are dependent on the aggregate themselves. For example, the stability of sequestered or cytoplasmic TDP-43 may be dependent on the nature of the aggregate. Furthermore, we do not know the mechanism by which H5 induced loss of GAPDH and TDP-43. This does not appear to be a global effect, as the levels of HNRNPA1 and TIAR were unaltered. As the H5 paradigm used here is unlikely to be representative of disease, we cannot be certain that any mechanism discovered for these proteomic changes would be relevant to FTLD or ALS.
Herein, we have provided the most thorough description of the alternative splicing of the Tardbp transcript. Our analysis of these transcripts and the autoregulatory capacity of TDP-43 provides insights into TDP-43 function, splicing, and potential mechanisms in FTLD and ALS.
MATERIALS AND METHODS
Animal husbandry
All procedures involving FVB mice and transgenic TDP-43 animals were conducted according to the National Institutes of Health guide for animal care and approved by the Institutional and Animal Care and Use Committee at University of Florida.
3′ and 5′ rapid amplification of cDNA ends
3′ and 5′ rapid amplification of cDNA ends (RACE) was performed using the Generacer kit (Invitrogen) using manufacturer's instructions and custom primers as detailed in Supplemental Table 1. PCR products from the second, nested PCR reaction were blunt-end ligated into pCR4-blunt and following standard transformation and selection techniques the plasmid was sequenced to identify PCR product.
RNA preparation and quantitative real-time PCR
Total RNA was isolated from dissected cortex or immortalized cell lines using TRIzol reagent (Life Technologies) and Pure Link RNA Mini Kit (Life Technologies). The resulting RNA purity and integrity was assessed by spectrophotometer (Nanodrop). Two microgram of RNA was used to synthesize cDNA using the high capacity cDNA Reverse Transcription Kit (Applied Biosystems). All samples were run in triplicate on the ABI 7900HT real-time PCR detection system using SYBR green PCR master mix (Applied Biosystems). Primer efficiencies (E) were calculated using E = 10(−1/slope), where slope was determined by plotting the Cq (quantification cycle) values against the log10 input of a cDNA dilution series. Single products were confirmed by melt curve analysis. A 1/10–1/50 dilution of each experimental cDNA sample was run. From this, the relative quantities (RQ) were calculated for each primer pair in each sample and normalization of the target performed using the geometric mean of at least two of the three reference genes gapdh, actb, and rps6 (murine transcripts) or GAPDH and ACTB (human transcripts) giving the Normalized Relative Quantity (NRQ) (Hellemans et al. 2007). Primer sequences are detailed in Supplemental Table 1, with the exception of RT-PCR primers for Figure 6C, which are identical to that of Tollervey et al. (2011).
Plasmid generation
Full-length human TDP-43 complementary cDNA in plasmid pEGFP-C1 (Zhang et al. 2009) was used as PCR template to generate N-terminally myc- and FLAG-tagged human TDP-43. Alternative isoforms of human TDP were amplified using cDNA from H4 neuroglioma cells. All mouse Tdp-43 isoforms were generated using cDNA from mouse cortex. Primers are detailed in Supplemental Table S1, with the exception of the forward primers for myc-hTDP-43 (5′-TACGGATCCCACCATGGAACAAAAACTCATCTCAGAAGAGGATCTGATGTCTGAATATATTCGGGTAACCGAA-3′), FLAG-hTDP (5′-TACGGATCCCACCATGGACTACAAGGACGACGATGACAAGATGTCTGAATATATTCGGGTAACCGAA-3′), FLAG-mTDP (5′-TACGGATCCCACCATGGACTACAAGGAC GACGATGACAAGATGTCTGAATATATTCGGGTAACAGAA-3′), and myc-mTDP (5′-TACGGATCCCACCATGGAACAAAAACTC ATCTCAGAAGAGGATCTGATGTCTGAATATATTCGGGTAAC AGAA-3′). Products were cloned into pcDNA3.1 (Life Technologies) or CTR0, in which expression is driven by the CMV early enhancer/chicken β actin (CAG) promoter (Rutherford et al. 2013).
Cell culture and transfection
N2a, HeLas3, HEK293T, and H4 cell lines were maintained in Dulbecco's Modified Eagle's Medium (Lonza) supplemented with 10% fetal calf serum (Sigma) and penicillin-streptomycin (Life Technologies). Transfections were performed with Lipofectamine 2000 (Sigma) or Fugene HD (Promega) using 250 ng of DNA per well of a 24-well plate following the manufacturer's guidelines. For low level transfection of H5, we used 250 ng of plasmid per well in a 24 well plate at a ratio of 2.5:1 lipofectamine:DNA. For high-level transfection, we used 1.5 µg of plasmid per well in a 24-well plate at a ratio of 2.5:1 lipofectamine:DNA. In both cases, complexes were formed in penicillin-streptomycin free media for 20 min and added to cells at >95% confluence.
Protein isolation, fractionation, Western blotting, and antibodies
Cortex from mice and human was homogenized in lysis buffer (50 mM Tris, 300 mM NaCl, 1% Triton X-100, 1 mM EDTA, and protease inhibitors and phosphatase inhibitors [Sigma]) at 6 mL/g tissue, and aliquots of homogenate stored at −80°C. Lysate was prepared via brief sonication, addition of SDS to 2% and centrifugation at 40Kg for 20 min at 4°C. For preparation of protein lysate from cell culture, cells were washed once in ice cold PBS, harvested, collected via brief centrifugation, and lysed in lysis buffer as above. In Figure 6, cells were lysed directly in urea buffer (30 mM Tris at pH 8.5, 4% CHAPS, 7M urea, 2M thiourea) following collection, and briefly sonicated. Ten microgram protein was loaded onto 10% or 15% Tris–glycine polyacrylamide gel (Novex). Following electrophoresis and transfer, nitrocellulose membranes were blocked with 5% milk in TBS (Tris-buffered saline), incubated with appropriate primary and HRP-conjugated secondary antibodies and visualized using ECL reagent (PerkinElmer). Images were captured on the ProteinSimple FluorChem E (ProteinSimple). Antibodies are listed in Supplemental Table 1.
To analyze solubility of TDP-43, cells from a 6-well plate were harvested in 100 µL of high salt buffer (10 mM Tris at pH 7.5, 5 mM EDTA, 10% sucrose, 1% triton, 0.5M NaCl, with protease and phosphatase inhibitors), incubated at 4°C for 15 min and clarified by centrifugation at 110Kg. The supernatant containing the high salt fraction was snap frozen on dry ice and the pellet washed and re-homogenized in 100 µL myelin floatation buffer (high salt buffer with 30% sucrose), followed by a brief sonication. This homogenization, centrifugation and fraction collection sequence was repeated with sarkosyl buffer (high salt buffer substituting 1% triton for 1% N-lauroyl-sarcosine, incubation 1 h at room temperature), and finally in 60 µL urea buffer (30 mM Tris at pH 8.5, 4% CHAPS, 7M urea, 2M thiourea, followed by brief sonication). Fifty microgram of high salt extract and equivalent volumes of all other fractions were analyzed by Western blotting as described above.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.5% triton for 10 min prior to incubation in blocking buffer (2% fetal calf serum/3% bovine serum albumin in PBS) for 45 min. Primary antibodies diluted in blocking buffer were incubated between 1 h and overnight and following extensive washing with 0.1% Triton-PBS were labeled with fluorescence-conjugated secondary antibodies (1:1000, Invitrogen Alexa Fluor molecular probes). Vectashield with DAPI (Vector Laboratories) was used to stain nuclei. Images were captured using an Olympus BX60 microscope or Olympus DSU-IX81 confocal microscope (Olympus).
Mass spectrometry
MS was performed by the University of Florida mass spectrometry core. The gel band was excised using a razor blade and digested with trypsin as previously described (Sheffield et al. 2006). Resulting lyophilized peptides were resuspended in loading buffer (3% acetonitrile, 0.1% acetic acid, 0.01% trifluoroacetic acid) and loaded onto a C18 capillary trap cartridge (LC Packings). They were then separated on a 15 cm nanoflow analytical C18 column (PepMap 75 μm id, 3 μm, 100 Å) at a flow rate of 300 nL/min on a nanoLC ultra 1D plus system (ABsciex). For this, solvent A composition was 3% acetonitrile (ACN) v/v, 0.1% acetic acid v/v and solvent B was 97% ACN v/v, 0.1% acetic acid v/v. A linear gradient from 3% to 40% of solvent B for 60 min, followed by an increase to 90% for 5 min (Silva-Sanchez et al. 2014) was used to separate peptides. The eluted peptides were sprayed into an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Inc.) and MS2 spectra were acquired in a data-dependent mode. An Orbitrap full MS scan (resolution: 3 × 104, mass range 300–2000 Da) was followed by 10 MS2 scans in the ion trap. These were performed via collision induced dissociation on the top 10 most abundant ions. Isolation window for ion selection was 3 Da. Normalized collision energy was set at 28%. Dynamic exclusion time was 20 sec (Li et al. 2012). Additionally, a parent list of predicted unique peptides for the C-terminus of TAR-DNA binding protein was included as targeted sequences (Supplemental Table 1). The acquired mass spectra were searched against a IPI mouse database (59534 entries, 12 Jul 2012) using Mascot 2.2 search engine (http://www.matrixscience.com) with the following parameters: tryptic peptides with 1 missed cleavage site, mass tolerance of precursor ion of 15 ppm and MS/MS ion of 0.8 Da, fixed carbamidomethylation of cysteine, variable methionine oxidation, asparagine, and glutamine deamination. Unambiguous identification was done using Scaffold software v. 3.3.2 (Proteome Software).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a generous donation from the Wilder Foundation, and funding from The University of Florida and the National Institutes of Health, National Institute of Neurological Disorders and Stroke (NIH/NINDS) 5R21NS071097-02 (J.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.047647.114.
REFERENCES
- Avendaño-Vázquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE. 2012. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev 26: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. 2008. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 121: 3778–3785. [DOI] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, et al. 2011. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 30: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembich S, Herzog JS, De Conti L, Stuani C, Avendaño-Vázquez SE, Buratti E, Baralle M, Baralle FE. 2014. Predominance of spliceosomal complex formation over polyadenylation site selection in TDP-43 autoregulation. Nucleic Acids Res 42: 3362–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandmeir NJ, Geser F, Kwong LK, Zimmerman E, Qian J, Lee VM, Trojanowski JQ. 2008. Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta Neuropathol 115: 123–131. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon A, Yang B, Knight J, Farnham IM, Zhang Y, Wuertzer CA, D'Alton S, Lin WL, Castanedes-Casey M, Rousseau L, et al. 2012. Neuronal sensitivity to TDP-43 overexpression is dependent on timing of induction. Acta Neuropathol 123: 807–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che MX, Jiang YJ, Xie YY, Jiang LL, Hu HY. 2011. Aggregation of the 35-kDa fragment of TDP-43 causes formation of cytoplasmic inclusions and alteration of RNA processing. FASEB J 25: 2344–2353. [DOI] [PubMed] [Google Scholar]
- D'Alton S, Altshuler M, Cannon A, Dickson DW, Petrucelli L, Lewis J. 2014. Divergent phenotypes in mutant TDP-43 transgenic mice highlight potential confounds in TDP-43 transgenic modeling. PLoS One 9: e86513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo V, Grossi E, Laneve P, Morlando M, Dini Modigliani S, Ballarino M, Bozzoni I, Caffarelli E. 2013. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol Neurobiol 48: 952–963. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. 2011. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem 286: 18664–18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, et al. 2011. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 121: 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Mieda-Sato A. 2012. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci 109: 3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ferraris JD, Yu D, Singh T, Izumi Y, Wang G, Gucek M, Burg MB. 2012. Proteomic analysis of high NaCl-induced changes in abundance of nuclear proteins. Physiol Genomics 44: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. 2010. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci 107: 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. 2009. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet 18: 3353–3364. [DOI] [PubMed] [Google Scholar]
- Parada GE, Munita R, Cerda CR, Gysling K. 2014. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res 42: 10564–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, Cannon A, Xu G, Golde TE, Shaw G, et al. 2013. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res 1524: 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield J, Taylor N, Fauquet C, Chen S. 2006. The cassava (Manihot esculenta Crantz) root proteome: protein identification and differential expression. Proteomics 6: 1588–1598. [DOI] [PubMed] [Google Scholar]
- Silva-Sanchez C, Chen S, Li J, Chourey PS. 2014. A comparative glycoproteome study of developing endosperm in the hexose-deficient miniature1 (mn1) seed mutant and its wild type Mn1 in maize. Front Plant Sci 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. 2007. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci 35: 320–327. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43; implications for neurodegenerative diseases. Nat Neurosci 14: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Reddy NM, Shen CK. 2002. Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci 99: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Wang IF, Bose J, Shen CK. 2004. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83: 130–139. [DOI] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. 2008. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 283: 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YF, Prudencio M, Hubbard JM, Tong J, Whitelaw EC, Jansen-West K, Stetler C, Cao X, Song J, Zhang YJ. 2013. The pathological phenotypes of human TDP-43 transgenic mouse models are independent of downregulation of mouse Tdp-43. PLoS One 8: e69864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes-Casey M, Ash P, et al. 2009. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci 106: 7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.