Abstract
Objective(s):
Development of molecules that specifically recognize cancer cells is one of the major areas in cancer research. Human epidermal growth factor receptor 2 (HER2) is specifically expressed on the surface of breast cancer cells. HER2 is associated with an aggressive phenotype and poor prognosis. In this study we aimed to isolate RNA aptamers that specifically bind to HER2 overexpressing TUBO cell line.
Materials and Methods:
Panel of aptamers was selected using cell-based systematic evolution of ligands by exponential enrichment (cell-SELEX).
Results:
Binding studies showed that selected aptamers can identify TUBO cell line with high affinity and selectivity. Our preliminary investigation of the target of aptamers suggested that aptamers bind with HER2 proteins on the surface of TUBO cells.
Conclusion:
We believe the selected aptamers could be useful ligands for targeted breast cancer therapy.
Keywords: Breast cancer, Cell-SELEX, RNA aptamer, TUBO cell line
Introduction
A variety of novel drug delivery systems have been developed to improve the selectivity of anticancer agents. Targeted drug delivery systems exploit differences between cancer and normal cells.
Breast cancer is one of the most common female cancers in the world and is the most common cause of cancer death in women aged 45–55 years (1, 2).
Human epidermal growth factor receptor-2 (HER2/erbB-2) belongs to a family of four transmembrane receptors involved in signal transduction pathways that regulate cell growth and differentiation. HER2 has an important role in the pathogenesis of breast cancer. Overexpression of HER2 is associated with malignancy and poor prognosis of breast cancer. One strategy for targeted cancer therapy is the use of anti-HER2 targeting agents (1, 3, 4).
Aptamers are single strand nucleic acids that fold in 3D structures (5, 6) and specifically bind to their targets such as proteins, nucleic acids, phospholipids and sugars with high affinity and selectivity (7, 8). Aptamers offer advantages over antibodies, which make them useful tools for the validation of targets. Aptamers are small and nonimmunogenic molecules that can be produced by chemical synthesis without the need for biological systems. Chemical production is cost effective and can be automated. Aptamers are stable in wide ranges of pH and temperature. They can also withstand organic solvents. The characterization and modification of aptamers is easier than antibodies (5, 9, 10).
Aptamers that bind to specific targets are generated by SELEX (the Systematic Evolution of Ligands by Exponential enrichment). During this process oligonucleotides with unique sequences are selected from a random pool.
SELEX was independently developed in 1990 by Lary Gold and Jack Szostak (11-13). In the SELEX procedure an oligonucleotide library is incubated with the target. Then oligonucleotides with affinity for the target are eluted, amplified and single stranded. SELEX cycles are repeated for several rounds until proper sequences that specifically recognize the target are obtained (14, 15).
The aim of this study was to find RNA aptamers targeting HER2-overexpressing TUBO cells using the Cell-SELEX strategy. The selected aptamers showed strong affinity to TUBO cells. Specific targeting of HER2 receptor by aptamers was further confirmed by flow cytometry.
Materials and Methods
Cell lines and culture condition
TUBO, a cloned cell line that overexpresses the rHER2/neu protein, was used as the target of selection. This cell line was kindly provided by Dr Pier-Luigi Lollini (Department of Clinical and Biological Sciences, University of Turin, Orbassano, Italy) and was cultured in Dulbecco’s Modified Eagle Medium, (DMEM) supplemented with 20% fetal bovine serum (FBS).
A murine colon carcinoma cell line, CT26, was purchased from the Pasteur Institute of Iran, and cultured in RPMI-1640 medium supplemented with 10% FBS. CT26 cells (rHER2/neu negative) were as a negative control to remove sequences that bind to both cell lines (16, 17). Cells were regularly subcultured to maintain exponential growth.
Other cell lines were used in this project, all purchased from the Pasteur Institute of Tehran, Iran, and maintained in cell culture medium under the recommended conditions.
Primers and library
The random library (5’ACC GAG TCC AGA AGC TTG TAG TAC T-N35-GCC TAG ATG GAG TTG AAT TCT CCC TAT AGT GAG TCG TAT TAC-3’) was synthesized by DNA synthesizer (PolyGen) and purified by gel purification (18-20). This library was amplified using forward primer (5’-GTAATACGACTCACTATAGGGAGAATTCAACTGCCAT -CTA-3’) and reverse primer (5’-ACCGAGTCCAGAA-GCTTGTAGT-3’). RNA library was transcribed from the PCR product using DuraScribe T7 transcription kit (Epicentre Technologies). RNase A resistant product is accomplished by replacing CTP and UTP with 2’-Fluorine-dCTP (2’-F-dCTP) and 2’-Fluorine-dUTP (2’-F-dUTP) in the DuraScribe in vitro transcription reaction. After purification, the RNA library was added to 300 µl binding buffer containing HEPES-NaOH (20 mM, pH 7.4), NaCl (150 mM), CaCl2 (1.5 mM), MgCl2 (0.5 mM), and 10% yeast tRNA (Sigma). To retain correct configurations, the RNA library was denatured at 90 °C for 1 min and snap-cooled on ice.
Cell SELEX
TUBO cells were dislodged from the flask after a short period of incubation with trypsin and then counted. The cells’ viability was assessed by Trypan blue assay. 5-10 million cells were centrifuged, washed 3 times with washing buffer (20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 1.5 mM CaCl2, 0.5 mM MgCl2) and resuspended in the binding buffer (washing buffer plus 10% yeast tRNA (Sigma).
Cells were incubated with a solution of library in the binding buffer at 4 °C for 45 min. After 2 washes, sequences that bound to TUBO cells were recovered by denaturation of RNA sequences as well as surface proteins at 95 °C for 5 min. Cells were precipitated and RNA sequences were retrieved by ethanol precipitation of the supernatant. The obtained RNA sequences were reverse-transcribed using the Cloned AMV first-strand cDNA synthesis kit (Invitrogen), and PCR-amplified. The purified PCR products were transcribed in vitro using the Dura Scribe T7 transcription kit (Epicentre Technologies). After third round of selection, counter selection was performed to subtract sequences with affinity for both the control and target cells. For negative selection, the RNA sequences eluted from TUBO cells were incubated with the CT26 cell line, and unbounded sequences were ethanol precipitated (19, 21, 22).
Cloning, sequencing, and structure analysis of selected aptamers
After 12 rounds of selection, the PCR amplified dsDNAs were cloned in to Escherichia coli DH5-α using the TOPO TA cloning kit (Invitrogen K4500-40). Individual white colonies were picked and cultured in a liquid LB (Lurai-Bertani) medium. After a brief centrifugation, plasmids were purified using Gene lute TM HP Fine-Minute Plasmid MiniPrep (Sigma Aldrich) and sequenced by Bioneer Company (23).
Sequences were aligned using the sequence alignment program Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). A phylogenic tree was constructed using the DNAMAN version 6 software (Lynnon Corporation) and the sequences were grouped. Representative sequences from different groups were selected as candidate aptamers for further characterization (21, 24).
Flow cytometry binding assay
To monitor the enrichment of the library from aptamers with affinity for TUBO cells, eluted aptamers after 1, 3, 5, 8, and 12 SELEX cycles were labeled by Cy-5. Aminohexyl-ATP was incorporated into the RNA structure during the Dura Scribe transcription reaction followed by post transcriptional labeling with Cy5-NHS ester.
Then the Cy-5 labeled RNA sequences were incubated with about 5×105 target and negative cells for 30 min on ice in the dark in 500 µl binding buffer in the presence of 10% FBS. After incubation, the cells were washed two times and suspended in binding buffer and analyzed by flow cytometry (BD FACSCalibur cytometer).
The flow cytometry data was analyzed using FCS Express 4 Flow Cytometry (De Novo Software, Los Angeles, CA).
Determination of aptamer selectivity
The selectivity of aptamers for TUBO cells was tested by incubation of aptamers with the following cells: human prostatic carcinoma cell line (PC3), transformed mouse embryonic fibroblast cell line (NIH3T3), human breast cancer cell line (SK-BR-3), human Burkitt’s lymphoma cell line (Raji), and murine colon adenocarcinoma cell line (C26).
Determination of secondary structure of aptamers
The secondary structure of selected aptamers was analyzed by free-energy minimization using the algorithm according to the method of Zuker in mfold web based software (http://mfold.-rna.albany.edu/?q=mfold)(25).
Determination of apparent dissociation constant of aptamers
The range of Cy5 labeled aptamers and control library were incubated with a constant number of TUBO cells. The mean fluorescent intensities of aptamer and library at each concentration were determined. All binding assays were performed in triplicate. The mean fluorescence intensity of the unselected library was subtracted from that of the corresponding aptamer with target cells. Then, apparent dissociation constants (Kd) for the aptamer-cell interaction were calculated by fitting the dependence of fluorescence intensity (Y) and the concentration of aptamers (X) into the one-site saturation equation Y = Bmax X/(Kd + X) using Prism version 5 (GraphPad Software, San Diego, CA). In this equation Bmax is the maximum specific binding with the same unit as Y (21).
Effect of trypsin treatment on binding of aptamers to TUBO cells
TUBO cells were incubated with trypsin for 8 min at 37 °C. To stop trypsin activity, cells were mixed with ice cold culture medium containing FBS. Then, the cells were quickly washed and centrifuged. After resuspension in the binding buffer, cells were incubated with aptamers. Binding of aptamer to treated cells was assessed by flow cytometry and compared with untreated TUBO cells as positive control (26, 27).
Effect of temperature and culture medium on the binding of aptamers to cells
All aptamers were isolated at 4 °C in binding buffer. To verify the ability of target recognition of aptamers at 37 °C, binding assay was performed at 37°C and in culture medium as described in the binding assay section (26, 28).
Verification of aptamer binding to extracellular domain of HER2
TUBO cells were prepared as described in the flow cytometry section. TUBO cells were incubated with anti-HER2 Affibody® molecules (Affibody AB). After two washes, cells were incubated with Cy5 labeled aptamers. TUBO cells were also incubated with Cy5 labeled aptamers as positive control.
Mean fluorescence intensity of aptamers binding to Affibody pretreated cells was determined and compared with aptamers binding in positive control.
Results
SELEX and aptamer enrichment monitoring
SELEX was used to screen aptamers specifically binding to the breast cancer cell line TUBO, with CT26 as counter selection. Following every two continuous rounds, binding assay was performed. The enrichment of aptamer pool was monitored by flow cytometry. As we progressed with SELEX, the fluorescence intensity gradually increased, while there was no significant change in the same experiment with the counter cells. Selection cycles were stopped when significant difference between the mean fluorescence intensities of the unselected library and the selected RNA pool was observed (Figure 1).
Figure 1.
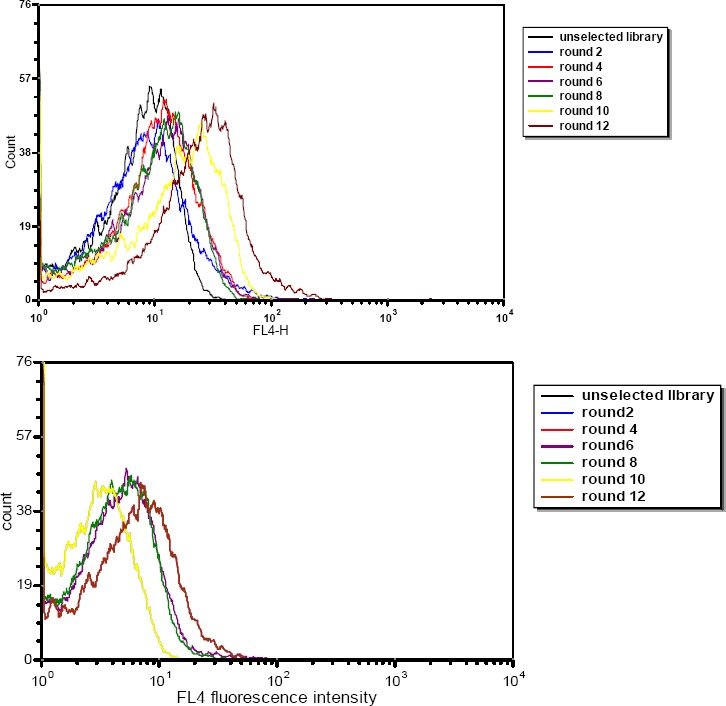
Enrichment of selected RNA pools of target TUBO (A) and control CT26 (B) during selection and monitored by flow cytometry
Cloning, sequencing and structure analysis of selected aptamers
After 12 rounds of selection the RNA pool was reverse transcribed into cDNA and amplified by PCR. The highly enriched pools were cloned into pTZ57R/T vector, and after culturing, positively inserted clones were picked, and plasmids were purified and sequenced using M13F (-20) and M13R (-40). Sequences were aligned and clustered based on their sequence homology (Figure 2). From the sequencing and alignment results TSA6, TSA7, TSA10, TSA12, and TSA14 were synthesized as potential aptamer candidates.
Figure 2.
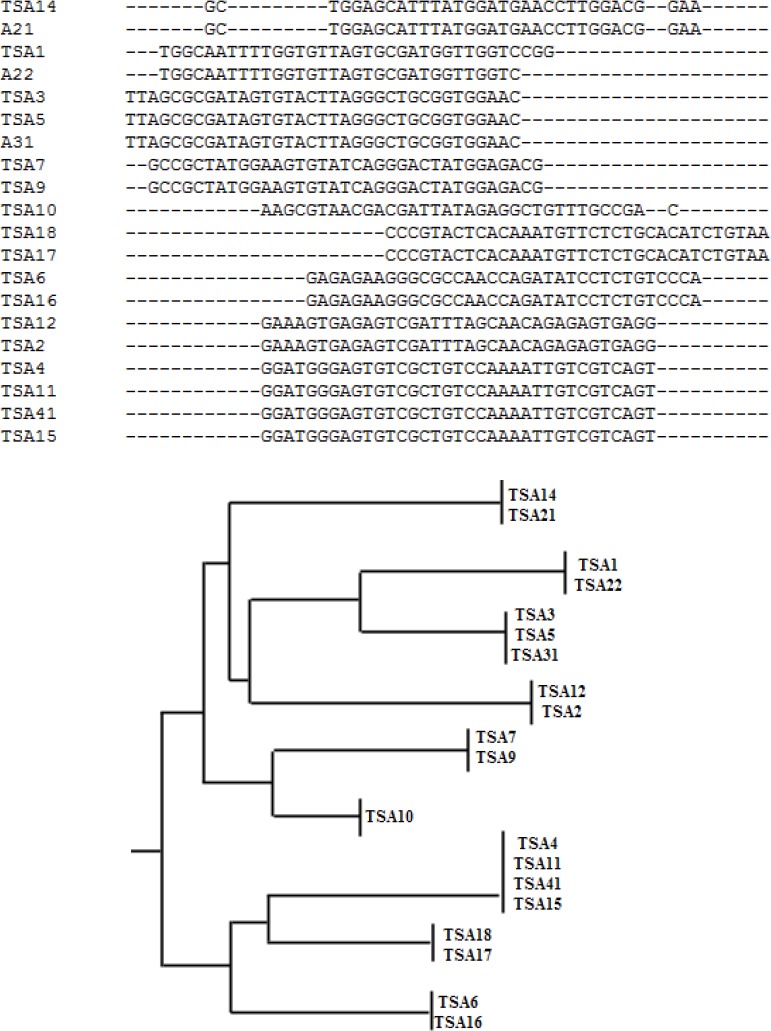
(A) Sample data of sequence alignment using Clustal Omega showing homologous families and nonhomologous sequences of potential aptamer sequences. (B) Phylogenic tree of selected sequence
Determination of aptamer selectivity
We investigated the binding affinity of selected aptamers for transformed mouse embryonic fibroblast cell line (NIH3T3), SKBR3 (hHER2+) breast cancer cell line, human prostate cancer cell line (PC3), Burkitt’s lymphoma cell line (Raji), and C26 murine colon carcinoma cell line (Table 1).
Table 1.
Specificity determination of TSA6, TSA7, TSA10, TSA12, and TSA14 aptamer
| Cell line | Disease | TSA6 | TSA7 | TSA10 | TSA12 | TSA14 |
|---|---|---|---|---|---|---|
| TUBO | mouse breast cancer | ** | *** | ** | ** | **** |
| CT26 | mouse colon carcinoma | * | * | * | - | * |
| PC3 | human prostate cancer | * | * | * | - | * |
| NIH3T3 | transformed mouse embryonic fibroblast cell line | * | * | * | * | * |
| SKBR3 | human breast cancer | * | ** | ** | ** | ** |
| Raji | human Burkitt’s lymphoma | * | * | * | * | * |
| C26 | mouse colon carcinoma | * | * | * | * | * |
Note: The mean fluorescence intensity (MFI) of TSA14 aptamer with TUBO cells was chosen as maximum intensity (****). The MFIs with other cells and other aptamers were therefore compared and accordingly assigned approximate binding intensity. A”-” indicates that the MFI is similar to MFI of unselected library with cell line
Although some levels of recognition were observed with these cell lines, affinity of aptamer candidates to TUBO cell line was much higher. There were no sequences that recognize TUBO cells exclusively. All selected aptamers had fair affinity to SKBR3 cell lines. As shown in Table 1 interaction of aptamers and normal cell line was like control cells. Affinity of aptamers for C26 cell line was measured, because it was closely related to control cell line.
TSA14 showed clearly stronger signal with TUBO cells compared to other sequences. Mean fluorescence intensity of TSA14 was about 2.5 times higher than other aptamers. TSA12 aptamer did not recognize CT-26, and PC3 cell lines and had minimum affinity for cell lines other than TUBO cells. According to this data, TSA14 and TSA12 were selected for further studies.
Determination of the secondary structure of aptamers
The secondary structure of RNA aptamers was predicted with the mfold web based software by calculation of minimum free energy (http://mfold.rna.albany.edu/?q=mfold). Figure 3, A and Figure 3, B show the predicted secondary structure of TSA12 and TSA14 aptamers. The free energy (ΔG) of the most stable structures of TSA12 and TSA14 was -8.29 and -9.68, respectively. Free energy was more negative for TSA14.
Figure 3.
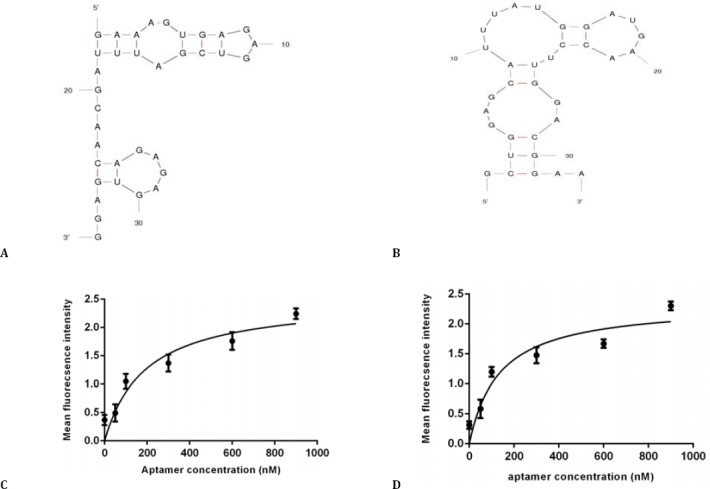
Software simulated secondary structure of TSA12 (A) and TSA14 (B) aptamers, binding curve of TSA12 aptamer (C) and TSA14 aptamer (D) with TUBO cells. Cells were incubated with varying concentrations of Cy5-labeled aptamer and unselected library in triplicate. Mean fluorescence intensity of each concentration was determined. The mean fluorescence intensity of the unselected library (background binding) was subtracted from the mean fluorescence intensity of corresponding aptamer. Using Prism, the apparent dissociation constant (Kd) of aptamer-cell interaction was obtained by fitting the dependence of fluorescence intensity of specific binding on the concentration of aptamers to the one-site saturation equation Y= Bmax X/(Kd + X)
The predicted structure of TSA14 was more complicated, having 3 loops.
Determination of aptamer binding affinity
Dissociation constant (Kd) of aptamer-TUBO cell interaction was determined. Kd of TSA12 and TSA14 were 191.9±21.77 nM and 133.9±12.78 nM, respectively (Figure 3, C and Figure 3, D). TSA14 had higher affinity for target cells.
Effect of trypsin treatment on binding of aptamer to TUBO cells
Figure 4, A shows that trypsin pretreatment decreased the fluorescence intensities compared to untreated cells. Trypsin treatment assay indicated that TSA14 aptamer bound to surface proteins that were digested by trypsin.
Figure 4.
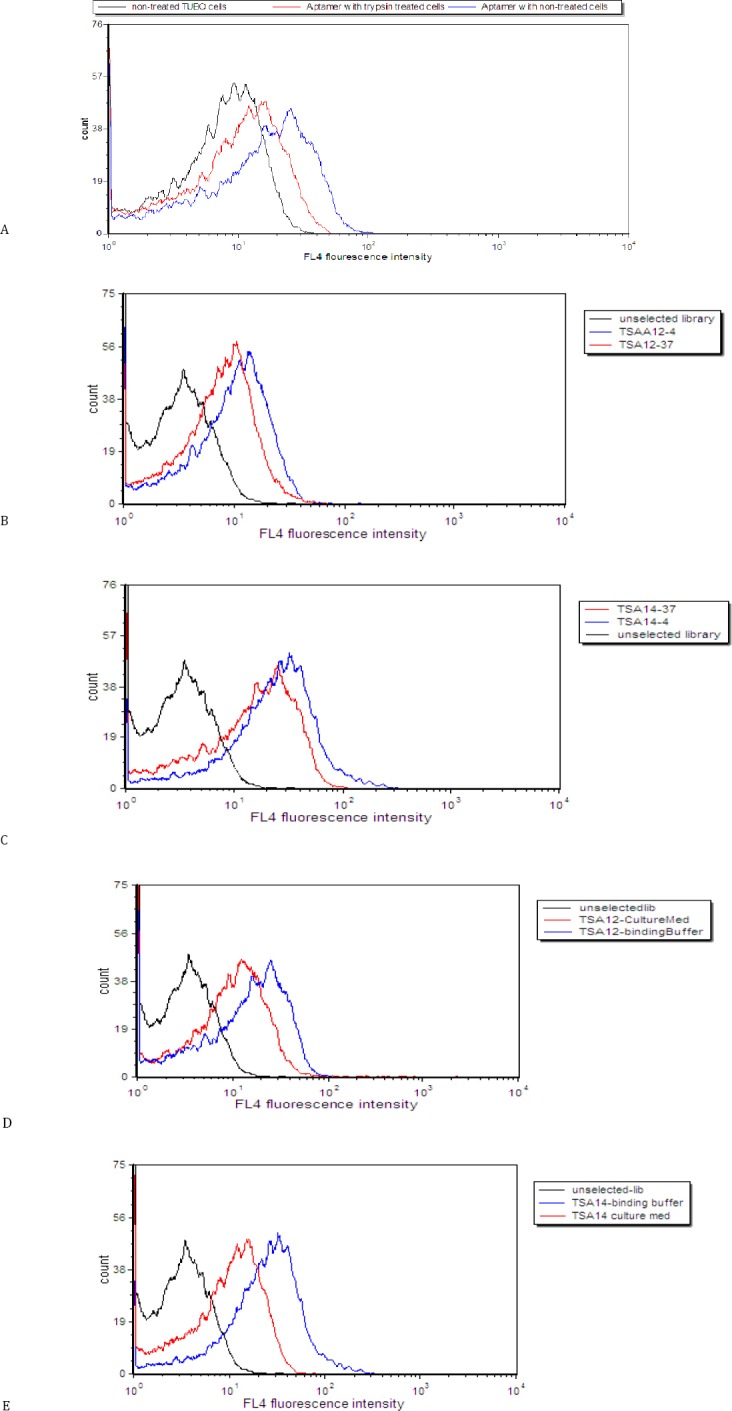
Effect of trypsin treatment on binding of aptamer to TUBO cells. Binding of unselected library to untreated cells (black histogram), binding of TSA14 aptamer to trypsin-treated cells (red histogram) and binding of TSA14 aptamer to untreated cells (blue histogram) (A). Assessment of the binding of TSA12 (B) and TSA14 (C) at 37 °C and 4 °C. Black (unselected library); blue (binding at 4°C) and red (binding at 37 °C). Assessment of the binding of TSA12 (D) and TSA14 (E) aptamers in culture medium. Black (unselected library); blue (binding in buffer medium) and red (binding in culture medium with FBS)
Effect of temperature and culture medium on the binding of aptamers
As shown in Figure 4, B and Figure 4, C the binding of aptamers changed slightly at 37 °C. Moreover, TSA12 and TSA14 showed reduced, but still significant binding to TUBO cells in culture medium (Figure 4, D and Figure 4, E).
Verification of aptamer binding to the extracellular domain of HER2
In order to confirm that aptamers bind to HER2 on the surface of TUBO cells, the external domain of HER2 was masked with anti-HER2 affibodies. Flow cytometry data showed that binding of Cy5-labeled aptamers decreased considerably after the incubation of TUBO cells with affibodies (Figure 5).
Figure 5.
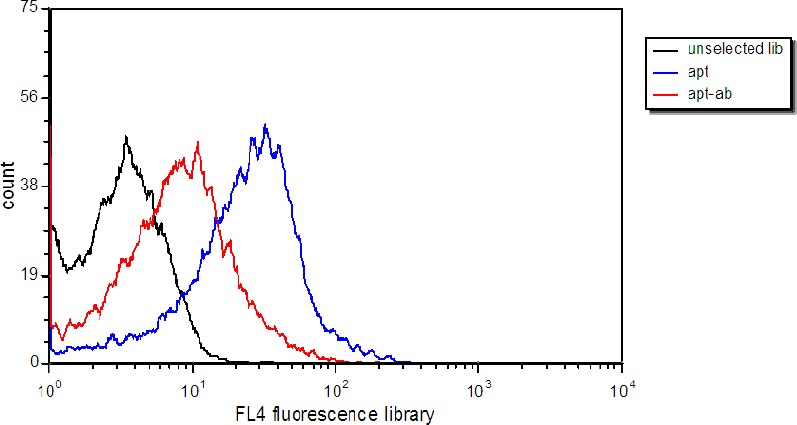
Verification of aptamer binding to extracellular domain of HER2. Selected aptamers were incubated with TUBO cells (blue), and with TUBO cells treated with anti-HER2 affibodies. Black histogram represents binding of unselected library to TUBO cells
Discussion
We successfully performed cell SELEX to isolate RNA aptamers that bind with high affinity and selectivity to TUBO cell line (rHER2/neu +). Human epidermal growth factor receptor 2 (HER2/neu), a 185-kDa tyrosine kinase receptor, is a proto-oncogene, and activating mutations of this gene can result in constitutive tyrosine kinase activity and uncontrolled proliferation (29-31). Her2/neu is over-expressed in 20-30% of breast cancers, typically as a result of gene amplification, and is associated with aggressive disease and a poor prognosis (32). TUBO cells are a murine cell line that overexpresses rHER2/neu protein (17).
Some efforts have previously been made to find aptamer molecules that specifically bind to HER2/neu (24, 33-35). Previous studies have used human cell lines to perform cell SELEX. Dastjerdi et al employed cell-SELEX procedure to generate an enriched pool DNA aptamers by using HER2-positive cells as the target for aptamer selection (36). Kim and Jeong developed an RNA aptamer against HER2 protein, and proposed that the selected aptamer could potentially be utilized as imaging agents for HER2-positive cancers (33). Giangrande et al used “cell-internalization SELEX” to select a series of RNA aptamers against HER2 that specifically recognized and were efficiently internalized by HER2-positive breast cancer cells (37). Here, we picked TUBO cells to proceed with the SELEX process. With no or limited access to nude mice facilities, the development of a proper targeting ligand to a rodent cell line, like TUBO cells, may facilitate subsequent in vivo studies in BALB/c mice. Since rHER2/neu and human HER2/neu are highly homologous (38), the SELEX products may be used as targeting ligand in hHER2/neu overexpressing cell lines.
RNA aptamers have advantages over antibodies and DNA aptamers; they can be chemically synthesized in short time; they are small and nonimmunogene; they are stable in harsh situations. Chemical modifications (e.g. 2’deoxy, 2’F, 2’NH3, 2’OMe) improve RNA aptamer stability against RNase in biological fluids like blood. Because of its single stranded nature, RNA can fold into many different three-dimensional shapes, allowing for tighter and more specific binding to target molecules. Moreover, RNA aptamers have smaller size compared to DNA aptamers of the same size, and thus pass cell membranes much easier than DNA aptamers (39, 40).
In this project we performed positive selection by retrieving sequences that bind to TUBO cells. After 3 rounds of selection, counter selection was performed using CT26 cell line (rHER2/neu negative)(16, 17). As both target and control cell lines were murine cancerous cells, it was assumed that they have common surface cancerous ligands with at least one exception for HER2 molecules on TUBO cells (17).
In each round, cell suspension was prepared by short incubation, about 1 min, of cell culture with trypsin. This treatment did not have a significant effect on the binding of aptamers. We noticed that enzyme free dissociation buffers, like enzyme free dissociation solution (Millipore), increased the number of dead cells in flow cytometry assay. To generate aptamers that specifically bind to target cells with high affinity, we gradually increased the stringency of selection conditions during the SELEX process: the volume of binding buffer and washing buffer were increased and the number of target cells and incubation time was reduced. Starting from the fourth round of SELEX, 10% FBS was added to the binding buffer and gradually increased up to 20% (21). After 12 rounds of selections, cloned and sequenced aptamers were tested for their ability to bind TUBO cells and other cancerous or normal cell line. The ultimate aim of this study was to design aptamers that can bind to the extracellular domain of HER2 protein. Hence it was important that selected aptamers could recognize SKBR3 cell line. All selected aptamers had fair affinity for SKBR3 cell lines. It could be due to the high percent of homology (about 90%) between the extra cellular domains of human HER2/neu and mouse HER2/neu (41). We selected TSA12 that had the most affinity for TUBO cells, and TSA12 that had the most specificity to TUBO cells for further studies. Then the secondary structures of TSA12 and TSA14 were determined with the mfold program. The predicted structure of TSA14 was more complicated, having 3 loops. Three hair pin motif in the secondary structure of TSA14 makes it more stable compared to TSA12. Hairpins serve as recognition motifs in RNA aptamer structures (42). Aptamers with hairpin structures have more affinity for their targets. TSA14 had higher affinity to target cells and had a more stable secondary structure. The obtained results corroborated the relationship between the structure and affinity of aptamers. It seems reasonable to assume that aptamers bind to cell surface proteins. This behavior has been reported in previous studies (26, 43, 44). Trypsin treatment assay is a preliminary test that has been used to study the binding of aptamers to cell surface proteins (26, 45-48). Trypsin pretreatment assay was performed to confirm that the interaction of designed aptamers with cell lines is through an interaction with cell surface proteins. The results showed decreased fluorescence intensities of trypsin pretreated cells compared to untreated cells. This means that TSA14 aptamer binds to surface proteins that are digested by trypsin.
In order to find aptamers that bind to the surface of target cells, Cell SELEX process was performed at 4 °C (21, 45). Since, it is necessary to test binding activity of aptamers at physiologic temperature.
We also assessed aptamer binding behavior in culture medium supplemented with FBS. The data showed that selected aptamers could be adopted in different conditions and still keep their binding ability to TUBO cells, which is important for in vivo studies.
As previously mentioned the purpose of this study is to produce aptamers that can recognize HER2 proteins on the surface of TUBO cells. In order to determine whether the target of the aptamers is a HER2 protein on the cell surface, anti-HER2 affibodies were incubated with TUBO cells and their binding of aptamers was analyzed by flow cytometry. The decrease in binding efficiency after incubation with affibodies strongly implied that aptamer targets on TUBO cells were most probably extracellular domain of HER2.
Conclusion
Whole cell SELEX was performed to find RNA aptamers against TUBO cell line. After 12 rounds of selection, a panel of aptamers was selected that had high affinity to target cells. Among them TSA14 aptamer had the most affinity for target cells. Different experiments showed that the interaction of TSA14 aptamer with TUBO cells is mediated by binding to the extracellular domain of HER2. Therefore, the TSA14 aptamer could be a candidate molecule for targeting the HER2 + tumors and merits further investigation.
Acknowledgment
The financial support of the Nanotechnology Research Center and Biotechnology Research Center, Mashhad University of Medical Sciences (MUMS) is gratefully acknowledged. This study was part of the PhD thesis of Seyedeh Alia Moosavian, which was completed in Nanotechnology Research Center, MUMS, Iran.
References
- 1.Incorvati JA, Shah S, Mu Y, Lu J. Targeted therapy for HER2 positive breast cancer. J Hematol Oncol. 2013;6:1–9. doi: 10.1186/1756-8722-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Wu M, Wang H, Xu G, Zhu T, Zhang Y, et al. Ezrin silencing by small hairpin RNA reverses metastatic behaviors of human breast cancer cells. Cancer Lett. 2008;261:55–63. doi: 10.1016/j.canlet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61:1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- 4.Pal S, Pegram M. HER2 targeted therapy in breast cancer. beyond Herceptin. Rev Endocr Metab Disord. 2007;8:269–277. doi: 10.1007/s11154-007-9040-6. [DOI] [PubMed] [Google Scholar]
- 5.Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ, Langer R, Farokhzad OC. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Proske D, Blank M, Buhmann R, Resch A. Aptamers--basic research, drug development, and clinical applications. Appl Microbiol Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 7.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 8.Cox JC, Rudolph P, Ellington AD. Automated RNA selection. Biotechnol Prog. 1998;14:845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 9.Farokhzad OC, Karp JM, Langer R. Nanoparticle-aptamer bioconjugates for cancer targeting. Expert Opin Drug Deliv. 2006;3:311–324. doi: 10.1517/17425247.3.3.311. [DOI] [PubMed] [Google Scholar]
- 10.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dua P, Kim S, Lee D-k. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54:215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Guo KT, Ziemer G, Paul A, Wendel HP. CELL-SELEX: Novel perspectives of aptamer-based therapeutics. Int J Mol Sci. 2008;9:668–678. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoltenburg R, Reinemann C, Strehlitz B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Liu C, Tan W. Aptamers generated by Cell SELEX for biomarker discovery. Biomark Med. 2009;3:193–202. doi: 10.2217/bmm.09.5. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Yang D, Schluesener HJ, Zhang Z. Advances in SELEX and application of aptamers in the central nervous system. Biomol Eng. 2007;24:583–592. doi: 10.1016/j.bioeng.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Penichet ML, Challita PM, Shin SU, Sampogna SL, Rosenblatt JD, Morrison SL. In vivo properties of three human HER2/neu-expressing murine cell lines in immunocompetent mice. Lab Anim Sci. 1999;49:179–188. [PubMed] [Google Scholar]
- 17.Jalali SA, Sankian M, Tavakkol-Afshari J, Jaafari MR. Induction of tumor-specific immunity by multi-epitope rat HER2/neu-derived peptides encapsulated in LPD Nanoparticles. Nanomedicine. 2012;8:692–701. doi: 10.1016/j.nano.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Hall B, Micheletti JM, Satya P, Ogle K, Pollard J, Ellington AD. In Curr Protoc Mol Biol. Vol. 9. John Wiley & Sons, Inc; 2009. Design, Synthesis, and Amplification of DNA Pools for In Vitro Selection; pp. 9.2.1–9.2.28. [DOI] [PubMed] [Google Scholar]
- 19.Mayer G, Piasecki S, Hall B, Ellington A. Nucleic Acid and Peptide Aptamers. Humana Press; 2009. Nucleic Acid Pool Preparation and Characterization; pp. 3–18. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 20.Ellington A. PJ. Protoc Mol Biol. Vol. 42. John Wiley & Sons, Inc; 1998. Purification of Oligonucleotides Using Denaturing Polyacrylamide Gel Electrophoresis; pp. 2.12.11–12.12.17. [Google Scholar]
- 21.Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 22.Sioud M, Meng L, Sefah K, Colon D, Chen H, O'Donoghue M, et al. In RNA Therapeutics. Vol. 629. Humana Press; Using Live Cells to Generate Aptamers for Cancer Study; pp. 353–365. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 23. http://eng.bioneer.com/home.aspx.
- 24.Kang HS, Huh YM, Kim S, Lee Dk. Isolation of RNA aptamers targeting HER-2-overexpressing breast cancer cells using cell-SELEX. Bull Korean Chem Soc. 2009;30:1827–1831. [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan W. DNA Aptamers as Molecular Probes for Colorectal Cancer Study. PLoS On. 2010;5:e14269. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang D, Wang J, Zhang W, Song Y, Li X, Zou Y, et al. Selection of DNA Aptamers against Glioblastoma Cells with High Affinity and Specificity. PLoS On. 2012;7:e42731. doi: 10.1371/journal.pone.0042731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akiyama T, Sudo C, Ogawara H, Toyoshima K, Yamamoto T. The product of the human c-erbB-2 gene: a 185-kilodalton glycoprotein with tyrosine kinase activity. Science. 1986;232:1644–1646. doi: 10.1126/science.3012781. [DOI] [PubMed] [Google Scholar]
- 30.Disis ML, Calenoff E, McLaughlin G, Murphy AE, Chen W, Groner B, et al. Existent T-cell and antibody immunity to HER-2/neu protein in patients with breast cancer. Cancer Res. 1994;54:16–20. [PubMed] [Google Scholar]
- 31.Neve RM, Sutterluty H, Pullen N, Lane HA, Daly JM, Krek W, et al. Effects of oncogenic ErbB2 on G1 cell cycle regulators in breast tumour cells. Oncogen. 2000;19:1647–1656. doi: 10.1038/sj.onc.1203470. [DOI] [PubMed] [Google Scholar]
- 32.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Scienc. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 33.Kim MY, Jeong S. In vitro selection of RNA aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther. 2011;21:173–178. doi: 10.1089/nat.2011.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dastjerdi K, Tabar GH, Dehghani H, Haghparast A. Generation of an enriched pool of DNA aptamers for an HER2-overexpressing cell line selected by Cell SELEX. Biotechnol Appl Biochem. 2011;58:226–230. doi: 10.1002/bab.36. [DOI] [PubMed] [Google Scholar]
- 37.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seavey MM, Pan ZK, Maciag PC, Wallecha A, Rivera S, Paterson Y, et al. A novel human Her-2/neu chimeric molecule expressed by Listeria monocytogenes can elicit potent HLA-A2 restricted CD8-positive T cell responses and impact the growth and spread of Her-2/neu-positive breast tumors. Clin Cancer Res. 2009;15:924–932. doi: 10.1158/1078-0432.CCR-08-2283. [DOI] [PubMed] [Google Scholar]
- 39.Ray P, White RR. Aptamers for Targeted Drug Delivery. Pharmaceuticals. 2010;3:1761–1778. doi: 10.3390/ph3061761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Germer K, Leonard M, Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol Biol. 2013;4:27–40. [PMC free article] [PubMed] [Google Scholar]
- 41.Disis ML, Grabstein KH, Sleath PR, Cheever MA. Generation of immunity to the HER-2/neu oncogenic protein in patients with breast and ovarian cancer using a peptide-based vaccine. Clin Cancer Res. 1999;5:1289–1297. [PubMed] [Google Scholar]
- 42.Svoboda P, Cara AD. Hairpin RNA: a secondary structure of primary importance. Cel Mol Life SciCMLS. 2006;63:901–908. doi: 10.1007/s00018-005-5558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer Directly Evolved from Live Cells Recognizes Membrane Bound Immunoglobin Heavy Mu Chain in Burkitt's Lymphoma Cells. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Jimenez E, Sefah K, Lopez-Colon D, Van Simaeys D, Chen HW, Tockman MS, et al. Generation of lung adenocarcinoma DNA aptamers for cancer studies. PLoS On. 2012;7:e46222. doi: 10.1371/journal.pone.0046222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z, Parekh P, Turner P, Moyer RW, Tan W. Generating aptamers for recognition of virus-infected cells. Clin Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ara MN, Hyodo M, Ohga N, Hida K, Harashima H. Development of a Novel DNA Aptamer Ligand Targeting to Primary Cultured Tumor Endothelial Cells by a Cell-Based SELEX Method. PLoS On. 2012;7:e50174. doi: 10.1371/journal.pone.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]


