Abstract
In the companion paper to this work, we have described development of a new type of hydrogen exchange (HX) mass spectrometry (MS) measurement that integrates Langmuir monolayers. With Langmuir monolayers, the lipid packing density can be reproducibly controlled and changed as desired. Analysis of HX in proteins that may undergo conformational changes as a function of lipid packing, for example conformational rearrangements after insertion into a lipid layer, are then possible. We previously used neutron reflection to characterize just such a conformational change in the myristoylated HIV-1 Nef protein (myrNef): at high lipid packing density, myrNef could not insert into the lipids and maintained a compact conformation adjacent to the monolayer whereas at lower lipid packing density, myrNef was able to insert N-terminal arm residues causing displacement of the core domain away from the monolayer. In order to locate where conformation may have been altered by lipid association, we applied the HX MS Langmuir monolayer method to myrNef associated with monolayers of packing densities identical to those used for the prior neutron reflection measurements. The results show that the N-terminal region and the C-terminal unstructured loop undergo conformational changes when associated with a low lipid density lipid monolayer. The results are not consistent with the hypothesis of myrNef dimerization upon membrane association in the absence of other myrNef binding partners. The HX MS Langmuir monolayer method provides new and meaningful information for myrNef that helps explain necessary conformational changes required for function at the membrane.
Keywords: Nef, deuterium, dynamics, membrane protein, monolayer
Interrogating conformational changes in proteins when they interact with a lipid membrane is highly desirable. A well known obstacle, however, to such structural characterization is the membrane itself, which is generally not compatible with structural studies and many biophysical measurements. In the highly likely event that high resolution structural methods such as X-ray crystallography are unable to provide information, alternative methods have been utilized that are less susceptible to interference from the membrane. We have used neutron reflection (NR) methods1–6 to study peptides and proteins at lipid monolayers (one leaflet/half of a lipid membrane). While NR is capable of resolving and modeling an overall, lipid-associated shape profile, it is silent to the finer details of protein motion/dynamics and local conformation. The opportunity to combine the local information provided by other methods with global structural information by NR (or also X-ray reflection) is very attractive. To this end, we developed (described in an accompanying paper7) hydrogen exchange (HX) mass spectrometry (MS) methods that use the same Langmuir monolayer trough system that is central to NR. This strategy was intended to combine information from NR and HX MS for a multifaceted and more comprehensive characterization of protein conformation at membranes. Overall shape, distance from the monolayer, and nuclear density are given by the NR measurements while protein dynamics, and location of protected/unprotected backbone amide hydrogen are given by the HX MS measurements.
HX MS Langmuir monolayer data are obtained using the same Langmuir trough used to obtain NR data. In this system, there can be very fine and highly reproducible control over the packing density of the lipid layer. Unlike other parameters (e.g., lipid composition, head-group charge, lipid tail chain length, etc.) lipid packing density is a parameter that is not easy to control in many membrane mimetics used for biophysical analyses. However, such control can be essential for monitoring conformational changes in proteins that are sensitive to lipid packing. In addition, it may also be possible to perform HX with tethered lipid bilayers, allowing a much wider range of inserted or membrane-associated proteins to be investigated.
In our recent analysis of the HIV-1 Nef protein, we observed a conformational transition that is sensitive to lipid packing5. Biological evidence has shown that membrane association is important for the cellular functions of the 25 kDa myristoylated Nef protein from HIV-1 (myrNef) including interaction with various signaling molecules also localized to the cytoplasmic face of the plasma membrane8–11 and interaction with and removal of CD4 receptors from the cell surface12. There is no full-length crystal or NMR structure for Nef due to its propensity to aggregate. While full-length Nef is too flexible for high-resolution structural analysis, portions of the protein have been characterized by X-ray crystallography and NMR and assembled into a model of the full-length protein13. Nef consists of a well folded, ordered core with two highly flexible regions: the N-terminal arm and the C-terminal disordered loop14. These flexible regions comprise nearly half of the protein and are hypothesized to play important roles in protecting or exposing binding sites14. HX MS, which is compatible with much more dilute concentrations than many other methods, was successfully used to investigate the solution conformations of both myrNef15 and nonmyristoylated Nef16. It was proposed early17,18 that membrane association alters Nef structure, but it was not until analysis with NR5 that direct experimental evidence for this idea was obtained. The NR studies showed that the N-terminal myristic acid and hydrophobic residues on the N-terminal arm inserted into the lipid layer and these interactions caused the core of myrNef to reposition from directly adjacent to the monolayer to a position approximately 70 angstroms away. Very importantly, however, was the discovery and characterization with neutron reflection of the conformational transition that was dependent on lipid packing density. When lipid packing density was low, myrNef was able to insert and rearrange its conformation but when lipid packing density was high, quite a different conformation was obtained.
Development and validation of the Langmuir monolayer HX MS method, as reported in an accompanying paper7, made it possible for us to study the conformational transition in myrNef in more detail and to compare the all-important functional conformation at a monolayer with the conformation in solution. The results reported here show that indeed, new and valuable information about myrNef at the membrane can be obtained from HX MS in Langmuir monolayers.
Experimental Procedures
Myristoylated Protein Expression and Purification
Myristoylated HIV-1 Nef (strain SF2) expression and purification were as described previously15. Briefly, myristoylated Nef (myrNef) was expressed using a pET-Deut vector (obtained from the Wilbold lab19) containing human N-myristoylatransferase 1 (hNMT1) in the first multiple cloning site and Nef with a C-terminal polyhistidine tag in the second multiple cloning site. Nef was isolated using Ni-NTA agarose (QIAGEN, Valencia, CA), washed and eluted as described previously15. Purity and proper myristoylation were confirmed by polyacrylamide gel electrophoresis (SDS-PAGE) and electrospray mass spectrometry.
Solution Hydrogen Exchange
Solution hydrogen exchange experiments were carried out at room temperature (22 °C) by diluting myrNef stock solutions in equilibration buffer (50 mM citric acid, 50 mM sodium phosphate, 150 mM NaCl, pH 6.0, H2O) 15-fold with labeling buffer (50 mM citric acid, 50 mM sodium phosphate, 150 mM NaCl, pD 6.0, 99.8% D2O). Both Nef buffers contained 1 mM dithiothreitol. Following dilution into D2O, samples were continuously labeled for predetermined times ranging from 10 seconds to 1 hour before being quenched to pH 2.6 using a 0 °C solution of quench buffer (0.8% formic acid and 0.8M guanidine hydrochloride in H2O). Quenched samples were digested on ice for 5 minutes by adding pepsin and aspergillopepsin (60 µg and 70 µg, respectively, dissolved in water). Digested samples were injected into a Waters nanoAcquity UPLC with HX technology20 (Milford, MA) for desalting, separation, and mass analysis.
Monolayer Hydrogen Exchange Experiments
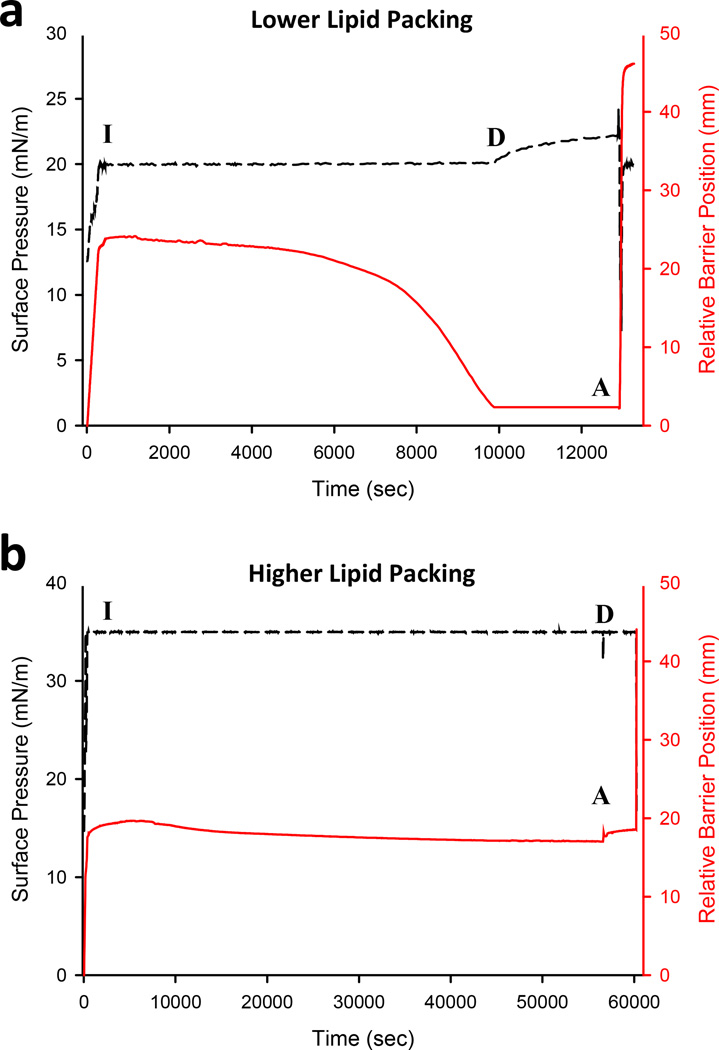
Monolayer hydrogen exchange experiments were performed as described in the accompanying paper7. All myrNef experiments were performed at room temperature (22 °C) and 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG) was used for the monolayer. MyrNef was injected under the monolayer to a final concentration of ~1 µM and the barrier position and pressure were monitored (FIGURE 1). Interaction of the protein with the Langmuir monolayer was allowed to proceed until the relative barrier position was less than 5 mm from the back edge of the trough (for inserting species, FIGURE 1A) or until it remained constant (for non-inserting species, FIGURE 1B). Then, 100 mLs of labeling buffer were rapidly circulated, protein isolated, and exchange quenched, as described7. The quenched sample was digested for 5 minutes at 0 °C (all parameters identical to solution HX experiments) before injection into the UPLC-MS system.
Figure 1.
Adsorption of myrNef to low lipid packing (A) and high lipid packing monolayers (B). Change in surface pressure (black) and barrier position (red) in the trough are shown. For all plots, “I” represents when protein was injected underneath the monolayer, “D” represents the subphase exchange with D2O, and “A” represents aspiration of the sample. For low lipid packing experiments (a), the surface pressure was held constant after addition of protein. The barrier gradually moved back as more protein associated and inserted residues into the monolayer. The barrier was stopped within a few millimeters from the end of the trough. The surface pressure rose slightly after the barrier was stopped indicating that protein was still associating with the monolayer. For high lipid packing experiments (b), Nef was unable to insert into the monolayer. As a result, the barrier did not gradually move back as in the low packing experiments. In all cases, aspiration dropped the surface pressure quickly and the barrier rapidly moved forward in order to reestablish the preset surface pressure.
Mass analyses and data processing
Peptide separation, mass analysis and data processing were performed as described in the accompanying paper7. The peptic peptides from myrNef are shown in the Supporting Information (FIGURE S1). As all experiments were performed under very similar experimental conditions, all deuterium levels are reported as relative21 and there were no corrections for back-exchange. The relative deuterium incorporation curves for the peptides of myrNef are provided in FIGURE S2.
Results and Discussion
Myristoylated Nef was subjected to HX both in the Langmuir trough system and in solution and the results were compared. Nef was studied at two Langmuir monolayer pressures: 20 mN/m where insertion occurs and 35 mN/m where insertion does not occur5. As described in Akgun et al.5, the barrier position reports on if peptide/protein interacts with the lipids in the monolayer. If there is insertion of proteinaceous material into the lipid layer, the monolayer becomes more crowded and increases the molecular packing density. To maintain constant pressure, the position of the barrier holding the monolayer in place must be adjusted (done automatically by computer). As expected, myrNef inserted into the monolayer at 20 mN/m (FIGURE 1A) but not at 35 mN/m (FIGURE 1B). These conditions are the same as those used for the previous NR experiments with myrNef5.
A total of 31 peptides were generated from digestion of myrNef that was aspirated from the Langmuir monolayer, resulting in 90% coverage of the protein backbone (Supporting information, FIGURE S1). Overlapping peptides were identified in nearly all areas of the protein and similar HX trends were observed in areas of redundancy (full dataset in FIGURE S2). Deuterium incorporation was measured for at least triplicate biological replicates of myrNef with a fresh monolayer spread for every deuterium labeling time. While the reproducibility of HX MS measurements in Langmuir monolayers was discussed in detail in the accompanying manuscript7, the error bars in the deuterium incorporation graphs for each peptide (FIGURE S2) demonstrate that reliable conclusions could be reached from triplicates measurements. The exchange of myrNef was measured at pH 6.0, a pH at which the functional activity of Nef, as measured by Hck activation22, was indistinguishable from that at pH 7.3 (FIGURE S3) or higher (as shown in Ref. 22).
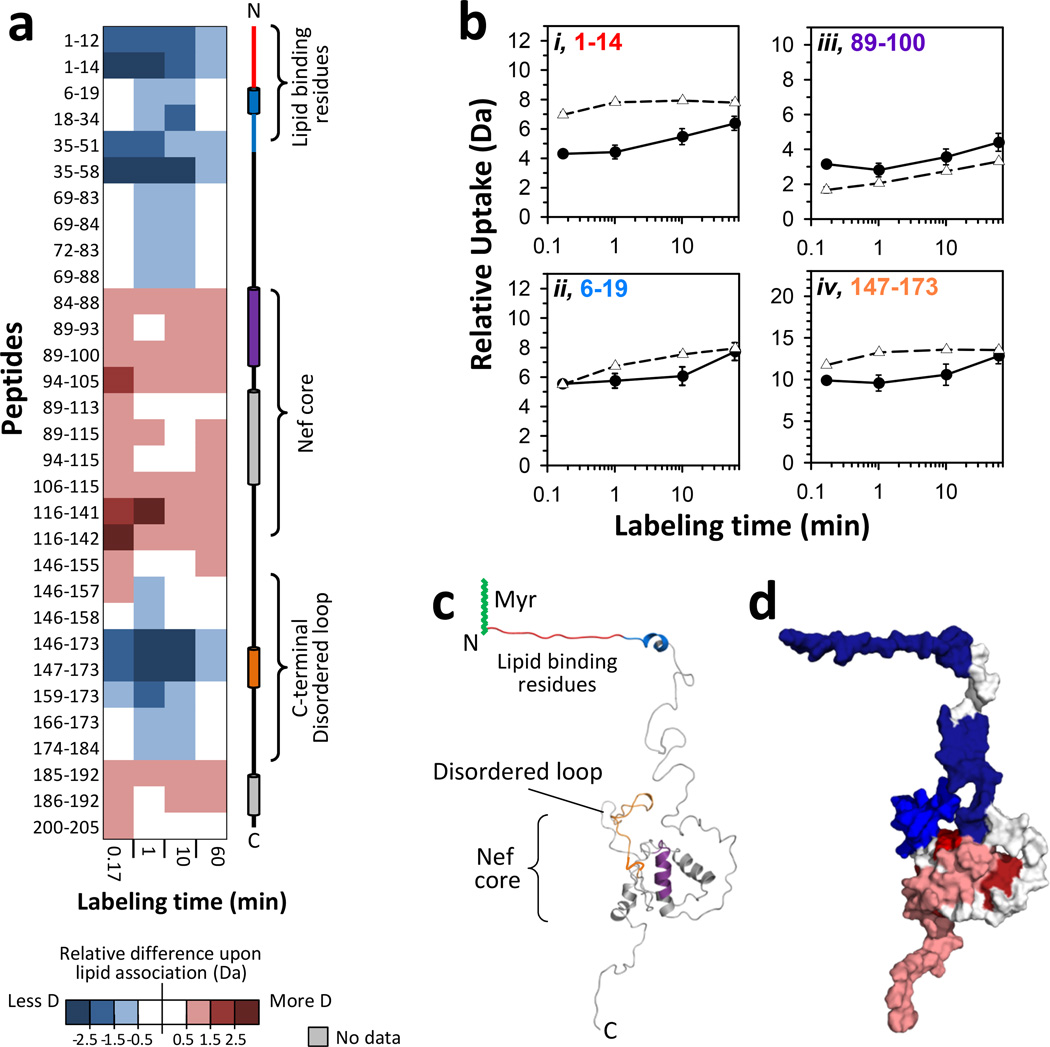
For the case of association with monolayers at a pressure of 20 mN/m, HX in myrNef increased in some regions and decreased in others relative to that measured for myrNef in solution (FIGURE 2; see also FIGURE S4). In solution, the core of myrNef did not incorporate much deuterium (e.g., FIGURE 2Biii) and after one hour of labeling remained fairly protected from labeling, consistent with a well folded structure with little protein dynamics as seen previously by HX MS15,16. Increased deuterium incorporation upon lipid association was observed principally in the core domain and some parts of the C-terminal region (FIGURE 2A). Lipid association did not appear to change the dynamics of the core as the slopes of the deuterium incorporation graphs were similar to those of myrNef in solution (see also Ref. 23). Lipid association exposed several backbone amide hydrogens in the Nef core: peptide 89–100 showed an increase of several Daltons (FIGURE 2Biii) as well as other peptides (e.g. residues 116–142) from within the Nef core (FIGURE 2A and FIGURE S2). In solution these residues were less deuterated perhaps due to interaction with residues of the N-terminal arm.
Figure 2.
Effects of monolayer association on myristoylated Nef. All results were obtained with a monolayer pressure of 20 mN/m. (a) Difference map comparing monolayer-associated myrNef HX versus myrNef HX in solution. Deuterium levels for the peptides indicated at the left were obtained from triplicate experiments (data in FIGURE S2). The average amount of deuterium for HX in solution was subtracted from the average amount of deuterium after HX in the trough (monolayer associated) and the value colored (positive values in reds, negative values in blues, as indicated – i.e. blue: less deuterium when with lipid, red: more deuterium when with lipid). Secondary structure elements in myrNef are displayed on the right. (b) Deuterium incorporation in four selected myrNef peptides for monolayer associated (-●-) HX and solution (-Δ-) HX. Error bars represent the spread of triplicate measurements. The residue numbers of each peptide are colored (red, blue, purple, orange) to match the colored secondary structure elements at the right in panel a. (c) Structural locations of peptides in panel B on the model of Nef which was first reported in 13 and here adapted from5. (d) Space filling model illustrating regions of increased HX (red) and decreased HX (blue) in myrNef when bound to the monolayer. These data are for 10 seconds of deuteration and myrNef has been colored according to the scale in panel a. See FIGURE S4 for deuterium levels mapped on the structural model of Nef at other exchange times, both in solution and at the monolayer.
Both the N-terminal myristoylated region and the C-terminal disordered loop of myrNef showed significant reduction in deuterium incorporation when the protein was associated with the Langmuir monolayer at 20 mN/m (FIGURE 2A, 2Bi,ii,iv). This protection from labeling suggests either contact with and burial into the lipid layer, structural creation and stabilization, or a combination of both. Protection from exchange in the first ~20 residues is consistent with the hypothesis that both the myristic acid and residues from the N-terminus including a positively charged region between residues 17–2214,24,25 insert into the membrane. Insertion of arm residues in addition to the myristate group is indicated by the large movement of the barrier (FIGURE 1A). In solution, residues next to the lipid binding region (e.g., peptides 35–51 and 35–58) were deuterated rapidly at ten seconds (FIGURES S2), suggesting high solvent accessibility or a lack of structure, but upon lipid association displayed 2–3 Dalton reductions in deuterium incorporation at ten seconds (FIGURE 2A). An α-helical sequence (αH2) is located within that span of residues, and we speculate that this helical region may become stabilized when the N-terminal arm separates from the core upon membrane binding. Reduced deuteration of the C-terminal disordered loop of lipid-associated myrNef (e.g., residues 147–173) suggests that some backbone amide hydrogens in the C-terminal loop become protected, perhaps by hydrogen bonding with the core of Nef, or become more stabilized. Arold and Baur predicted that upon binding to the membrane, Nef would adopt a “signaling” conformation where the C-terminal loop wrapped around the core domain14. Interactions with the core domain, they argued, would avoid rapid endocytosis and removal from the membrane while other events such as such as phosphorylation or binding to another protein could trigger the loop to become exposed. The observed protection of the C-terminal loop in our HX MS data agrees with this earlier prediction.
Recently a structure of HIV-1 Nef in complex with the AP-2 clatherin adaptor complex was reported that contained the loop region26. In that structure two helical regions within the loop are apparent, a helix from residues 150–157 (αH4) and another single turn helix from 167–170 (αH5). αH5, along with a series of turns at the C-terminal end of the loop (residues 171–179) are located between AP-2 α-σ2 and the Nef core, with αH5 packing against the β-sheet of the core and the turn-rich section of the loop from 171–179 anchored by a hydrogen bond between Asp174 and Glu104 and by a salt bridge between Asp175 of the loop and Nef Arg134 of the core β-sheet. It was concluded that these interactions between loop and core play an important role in organizing this region of the loop. Our HX results are consistent with interactions between loop and core upon membrane binding when the N-terminal arm separates from the core.
In addition to providing new insight into the disposition of the C-terminal loop, these HX MS data strongly suggest that myrNef does not dimerize through its core upon membrane binding, as the residues on the core domain show more deuterium uptake relative to the solution form, rather than less deuterium as a result of dimer-induced protection. Dimerization of Nef has been identified in cells and shown to be critical for downregulation of CD4 receptors27. We have seen no evidence of dimer formation in previous solution HX MS measurements of either myrNef15 or nonmyristoylated Nef16. Crystal structures of truncated Nef as a dimer in complex with SH3 and SH3-SH2 domains of Src family kinases have been reported8,28. The dimerization interface between the two core domains is different in these two structures, consistent with the hypothesis that Nef dimerization is driven by interaction with the host protein and not by an inherent affinity of the core domain for itself. The present HX MS data, showing strong evidence that the core regions of Nef do not come together in a dimer when associated with the monolayer, support this host-protein-dependent dimerization hypothesis.
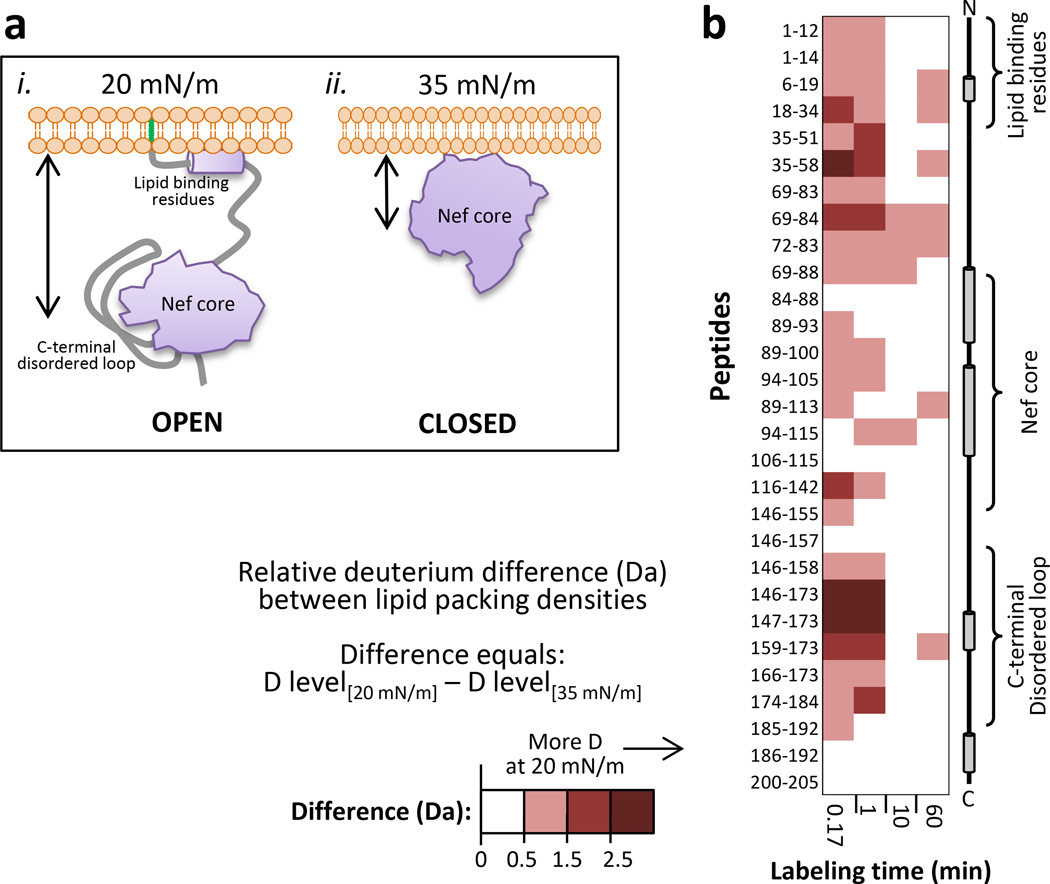
With the Langmuir trough system, the pressure of the monolayer is measured continuously (FIGURE 1) and can be controlled by manipulating the barrier position. HX can therefore be done at various pressures that can be generated very reproducibly. We used this strategy to compare HX in myrNef when associated with a DPPG monolayer with a pressure of 20 mN/m (as above, FIGURE 2) versus a monolayer with a pressure of 35 mN/m. At higher lipid packing density, the arm of myrNef is unable to insert into the monolayer and myrNef remains in a closed conformation directly against the lipid headgroups, as shown previously by neutron reflection5. A model of myrNef conformation with either low (20 mN/m) or high (35 mN/m) lipid packing density, based on the neutron reflection data, is shown in FIGURE 3A. At higher lipid packing density, residue insertion at the monolayer was prohibited and greatly altered HX of myrNef; deuterium incorporation was significantly higher throughout the protein at 20 mN/m versus 35 mN/m (FIGURE 3B). In addition, the HX of myrNef at the 35 mN/m monolayer was not identical to that of myrNef in solution (full dataset in Supporting Information, FIGURE S2). There was less deuterium in residues 1–83 and 146–184 of myrNef associated with the 35 mN/m monolayer compared with myrNef in solution, implying stabilization of these parts of the structure, protection by the lipid layer, conformational rearrangements, or all three. The deuteration levels of residues 84–145 and 185–210 were largely the same at the two monolayer pressures. MyrNef with the 20 mN/m monolayer had deuterium levels in residues 1–83 and 146–184 that were intermediate between the 35 mN/m monolayer and solution HX measurements. Our interpretation of these results is that when myrNef associates with the monolayer at higher pressures (35 mN/m) through interaction of the positively-charged residues (17–22) with the negatively-charged headgroups, the arm partially dissociates from the core allowing the C-terminal loop to associate with the core. Yet because the lipid packing is too high for αH1 (residues 6–22) to insert, myrNef remains in a closed conformation directly against the lipid layer in an orientation such that the N-terminal portion (residues 30–84) and the C-terminal loop are occluded from solvent by proximity to the monolayer. When the monolayer pressure is lower (20 mN/m) and αH1 can insert into the monolayer, the N-terminal arm releases completely from the core and the conformation becomes extended (as on the left of FIGURE 3A). In that case more deuterium can be incorporated into the N-terminal region and the C-terminal loop (FIGURE 3B), although not as much as for the solution form due to interaction of the loop with the core in the monolayer-bound state.
Figure 3.
Differences in myrNef due to monolayer packing density. (a) Cartoon representation of the myristoylated Nef structure associated with (i, left.) low lipid packing density of 20 mN/m or (ii., right) high lipid packing density of 35 mN/m. The black arrows represent core domain distance from the lipids. The myristoyl moiety is shown in green. This representation is based data in Akgun et al.5. (b) Difference map comparing myrNef HX at monolayer pressures of 20 or 35 mN/m. Deuterium levels for the peptides indicated at the left were obtained from triplicate experiments (data in FIGURE S2 and structural locations in FIGURE S4). The average amount of deuterium after HX using a monolayer pressure of 35 mN/m was subtracted from the average amount of deuterium after HX using a monolayer pressure of 20 mN/m and the value colored (positive values in reds, as indicated – i.e., red: more deuterium when with lipid). Secondary structure elements in myrNef are displayed on the right.
Conclusions
The newly developed method for analyzing conformational features of membrane-associated peptides and peripheral membrane proteins combines HX MS and Langmuir monolayers. An advantage of using the Langmuir monolayer system is that the lipid packing density can be controlled and reproduced from monolayer to monolayer. For proteins that undergo conformational changes as a result of lipid packing density, this is a very valuable feature. We showed how packing density could be altered by and result in different conformations of the myrNef protein. Our myrNef Langmuir monolayer HX exchange data support and expand upon previous conformational studies by neutron reflection5. In particular these data provide the first direct evidence that that the C-terminal loop is occluded from solvent in the monolayer-associated and open conformation form (as predicted for the “signaling” form by Arold and Baur14) and that myrNef does not dimerize upon associating with membranes. The latter suggests that dimerization of myrNef is driven by interaction with host proteins, something that could now be examined directly with the HX MS Langmuir system.
Another major advantage on HX MS at Langmuir monolayers is the opportunity to combine global structural information from techniques such as neutron or X-ray reflection with more local information from HX. Our Langmuir monolayer HX MS method was designed to be integrated with a neutron or X-ray reflection workflow. In such a scheme, protein association with monolayers can first be investigated with the same trough described here using neutron or X-ray reflection, the profile of the protein with respect to the lipid layer obtained, packing density of the monolayer modulated (if desired) and the impact on conformation monitored. Once an interesting conformation is identified by neutron or X-ray reflection (or at any point, conformation, or condition such as packing density), the protein could be labeled with deuterium right at the reflectometer and the sample passed to a nearby UPLC-MS system for HX measurement. We believe there are significant benefits to such a strategy and the presented data provide strong justification for interrogating membrane protein association using both neutron reflection and HX MS.
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM086507 and GM101135 (to J.R.E.) and AI102724 (to T.E.S.), and a research collaboration with the Waters Corporation. Sandia National Laboratories is a multi-program laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
Supporting Information
Supporting information available: This material is available free of charge via Internet at http://pubs.acs.org.
References
- 1.Kent MS, Yim H, Murton J, Satija S, Majewski J, Kuzmenko I. Biophys J. 2008;94:1–13. doi: 10.1529/biophysj.107.113498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kent MS, Yim H, Sasaki DY, Satija S, Majewski J, Gog T. Langmuir. 2004;20:2819–2829. doi: 10.1021/la036207y. [DOI] [PubMed] [Google Scholar]
- 3.Kent MS, Yim H, Sasaki DY, Satija S, Seo YS, Majewski J. Langmuir. 2005;21:6815–6824. doi: 10.1021/la047433q. [DOI] [PubMed] [Google Scholar]
- 4.Kent MS, Yim H, Murton JK, Sasaki DY, Polizzotti BD, Charati MB, Kiick KL, Kuzmenko I, Satija S. Langmuir. 2008;24:932–942. doi: 10.1021/la700940x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akgun B, Satija S, Nanda H, Pirrone GF, Shi X, Engen JR, Kent MS. Structure. 2013;21:1822–1833. doi: 10.1016/j.str.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent MS, Murton JK, Sasaki DY, Satija S, Akgun B, Nanda H, Curtis JE, Majewski J, Morgan CR, Engen JR. Biophys J. 2010;99:1940–1948. doi: 10.1016/j.bpj.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirrone GF, Vernon BC, Kent MS, Engen JR. Anal Chem. 2015 doi: 10.1021/acs.analchem.5b01724. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 9.Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. Structure. 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 10.Manninen A, Hiipakka M, Vihinen M, Lu W, Mayer BJ, Saksela K. Virology. 1998;250:273–282. doi: 10.1006/viro.1998.9381. [DOI] [PubMed] [Google Scholar]
- 11.Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 12.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 13.Geyer M, Peterlin BM. FEBS Lett. 2001;496:91–95. doi: 10.1016/s0014-5793(01)02394-8. [DOI] [PubMed] [Google Scholar]
- 14.Arold ST, Baur AS. Trends Biochem Sci. 2001;26:356–363. doi: 10.1016/s0968-0004(01)01846-1. [DOI] [PubMed] [Google Scholar]
- 15.Morgan CR, Miglionico BV, Engen JR. Biochemistry. 2011;50:3394–3403. doi: 10.1021/bi200197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochrein JM, Wales TE, Lerner EC, Schiavone AP, Smithgall TE, Engen JR. Biochemistry. 2006;45:7733–7739. doi: 10.1021/bi060438x. [DOI] [PubMed] [Google Scholar]
- 17.Dennis CA, Baron A, Grossmann JG, Mazaleyrat S, Harris M, Jaeger J. Proteins. 2005;60:658–669. doi: 10.1002/prot.20544. [DOI] [PubMed] [Google Scholar]
- 18.Breuer S, Gerlach H, Kolaric B, Urbanke C, Opitz N, Geyer M. Biochemistry. 2006;45:2339–2349. doi: 10.1021/bi052052c. [DOI] [PubMed] [Google Scholar]
- 19.Gluck JM, Hoffmann S, Koenig BW, Willbold D. PLoS One. 2010;5:e10081. doi: 10.1371/journal.pone.0010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wales TE, Engen JR. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 22.Emert-Sedlak L, Kodama T, Lerner EC, Dai W, Foster C, Day BW, Lazo JS, Smithgall TE. ACS Chem Biol. 2009;4:939–947. doi: 10.1021/cb900195c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan CR, Engen JR. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1706s58. Chapter 17, Unit 17 16 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentham M, Mazaleyrat S, Harris M. J Gen Virol. 2006;87:563–571. doi: 10.1099/vir.0.81200-0. [DOI] [PubMed] [Google Scholar]
- 25.Gerlach H, Laumann V, Martens S, Becker CF, Goody RS, Geyer M. Nat Chem Biol. 2010;6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 26.Ren X, Park SY, Bonifacino JS, Hurley JH. Elife. 2014;3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poe JA, Smithgall TE. J Mol Biol. 2009;394:329–342. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarado JJ, Tarafdar S, Yeh JI, Smithgall TE. J Biol Chem. 2014;289:28539–28553. doi: 10.1074/jbc.M114.600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.