Abstract
Duplication of the genome in mammalian cells occurs in a defined temporal order referred to as its replication-timing (RT) program. RT changes dynamically during development, regulated in units of 400–800 kb referred to as replication domains (RDs). Changes in RT are generally coordinated with transcriptional competence and changes in subnuclear position. We generated genome-wide RT profiles for 26 distinct human cell types, including embryonic stem cell (hESC)-derived, primary cells and established cell lines representing intermediate stages of endoderm, mesoderm, ectoderm, and neural crest (NC) development. We identified clusters of RDs that replicate at unique times in each stage (RT signatures) and confirmed global consolidation of the genome into larger synchronously replicating segments during differentiation. Surprisingly, transcriptome data revealed that the well-accepted correlation between early replication and transcriptional activity was restricted to RT-constitutive genes, whereas two-thirds of the genes that switched RT during differentiation were strongly expressed when late replicating in one or more cell types. Closer inspection revealed that transcription of this class of genes was frequently restricted to the lineage in which the RT switch occurred, but was induced prior to a late-to-early RT switch and/or down-regulated after an early-to-late RT switch. Analysis of transcriptional regulatory networks showed that this class of genes contains strong regulators of genes that were only expressed when early replicating. These results provide intriguing new insight into the complex relationship between transcription and RT regulation during human development.
During development, complex transcriptional and epigenetic networks are established that are necessary for lineage specification and maintenance of cellular identity. Despite remarkable progress in characterizing dynamic changes in the transcriptome and epigenome during cell fate specification (Gifford et al. 2013; Xie et al. 2013; Dixon et al. 2015; Roadmap Epigenomics Consortium et al. 2015; Tsankov et al. 2015), mechanisms that regulate large scale spatial organization of the genome and its developmental consequences are still poorly understood.
All eukaryotes duplicate their genomes in a defined temporal order known as the replication-timing (RT) program (Hiratani et al. 2009; Pope and Gilbert 2013). Proper regulation of RT is essential for genome stability (Donley et al. 2013; Neelsen et al. 2013; Alver et al. 2014), and abnormal RT programs have been identified in cancer cells (Ryba et al. 2012). In mammals, cell fate commitment is accompanied by dynamic changes in RT in units of 400–800 kb known as replication domains (RDs) (Hiratani et al. 2008, 2010; Hansen et al. 2010; Ryba et al. 2010). RT is closely aligned with spatial organization of chromatin in the nucleus; early and late RDs reside in distinct nuclear compartments (Nakamura et al. 1986; Nakayasu 1989; O'Keefe et al. 1992), and cytogenetic visualization of pulse-labeled DNA synthesis reveals distinct punctate replication foci whose structure remains stable for many cell cycles (Jackson and Pombo 1998; Ma et al. 1998; Dimitrova and Gilbert 1999; Berezney et al. 2000; Sadoni et al. 2004). More recently, chromatin conformation methods that map long-range chromatin interactions (Lieberman-Aiden et al. 2009) have revealed that chromosomes consist of topologically associating domains (TADs) that correspond to units of RT regulation (Pope et al. 2014), whereas the interactions between TADs form two distinct subnuclear compartments that correspond to the early and late replicating segments of the genome within any given cell type (Ryba et al. 2010; Yaffe et al. 2010; Dixon et al. 2012; Moindrot et al. 2012). Hence, RT constitutes a very informative functional readout of large-scale chromatin organization across distinct cell types and its regulation during development.
Early replication is globally associated with active gene expression in all multicellular organisms (Schübeler et al. 2002, 2004; MacAlpine et al. 2004; Woodfine et al. 2004; Huvet et al. 2007; Desprat et al. 2009; Hiratani et al. 2009; Schwaiger et al. 2009; Maric and Prioleau 2010; Lubelsky et al. 2014), and developmentally regulated changes in RT are generally coordinated with transcriptional competence (Zhou et al. 2002; Hiratani et al. 2008, 2010; Desprat et al. 2009; Schultz et al. 2010; Yue et al. 2014). However, causal relationships between RT and gene expression remain a long-standing puzzle. Previous studies during early mouse development found coordinated changes in transcription and RT, but could not distinguish which changes first (Hiratani et al. 2010). Recently developed methods for human embryonic stem cell (hESC) differentiation (Schulz et al. 2012; Menendez et al. 2013) allow a highly synchronous derivation of distinct lineages and provide a unique opportunity to study the mechanisms that regulate the establishment of cell-type-specific RT programs and its relationship to differential gene expression, pluripotency, and lineage specification.
Here, we have generated genome-wide RT and transcriptome data from 26 distinct human cell types representing each of the three embryonic germ layers and neural crest including several key intermediate stages. This study constitutes the most comprehensive characterization of dynamic changes in the temporal order of replication and gene expression during human development and identifies lineage-specific RT programs and genes that change RT during distinct lineage differentiation pathways. In contradiction to all prior literature, two-thirds of genes that switched RT were transcriptionally active and late replicating in at least one cell type. Moreover, these genes were significantly more central in transcriptional regulatory networks than the smaller class of genes that were only expressed when early replicating. Taken together, these results support a hierarchical model that links global chromatin organization and RT control with gene regulatory networks during human development.
Results
Genome-wide RT programs of human cell types
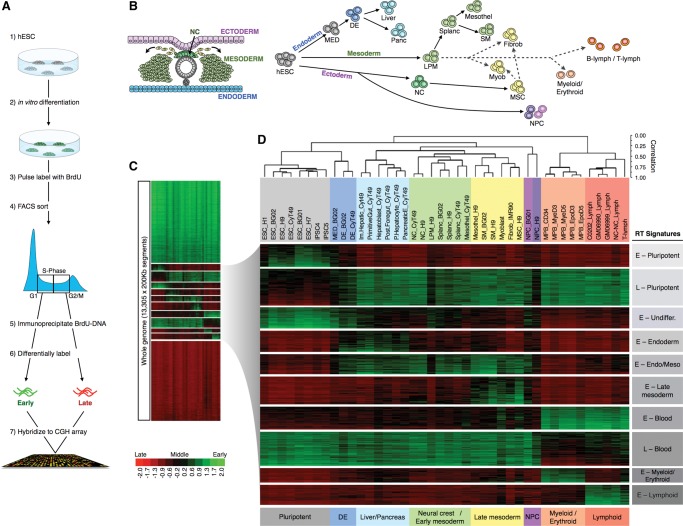
We obtained genome-wide RT profiles as previously described (Hiratani et al. 2008; Ryba et al. 2011a). Briefly, cells were labeled with BrdU, retroactively synchronized into early and late S-phase fractions by flow cytometry, and BrdU-substituted DNA from early and late S-phase populations was immunoprecipitated, differentially labeled, and cohybridized to a whole-genome oligonucleotide microarray (Fig. 1A). To compare all 84 data sets (Supplemental Table 1) representing 26 distinct cell types, we expressed RT profiles as numeric vectors of 13,305 average RT ratios for nonoverlapping 200-kb windows across the genome, excluding sex chromosomes and long stretches of repetitive DNA (see Methods). Correlation analysis confirmed close correspondence of technical and biological replicates (independently differentiated from hESC), which were averaged for subsequent analysis (Supplemental Fig. 1). We tested distinct combinations of thresholds for defining genomic segments that change RT during development (“switching RT regions”) and chose parameters that best distinguished cell types by RT (Supplemental Fig. 2). Switching regions were defined as being ≥+0.3 (Early) in at least one cell type and ≤−0.3 in at least one cell type. Using these criteria, which are relatively stringent compared to prior cut-offs (Hansen et al. 2010; Hiratani et al. 2010; Farkash-Amar et al. 2012), 30.5% of the genome was selected as changing RT during differentiation.
Figure 1.
Genome-wide RT patterns are lineage specific. (A) Genome-wide profiling of RT protocol. (B) Schematic diagram showing the three germ layers and the neural crest during the early stages of human development and differentiation pathways of the distinct cell types analyzed. Solid arrow lines depict the in vitro differentiation pathways of the distinct cell types from hESCs; dashed arrows depict the embryonic origin of the cell types not derived from hESCs (primary cells and cell lines). (C) RT changes across the different lineages. The whole genome was divided into 13,305 windows of 200 kb and their average RT ratios [=log2(Early/Late)] were compared across the human lineages. Heat map shows the RT ratios. (D) Hierarchical clustering of the distinct human lineages and k-means of switching 200-kb windows. Switching 200-kb segments were identified as being early replicated in at least one cell type (RT log2 ratio ≥+0.3) and late replicated in at least one other cell type (RT log2 ratio ≤−0.3) and analyzed by hierarchical clustering of the cell types. Branches of the dendrogram were constructed based on the correlation values between distinct cell types (distance = correlation value −1). A correlation threshold of >0.5 was used to color label the major groups of cell types. Specific clusters of cell types are indicated at the bottom: pluripotent, definitive endoderm, liver and pancreas, neural crest and early mesoderm, late mesoderm and fibroblasts, NPC, myeloid, and lymphoid. k-means clustering of switching segments defined the RT signatures labeled in gray boxes. The sex chromosomes were removed from the analysis to discard gender differences. (NC) neural crest; (MED) mesendoderm; (DE) definitive endoderm; (LPM) lateral plate mesoderm; (Splanc) splanchnic mesoderm; (Mesothel) mesothelium; (SM) smooth muscle; (Myob) myoblasts; (Fibrob) fibroblasts; (MSC) mesenchymal stem cells; (NPC) neural progenitor cells.
To analyze developmental transitions in RT, we removed genome segments that were invariant between cell types (“RT-constitutive regions”) and performed hierarchical and k-means clustering analysis of the switching segments (Fig. 1C,D). As expected, cell types clustered according to their developmental lineages (Fig. 1B,D). Importantly, normal primary cells extracted from healthy donors (myoblasts, T-lymphocytes), cell types differentiated in vitro from primary cells (hESCs and mobilized peripheral blood), and cell lines (fibroblasts IMR90 and EBV immortalized B-lymphocytes) clustered with their related cell types according to their developmental origin rather than their derivation history (Fig. 1D). Interestingly, both neural crest (NC), a uniquely vertebrate tissue derived from ectoderm, and NC-derived mesenchymal stem cells (MSC), which can be differentiated into cell types typically derived from mesoderm, clustered with mesoderm cell types, suggesting a close alignment of chromosome architecture in NC with mesoderm tissues (Fig. 1D). Also, the RT program of blood cells was substantially different from other cell types, with lymphoid and myeloid/erythroid lineages forming distinct subclusters (Fig. 1D).
RT signatures distinguish the major human lineages
k-means clustering of RT switching regions identified specific clusters of 200-kb windows replicating at times unique to specific lineages, which we refer to as RT signatures (Fig. 1D). Features of these signatures are highlighted below.
An RT signature for pluripotency
Regions from the E–Pluripotent RT signature were replicated early in hES and iPS cells and late in all differentiated cell types (Fig. 1D). These segments included genes known to be involved in maintaining pluripotency such as DPPA2, DPPA4, and ZFP42 (Supplemental Table 2) and were significantly enriched for genes whose promoters contain binding sequences for pluripotency transcription factors POU5F1, NANOG, and SOX2 (Supplemental Fig. 3). We also identified an RT signature (E–Undifferentiated) of regions uniquely early replicating in hESC and early stages of endoderm and mesoderm (Fig. 1D); these segments were enriched for genes involved in cell fate specification as well as genes with target sequences for NANOG, SOX2, and POU5F1 (Supplemental Fig. 3).
Endoderm RT signature
Genomic regions from the E–Endoderm signature (Fig. 1D) replicated early only in endoderm cell types and were enriched for genes from the FOXA1 transcriptional network (Supplemental Fig. 3), including key activators in endoderm differentiation such as FOXA2, GSC, and PBX1 (Supplemental Table 2).
Late mesoderm RT signature
Regions from the E–Late mesoderm signature replicated early only in late mesoderm (mesothelium, smooth muscle, myoblast, and fibroblast) and mesenchymal stem cells (Fig. 1D) and were enriched for genes involved in blood vessel, vascular, and muscle development as well as genes necessary for wound healing (Supplemental Fig. 3).
Blood RT signatures
We identified four distinct RT signatures for hematopoietic cell types (Fig. 1D): (1) regions replicated earlier in all hematopoietic cell types (E–Blood); (2) regions only late in all hematopoietic cells (L–Blood); (3) regions only early in the myeloid lineage (E–Myeloid); and (4) regions only early in the lymphoid lineage (E–Lymphoid). The E–Blood RT signature contains genes involved in immune system and hemostasis pathways. The E–Myeloid RT signature was highly enriched in genes associated with signal transduction by GPCR, which plays a critical role during inflammation and immune processes, whereas the E–Lymphoid RT signature was enriched in genes involved in the regulation of lymphocyte activation (Supplemental Fig. 3).
RT switching regions have distinct sequence composition
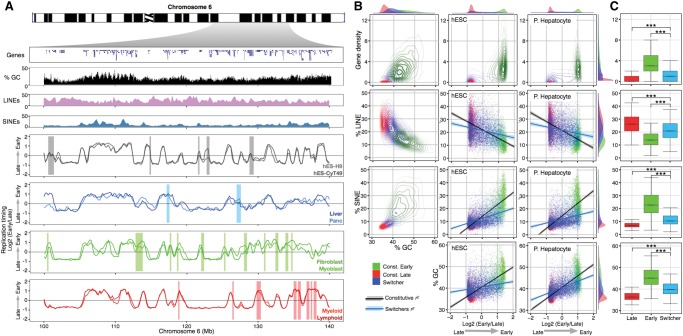
We next examined whether RT-switching regions and RT-constitutive regions have distinct DNA sequence composition (Fig. 2). Consistent with previous reports in mouse (Hiratani et al. 2008) and human (Hansen et al. 2010) cell types, constitutively early replicating regions were gene and GC-rich, with a lower density of LINEs and higher density of SINEs compared to constitutively late replicating regions, whereas RT-switching regions have intermediate sequence composition that exhibits a lower correlation with RT (Fig. 2B). However, unlike early mouse development, where the correlation of RT to GC and LINE content significantly increases during differentiation (Hiratani et al. 2004, 2008, 2010), correlations of DNA sequence features to RT did not change significantly between the cell types analyzed (Fig. 2B,C; Supplemental Fig. 4), including cell types very similar to the previously reported mouse cell types.
Figure 2.
RT switching segments have a distinct sequence composition. (A) Characterization of a genomic region from Chromosome 6. Top panels display the gene density, GC content, and densities of interspersed repetitive sequences (LINEs and SINEs). Below are the RT profiles of the same genomic region in distinct cell types. Bars in each plot highlight segments of the Chromosome 6 replicating differentially in specific cell types. (B) Density contours were plotted for gene density, GC content, and densities of LINEs and SINEs. Correlations of distinct genomic features were calculated against RT from hESC and hepatocytes. The top and right side histograms depict the density distribution of each feature. (C) Box plots of gene density, GC content, densities of LINEs and SINEs, and GC content of constitutive (Early and Late) and switching 200-kb regions: (***) P < 0.001.
Temporal consolidation and deconsolidation of RDs and ‘stemness’
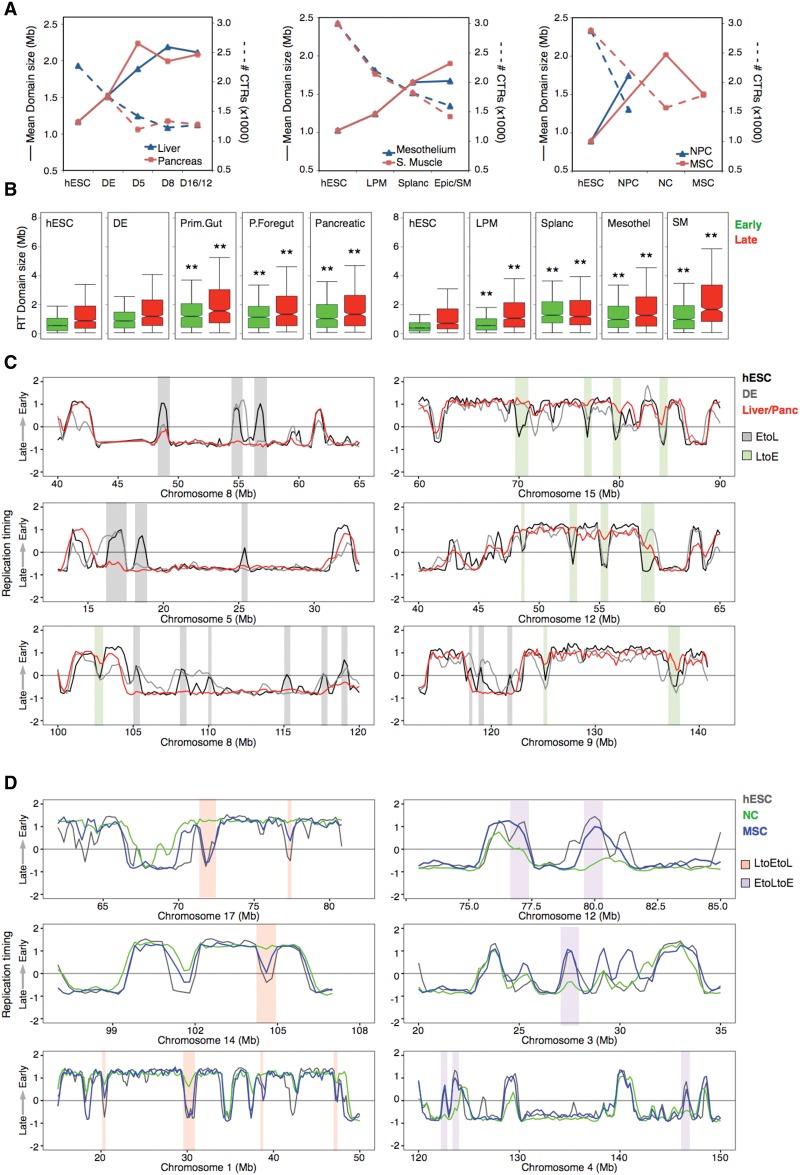
During differentiation of mouse and human ESCs, small differentially replicated RDs consolidate into larger regions of tandem RDs that replicate at similar times during S phase, termed constant timing regions (CTRs) (Hiratani et al. 2008; Ryba et al. 2010). We analyzed changes in global CTR organization during the intermediate stages of hESC differentiation toward endoderm, mesoderm, and ectoderm (see Methods; Fig. 3). Consistent with prior observations, a significant increase in the average CTR size together with a drop in the number of domains was observed in all differentiation pathways (Fig. 3A), both for early and late replicating regions (Fig. 3; Supplemental Fig. 5). Intriguingly, domain consolidation from ESCs to neural crest (NC) cells was followed by deconsolidation of both early and late domains on all chromosomes when NC cells were differentiated to mesenchymal stem cells (MSC) (Fig. 3; Supplemental Fig. 5) and recovered a portion of the ESC-like RT organization (Fig. 3D), suggesting that more discordant replication of adjacent RDs is a property of multipotency.
Figure 3.
Replication domain (RD) reorganization during hESC differentiation. (A) Number and sizes of constant timing regions (CTRs) in the distinct pathways. Solid lines depict the average size of the CTRs, whereas dashed lines depict the total number of CTRs in each cell type. (B) Sizes of early and late replicating CTRs during differentiation toward pancreas, mesothelium, and smooth muscle: (**) P ≤ 0.01 compared to hESC. (C) Representative RD consolidation during hESC differentiation. (D) Deconsolidation of RD during MSC differentiation.
Dynamic regulation of RT at the TSS during hESC differentiation
To understand better how RT of coding genes is regulated during differentiation, we focused on transcription start sites (TSS). We obtained RT ratios at the TSSs of 27,544 RefSeq genes (hg19 assembly), which were strongly correlated between replicates and averaged (Supplemental Fig. 6). RefSeq genes were classified into RT-constitutive and RT-switching genes using the same criteria as for 200-kb windows (described previously). Hierarchical clustering of the human lineages and k-means of the switching genes (Supplemental Fig. 7) confirmed results obtained using the whole genome, and genes present in each RT signature overlapped with the associated genes from the clusters of 200-kb windows (Fig. 1). As expected (Hiratani et al. 2008; Yokochi et al. 2009), most genes were replicated during the first half of S-phase (74.7%) across all cell types independent of their transcriptional activity (Fig. 4A; Supplemental Figs. 8, 9). In contrast, the subset of genes that switch RT during development was almost equally represented in early and late S-phase in all cell types analyzed (Supplemental Figs. 8, 9), implying that a similar number of genes change Early to Late (EtoL) and Late to Early (LtoE) during differentiation (Fig. 4A).
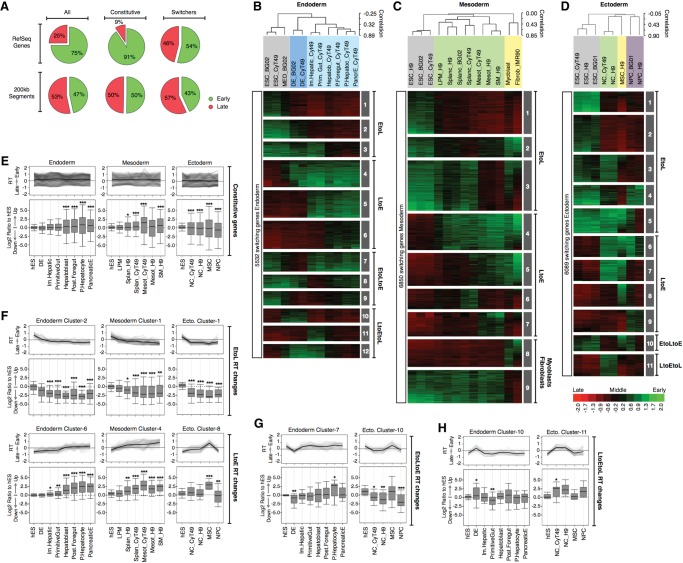
Figure 4.
Dynamic changes in RT and transcriptional activity during hESC differentiation. (A) Frequency distributions of 200-kb segments and RefSeq genes in early and late S-phase fractions. The first row includes a pool of 200-kb or RefSeq genes (All) across all cell types; the second row includes only the RT-constitutive regions (i.e., does change RT in any cell type); and the third row includes only the regions or genes switching RT at least in one cell type. RT changes during hESC differentiation toward endoderm (B), mesoderm (C), and ectoderm (D). Averaged RT values at the TSS of 27,544 RefSeq genes were obtained; the switching genes were extracted according to the same criteria of Figure 1 and analyzed by hierarchical clustering and k-means. The heat map shows the RT ratios [=log2(Early/Late)]. Branches of the dendrogram were constructed based on the correlation values (distance = correlation value −1), and distinct correlation thresholds were used to color label the major nodes. The sex chromosomes were removed from the analysis to discard gender differences. The k-means clusters are shown as numbered gray boxes. (E–H) Transcriptional regulation and its relationship with RT dynamics for RT-constitutive genes (E), genes switching EtoL and LtoE (F), EtoLtoE (G), and LtoEtoL (H). Note that in E, some genes appear to switch RT but do not meet the RT-switching cutoff used in this study. (F–H) Genes with a fold difference ≥6 in expression level compared with hESC at any differentiation stage were classified into the RT clusters from panels B to D to identify the kinetics of regulation. Line graphs depict the dynamics in RT of each cluster, and box plots were used to display the transcriptional ratios against expression in hESC: (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 compared to hESC.
When we analyzed separate differentiation pathways and their intermediate stages individually (Fig. 4B–D), distinct patterns of RT regulation emerged. First, consistent with mouse studies (Hiratani et al. 2010), many genes that switch from early to late replication (EtoL) during loss of pluripotency do so in all differentiation pathways, whereas genes switching from late to early (LtoE) are lineage specific and include regulators of key developmental transitions (Supplemental Fig. 10). However, analysis of differentiation intermediates identified genes switching transiently (EtoLtoE and LtoEtoL) during differentiation in specific lineages (Fig. 4B–D). Among the genes with transient changes LtoEtoL in RT were key regulators such as SOX17, a gene known to be involved in the differentiation toward endoderm cell types, whose expression is restricted to the transition stage of definitive endoderm (Supplemental Fig. 10).
RT and transcription are highly coordinated during hESC differentiation
To analyze the relationship between RT and transcriptional regulation during differentiation, we obtained the transcriptomes of distinct cell types using HumanHT-12 v4 Illumina bead-based arrays. Analysis of specific markers confirmed the expression levels detected by array hybridization and the homogeneity of the distinct intermediate cell types (Supplemental Fig. 11; Supplemental Methods). Next, genes that were induced or repressed during differentiation were classified by their RT changes (the cluster IDs identified in Fig. 4B–D) and the kinetics of their transcription changes during differentiation. Transcription changes for RT-constitutive genes did not show enrichment for either induction or repression (Fig. 4E), whereas genes switching RT showed highly coordinated kinetics of RT and transcriptional changes in all differentiation pathways for all RT regulation patterns and cluster IDs (Fig. 4F–H).
Loss of correlation between gene expression and RT during differentiation
Perhaps the strongest correlation between transcription and early RT has been demonstrated by comparing the probability of transcription of genes (i.e., on or off) and their RT, something observed in all Drosophila (Schübeler et al. 2002; MacAlpine et al. 2004), mouse (Hiratani et al. 2008, 2010), and human (Woodfine et al. 2004) cell types examined. To investigate the degree to which this correlation is consistent across different cell types, genes were scored as either expressed (log2 values >7.5) or not expressed. Surprisingly, we observed that the correlation between early replication and gene expression declined during differentiation in every pathway (Fig. 5A; Supplemental Fig. 12). Moreover, although the correlation of RT and gene expression remained constant for RT-constitutive genes, RT-switching genes showed a weaker correlation that was almost completely lost during differentiation. Prior reports did not observe this loss in correlation during differentiation likely because they focused on neural lineages (Hiratani et al. 2008, 2010; Ryba et al. 2010), which were the least affected by differentiation in our hands. Hence, during differentiation, a decrease in correlation between early RT and transcription occurs that is driven by the behavior of RT-switching genes. These results raise a conundrum, since Figure 4 demonstrates a strong temporal correlation between RT and transcription for these same genes.
Figure 5.
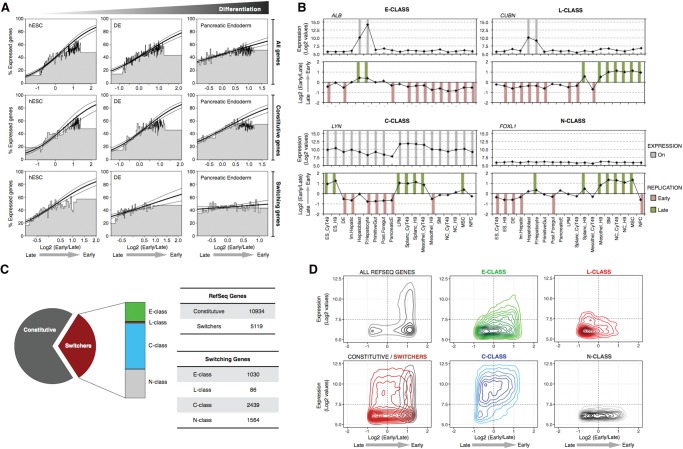
Switching genes and transcriptional regulation. (A) Correlation between early replication and the probability of expression changes during differentiation. Genes were scored as either expressed or not expressed and ranked by their RT ratio and divided into bins of 100 genes, the height of which represents the percentage of expressed genes within each bin. Logistic regression (inner line) and 95% confidence intervals (outer lines) reveal the correlation strength. Top row graphs include all genes, the middle includes only the RT-constitutive genes (i.e., do not change RT), and the bottom row includes only the genes that change RT during differentiation from hESC to pancreatic endoderm. (B) Distinct classes of switching genes. Transcriptional activity (top graphs) and RT (bottom graphs) of exemplary genes representing the distinct kind of switching classes: E-class genes are only expressed when early replicating (ALB); L-class genes can be expressed only when late replicating (CUBN); C-class genes can be transcribed whether early or late replicating (LYN); and N-class are genes with no transcriptional activity detected in the cell types profiled (FOXL1). Gray columns show the “On” calls for gene expression, and green/red columns show the early/late calls for RT according to the established thresholds (see Methods). (C) Frequencies of the distinct kind of genes based on their RT dynamics during differentiation. RT-constitutive genes were classified as O-class. (D) Density plots of all RefSeq genes, RT-constitutive and RT-switching genes, and the distinct categories of RT-switching genes according to their RT and transcriptional level.
Distinct classes of RT-switching genes
Next, we classified the RT-switching genes into distinct categories (Fig. 5B). E-class genes can be expressed only when they replicate early (no exceptions), whereas C-class were transcribed when late replicating in at least one cell type. L-class refers to a small number of RT-switching genes that were expressed only when replicated late. Finally, N-class genes were those that showed no detectable transcription activity in any profiled cell type and are presumably a mixture of C-, E-, and L-class. The RT-constitutive genes (that do not change RT) were classified as O-class genes. Interestingly, the C-class constituted the majority (47.6%) of RT-switching genes (Fig. 5C). C-class genes were expressed significantly higher than E- or L-class, and at comparable levels to O-class genes, regardless of their RT (Fig. 5D) and independent of cell type (Supplemental Fig. 13). We previously demonstrated that the promoters of genes that were late replicating and expressed during mouse differentiation had a high CpG content (Hiratani et al. 2010), a property that has been implicated in differential gene expression (Mikkelsen et al. 2007; Weber et al. 2007). However, we did not find significant differences in the CpG content of these distinct classes of genes in humans (Supplemental Fig. 14). A summary of the numbers of genes in each class and their RT regulation pattern (EtoL, LtoE, EtoLtoE, LtoEtoL) in different lineages is shown in Supplemental Figure 15.
C-class gene transcription is frequently coordinated with RT
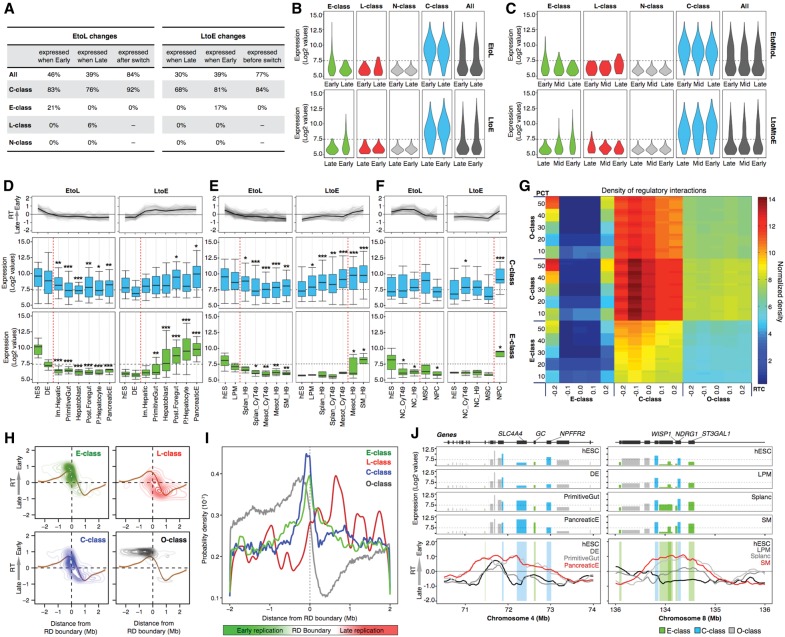
The results above demonstrate highly coordinated changes in transcription and RT for C-class genes (Fig. 4F–H), despite a lack of correlation between RT and gene expression (Fig. 5). One explanation for this discrepancy is if C-class genes are only highly transcribed in cell lineages in which the genes switch to early RT, but their expression in intermediate stages precedes or follows the RT switch. To investigate this possibility, we subclassified the RT changes as either gradual, with middle replication (EtoMtoL or LtoMtoE) (RT log2 ratio >−0.3 and <+0.3) identified in an intermediate stage (25% of cases), or abrupt, without a middle replication intermediate (75% of cases). We then restricted our analysis of changes in C-class transcription to the cell type intermediates just before and after abrupt versus gradual RT switches. This resulted in a complete absence of correlation of early replication to transcription (Fig. 6A–C). Next, we analyzed the kinetics of transcriptional and RT changes during differentiation in the three main germ layers. C-class genes followed the same dynamic changes in gene expression as E-class genes, except that transcriptional up-regulation often preceded the LtoE switch while a decrease in transcription often occurred after the EtoL RT switch (Fig. 6D–F; Supplemental Fig. 16). Thus, the subset of C-class genes that change transcription were induced when late, primarily during a window of differentiation that closely precedes or follows the RT switch.
Figure 6.
Gene expression regulation and RT changes. (A) RT changes were collected from all transition stages in all differentiation pathways, and the percentages of expressed genes in each stage (before and after the RT switch) were obtained. The first columns in EtoL and LtoE changes contain the percentages of genes expressed calculated from the total number of switching genes in each category. The last columns contain the percentages of genes expressed before/after the RT switch. EtoL changes = 3567 genes, and LtoE changes = 3098 genes. (B) Expression levels of the distinct classes of RT-switching genes during the EtoL (upper) and LtoE (lower) RT changes. (C) Expression levels of the distinct classes of RT-switching genes during the EtoMtoL (upper) and LtoMtoE (lower) RT changes. Transcriptional values were obtained from all respective RT changes occurring in between all intermediate stages of all differentiation pathways. D–F illustrate the transcriptional regulation and RT dynamic changes in endoderm (D), mesoderm (E), and ectoderm (F) differentiation per RT switching class. The genes with a fold difference ≥6 compared with hESC were classified into the RT clusters from Figure 4B–D and separated in the distinct categories of RT-switching genes to identify the kinetics of regulation. Line graphs depict the dynamics in RT of each cluster, and box plots were used to display the transcriptional levels across the differentiation pathways: (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 compared to hESC. Specific stages of differentiation are shown at the bottom. (G) Densities of regulatory interactions among E-class, C-class, and O-class genes. The probability of inter-class regulation was calculated by defining each class of genes by different values of RT cutoff (RTC), stringency (PCT), and switch cutoff (SC). Each PCT row is a composite of six rows representing SC values of 0.5 to 1.0 from the bottom up. Each cell represents the probability of the x-axis gene class regulating the y-axis gene class, based on the corresponding values of RTC, PCT, and SC. H-I Switching genes are preferentially located at the TTRs. Densities of the TSS from the distinct categories of genes were calculated as a function of their distance to the early replication boundary and plotted according to their RT (H) or probability densities (I). (J) Induction of C-class genes precedes EtoL RT changes and induction of E-class genes. Expression levels (top graphs) and RT changes (bottom graphs) of representative genomic regions during differentiation.
One concern was that high expression of a gene in a small percentage of cells replicating that gene early would be mis-classified as a C-class gene. This seemed unlikely given the homogeneity of markers for each intermediate stage (Supplemental Fig. 11; Supplemental Methods; Schulz et al. 2003, 2004, 2012; Menendez et al. 2011, 2013) and the fact that similar results were observed in all lineages. Moreover, some of the C-class genes were expressed to similarly high levels in all cell types across many switches in RT (Supplemental Fig. 17A), so it would be unlikely to have contaminating cells in every one of these cell types. More directly, for those C-class genes for which transcriptional up-regulation preceded the LtoE switch (Supplemental Fig. 17C) or those for which a decrease in transcription occurred after the EtoL RT switch (Supplemental Fig. 17B), we frequently saw that the level of induction of transcription reached or remained at its maximum level before or after the RT switch, respectively. If expression were due to contaminating early replicating cells, transcription would continue to increase with the increase in the percentage of early replicating cells, and this was not the case. The difference between C-class and E-class was also not due to the level of transcriptional induction because many E-class were induced to similar levels as the C-class genes (Supplemental Fig. 18).
An alternative explanation for C-class gene behavior could be asynchronous replication between gene homologs, with one homolog replicating early and expressed. However, very few loci in mammalian genomes display asynchronous replication, and so far all of them have been linked to either X Chromosome inactivation in female cells or to parental imprinting/monoallelic gene expression (Reik and Walter 2001; Goldmit and Bergman 2004; Gimelbrant et al. 2007; Farkash-Amar et al. 2008; Stoffregen et al. 2011; DeVeale et al. 2012; Koren et al. 2014; Mukhopadhyay et al. 2014; Donley et al. 2015). Moreover, direct comparison of known asynchronously replicated or monoallelically expressed genes to our categories of genes revealed that such genes are mostly constitutively replicated (Supplemental Fig. 19). It is also unlikely that C-class genes are becoming asynchronous uniquely in the lineages where we see their up-regulation, because we can detect a significant shift in replication timing even of a single homolog (e.g., the inactivated X Chromosome) (Hiratani et al. 2010), and 75% of the replication timing shifts we detected were single abrupt shifts without intermediate replication times. Together, these results are most consistent with the conclusion that a high level of transcription for many (not all) C-class genes is coordinated with early RT but can precede or follow the RT switch.
C-class genes are key regulators in TRNs
The fact that RT switches are frequently preceded by induction of C-class genes (Fig. 5) and followed by induction of E-class genes (Fig. 6D–F) suggests the hypothesis that C-class might drive the RT switches required for expression of E-class genes. One prediction of this hypothesis is that C-class genes should have a higher hierarchy in transcriptional regulatory networks (TRNs). We used available human TRNs (Gerstein et al. 2012) to compute the inter-class regulation probability and found that C-class genes are most central and regulate all three classes of genes (C-class, E-class, and O-class) (Fig. 6G). Notably, E-class genes are least central and regulated mainly by C-class genes (Fig. 6G). Together these results suggest a central role for C-class genes in the regulatory interactions during differentiation and a subordinate role for E-class. Further experimentation will be necessary to determine whether this is a direct regulatory role mediated by an RT switch.
Developmentally regulated changes in RT occur at RD boundaries
To better understand the differences between distinct classes of genes, we analyzed their positions relative to the boundaries of the RDs. First, we identified timing transition regions (TTRs) in all the cell types profiled based on changes in the slope of RT profiles, using a previously described algorithm (Pope et al. 2014; see Methods), and aligned the TSS of each category of genes to the RD boundaries (defined as the earliest replicating border of TTRs) (Pope et al. 2014). As expected, RT-constitutive genes (O-class) were highly enriched throughout early replicating RDs; in contrast, we found that switching genes were enriched at the RD boundaries (Fig. 6H,I). L-class genes were preferentially located in late domains. No clear differences were observed between C-class, E-class, and N-class genes—all three categories of switching genes were highly enriched at RD boundaries (Fig. 6H,I). These results are consistent with previous evidence suggesting that changes in RT might be associated with differential activation of replication origins in close proximity to TTRs (Norio et al. 2005).
Discussion
We have generated genome-wide RT profiles for 26 different cell types and distinct intermediate stages of endoderm, mesoderm, ectoderm, and neural crest differentiation, permitting changes in transcription and RT to be tracked during transitions between intermediate cell types in several cell lineages. In addition to providing a resource of genomic segments subject to RT regulation during development, our results revealed that the longstanding correlation between early replication and transcription does not hold for most “RT-switching genes,” which are up-regulated just prior to a switch from late to early replication and down-regulated just after the switch from early to late replication. We also identify a class of genes that are expressed only in cell types where they are replicated late. These results reveal previously unappreciated complexities in the relationship between RT and gene expression.
RT and transcriptional control
Changes in RT occur in segments of 400–800 kb known as replication domains (RDs) that consist of stable structural units of chromosomes detected as self-interacting structural domains by Hi-C and referred to as TADs (Pope et al. 2014). Genes within RD/TADs can be classified into those that switch RT during differentiation (RT-switching genes) and those that do not (RT-constitutive genes or O-class). Here, we examined the relationship between RT and transcription in each of these categories of genes. Almost all RT-constitutive genes are early replicating, highly expressed, and show a much greater breadth of expression (expressed in more tissues) than the rare constitutively late genes. RT-constitutive RD/TADs also exhibit a strong correlation between early replication and their chromatin nuclease sensitivity (Takebayashi et al. 2012a,b). RT-constitutive genes thus adhere to the longstanding notion that early replication is associated with open chromatin and transcriptional activity, albeit late replication does not preclude a low level of transcription for a small number of genes. Approximately one-third of RT-switching genes were expressed uniquely when early replicating (E-class). However, the majority of RT-switching genes were expressed to very high levels, even when late replicating (C-class), and showed a breadth of expression similar to O-class (RT-constitutive genes). Interestingly, RT-switching RD/TADs are also among the most resistant to nuclease attack in the genome, regardless of RT (Takebayashi et al. 2012a,b). Findings with developmental-regulated RDs then, which represent a substantial fraction of the genome (as stringently defined here, they encompass >30% of the genome), indicate that late replication and closed chromatin are not incompatible with high levels of transcription, challenging the longstanding notion we demonstrate has been driven primarily by constitutive RDs.
Unlike prior studies of mouse ESC differentiation (Hiratani et al. 2004, 2008, 2010), the hESC differentiation systems described here are sufficiently synchronous and homogeneous to permit us to track RT and transcription during intermediate stages of differentiation. First, we found that the correlation between RT and the probability of a gene being called “ON” or “OFF” dropped considerably during differentiation to almost no correlation in most cell types. This loss of correlation was accounted for largely by the C-class genes, which can be expressed in many broad cell types when replicated late. However, RT and gene expression changes were generally coordinated for all genes in all tracked lineages. Despite their ability to be transcribed at high levels when late replicating, transcription of C-class genes was indeed still coordinated with RT, but increased just prior to an LtoE switch and was down-regulated just after an EtoL switch. Frequently, the induction of E-class genes, which follows LtoE RT changes, is preceded by up-regulation of C-class genes from within the same developmental RD. Two representative examples are shown in Figure 6J: C-class genes are expressed at low levels in late replicating regions, they are up-regulated during differentiation just prior to an LtoE switch, after which the E-class genes—GC, ST3GAL1, and WISP1—begin transcription.
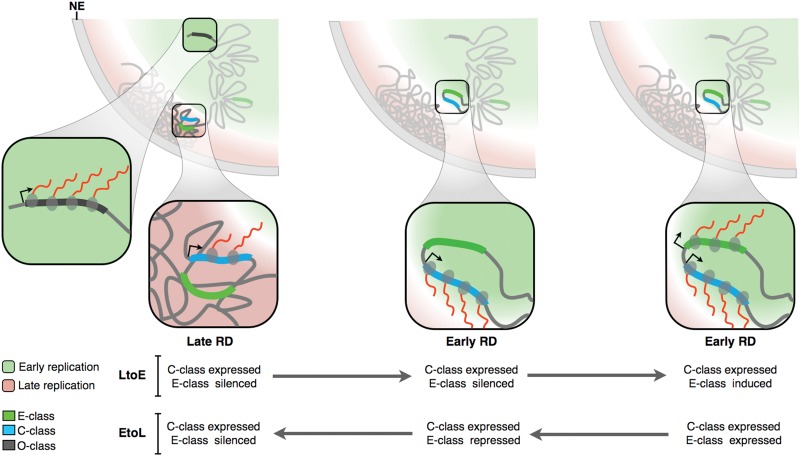
RT has been linked with the nuclear organization of chromatin, with early replicated regions located at the nuclear interior and late replicating regions associated with the nuclear lamina (Hiratani et al. 2010; Peric-Hupkes et al. 2010). The transcriptional potential of C-class genes regardless of their RT (Fig. 5) and their preferential position within TTRs (Fig. 6H,I), together with their central position in the TRNs (Fig. 6G), suggest an intriguing model of RT and gene expression regulation during cell fate specification that helps to understand the complex relationship between RT and transcription (Fig. 7). In this model, early RDs are located at the nuclear interior in closer proximity to regions of active transcription. In contrast, late RDs are positioned close to the periphery, in a compartment that is less permissive for transcription. Genes located at the early RDs can be expressed at high levels at any stage of differentiation. However, only C-class genes can be expressed to high levels when located within late RDs. This may have to do with specialized features of the regulatory elements of these genes allowing them to be expressed within late replicating chromatin. Alternatively, regulatory elements of C- and E-class genes may reside in different 3D orientations within the RD/TADs. In this model (Fig. 7), up-regulation of the C-class gene triggers an LtoE change in RT and movement to the nuclear interior, which permits the E-class gene to be expressed. This model makes the testable prediction that preventing C-class gene induction will prevent early replication and E-class gene expression.
Figure 7.
RT and transcriptional control. Early replication occurs in the nuclear interior, while late replicated regions are positioned at the periphery. Two RDs illustrate the differences in organization of early and late replicated regions. Transcriptional activity (red lines) and chromatin organization of E- versus C-class genes are represented in the expanded pictures during EtoL and LtoE RT changes (see text for details).
Methods
Cell culture and hESC differentiation
hES cell lines (CyT49 and H9) were differentiated using defined media specific for each pathway (Schulz et al. 2003, 2004, 2012; Menendez et al. 2011, 2013). See Supplemental Methods for differentiation protocols.
Purification of normal hematopoietic progenitors and in vitro differentiation toward myeloid/erythroid lineages
CD34+ cells were provided by the Yale Center for Excellence in Molecular Hematology, stimulated to proliferate and differentiate in vitro toward myeloid and erythroid lineages (see Supplemental Methods) according to established protocols (Mayani et al. 1993; Heike and Nakahata 2002; Manz et al. 2002; Mahajan et al. 2009; Migliaccio et al. 2009).
Genome-wide RT profiling
Genome-wide RT profiles were constructed as previously described by array hybridization (Hiratani et al. 2008; Ryba et al. 2011a). Previously, we have shown that array hybridization (Repli-chip) and NGS (Repli-seq) produce indistinguishable results (Supplemental Fig. 20; Pope et al. 2014; Yue et al. 2014). Published RT data sets from hESC (cell lines BG01, BG02, H7, and H1), iPSC, NPC (derived from hESC BG01), mesendoderm and DE (derived from hESC BG02), smooth muscle (from hESC BG02), myoblasts, and fibroblast and lymphoid cells were included in the analysis (Ryba et al. 2010, 2011b, 2012; Pope et al. 2011). RT data sets were normalized using the limma package (Smyth 2005) in R (R Core Team 2015) and rescaled to equivalent ranges by quantile normalization.
Clustering analysis
Hierarchical and k-means clustering analysis were performed using Cluster 3.0 (de Hoon et al. 2004). Uncentered correlation metrics and average linkage were used for clustering analysis in Figures 1 and 3, but standard correlation and Euclidean distances produced similar results (data not shown). Heatmaps and dendrograms were generated in JavaTreeView (Saldanha 2004). Sex chromosomes as well as pericentromeric and subtelomeric regions were removed from the analysis.
Gene expression analysis
Total RNA was extracted, labeled, and hybridized to HumanHT-12v4 Expression BeadChips (Illumina, Inc.). For detailed gene expression analysis, see Supplemental Methods.
Identification of constant timing regions (CTRs)
Segmentation of genome-wide RT profiles into constant timing regions (CTRs) was performed as previously described (Hiratani et al. 2008; Ryba et al. 2011a) using DNAcopy, version 1.36.0 (Venkatraman and Olshen 2007; http://bioconductor.org/packages/release/bioc/html/DNAcopy.html), in R to identify the regions with uniform y-axis values. To account for the dependency of segmentation on slight differences in autocorrelation of neighboring probes between data sets, we applied Gaussian noise to equalize their autocorrelation (ACF) before segmentation.
Identification of RD boundaries
Loess smoothed RT profiles were obtained from each quantile-normalized data set, and RT boundaries were obtained as previously described (Pope et al. 2014; Yue et al. 2014). Briefly, RD boundaries were identified as the early border of transitions between relatively earlier and later replicating regions (TTRs) in individual data sets with a slope above ±2.75×10−6 RT units/bp at the early border and with a sustained slope above ±1×10−6 RT units/bp for at least 250 kb.
TRN analysis
We used RT and gene expression data to classify genes into E-class, C-class, and O-class based on the values of three variables: replication time (RTC) and switch (SC) cutoffs, defined as in Supplemental Figure 2; and stringency cutoff (PCT), defined as the percentage of cell types in which the gene is expressed. We then used transcription regulatory networks (Gerstein et al. 2012) to compute the inter-class regulation probability. For each pair of classes, we count the number of edges connecting genes from each class. We divided this number by the number of possible edges, which is the product of the class sizes.
Data access
Replication timing and transcriptome data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE63428 and GSE63592, to ENCODE portal (https://www.encodeproject.org/), and are also publicly downloadable and graphically displayed at http://www.replicationdomain.org (Weddington et al. 2008).
Supplementary Material
Acknowledgments
We thank B.D. Pope for sharing the RD boundaries identification algorithm, D. Schübeler and M. Weber for sharing the CpG content gene classification, and D. Vera for helpful discussions. This work was supported by National Institutes of Health (NIH) grants GM083337 (D.M.G.) and GM085354 (S.D., D.M.G.) and a post-doctoral fellowship from CONACyT, Mexico to J.C.R.M.
Author contributions: Conception and design: J.C.R.M. and D.M.G.; myeloid and erythroid differentiation: Z.L. and S.W.; endoderm differentiation: A.J.R. and T.C.S.; mesoderm and ectoderm differentiation: L.M., M.J.K., and S.D.; flow cytometry: R.A.D.; RT data sets: J.C.R.M. and T.S.; transcriptome data sets: K.N. and J.F.L.; data analysis and interpretation: J.C.R.M., K.N., Q.B., J.Z., H.G., and T.K.; manuscript writing: J.C.R.M. and D.M.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.187989.114.
References
- Alver RC, Chadha GS, Blow JJ. 2014. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst) 19: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut CL, Thierry-Mieg J, Bouhassira EE. 2009. Predictable dynamic program of timing of DNA replication in human cells. Genome Res 19: 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B, van der Kooy D, Babak T. 2012. Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet 8: e1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N, Stoffregen EP, Smith L, Montagna C, Thayer MJ. 2013. Asynchronous replication, mono-allelic expression, and long range cis-effects of ASAR6. PLoS Genet 9: e1003423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N, Smith L, Thayer MJ. 2015. ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet 11: e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I. 2008. Global organization of replication time zones of the mouse genome. Genome Res 18: 1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash-Amar S, David Y, Polten A, Hezroni H, Eldar YC, Meshorer E, Yakhini Z, Simon I. 2012. Systematic determination of replication activity type highlights interconnections between replication, chromatin structure and nuclear localization. PLoS One 7: e48986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153: 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. 2007. Widespread monoallelic expression on human autosomes. Science 318: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Goldmit M, Bergman Y. 2004. Monoallelic gene expression: a repertoire of recurrent themes. Immunol Rev 200: 197–214. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heike T, Nakahata T. 2002. Ex vivo expansion of hematopoietic stem cells by cytokines. Biochim Biophys Acta 1592: 313–321. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Leskovar A, Gilbert DM. 2004. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proc Natl Acad Sci 101: 16861–16866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schübeler D, Gilbert DM. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi SI, Lu J, Gilbert DM. 2009. Replication timing and transcriptional control: beyond cause and effect—part II. Curr Opin Genet Dev 19: 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, Thermes C. 2007. Human gene organization driven by the coordination of replication and transcription. Genome Res 17: 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Handsaker RE, Kamitaki N, Karlic´ R, Ghosh S, Polak P, Eggan K, McCarroll SA. 2014. Genetic variation in human DNA replication timing. Cell 159: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. 2014. DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. 1998. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodríguez HK, Bell SP. 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan MC, Karmakar S, Newburger PE, Krause DS, Weissman SM. 2009. Dynamics of α-globin locus chromatin structure and gene expression during erythroid differentiation of human CD34+ cells in culture. Exp Hematol 37: 1143–1156.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz MG, Miyamoto T, Akashi K, Weissman IL. 2002. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci 99: 11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric C, Prioleau MN. 2010. Interplay between DNA replication and gene expression: a harmonious coexistence. Curr Opin Cell Biol 22: 277–283. [DOI] [PubMed] [Google Scholar]
- Mayani H, Dragowska W, Lansdorp PM. 1993. Lineage commitment in human hemopoiesis involves asymmetric cell division of multipotent progenitors and does not appear to be influenced by cytokines. J Cell Physiol 157: 579–586. [DOI] [PubMed] [Google Scholar]
- Menendez L, Yatskievych TA, Antin PB, Dalton S. 2011. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc Natl Acad Sci 108: 19240–19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez L, Kulik MJ, Page AT, Park SS, Lauderdale JD, Cunningham ML, Dalton S. 2013. Directed differentiation of human pluripotent cells to neural crest stem cells. Nat Protoc 8: 203–212. [DOI] [PubMed] [Google Scholar]
- Migliaccio AR, Whitsett C, Migliaccio G. 2009. Erythroid cells in vitro: from developmental biology to blood transfusion products. Curr Opin Hematol 16: 259–268. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A. 2012. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res 40: 9470–9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Lajugie J, Fourel N, Selzer A, Schizas M, Bartholdy B, Mar J, Lin CM, Martin MM, Ryan M, et al. 2014. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet 10: e1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165: 291–297. [DOI] [PubMed] [Google Scholar]
- Nakayasu H. 1989. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol 108: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IMY, Mijic S, Herrador R, Zellweger R, Ray Chaudhuri A, Creavin KD, Blow JJ, Lopes M. 2013. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev 27: 2537–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z. 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 20: 575–587. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol 116: 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Gilbert DM. 2013. The replication domain model: regulating replicon firing in the context of large-scale chromosome architecture. J Mol Biol 425: 4690–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Tsumagari K, Battaglia D, Ryba T, Hiratani I, Ehrlich M, Gilbert DM. 2011. DNA replication timing is maintained genome-wide in primary human myoblasts independent of D4Z4 contraction in FSH muscular dystrophy. PLoS One 6: e27413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Reik W, Walter J. 2001. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2: 21–32. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM. 2011a. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc 6: 870–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM. 2011b. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput Biol 7: e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM. 2012. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res 22: 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D. 2004. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117: 5353–5365. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442. [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SS, Desbordes SC, Du Z, Kosiyatrakul S, Lipchina I, Studer L, Schildkraut CL. 2010. Single-molecule analysis reveals changes in the DNA replication program for the POU5F1 locus upon human embryonic stem cell differentiation. Mol Cell Biol 30: 4521–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TC, Palmarini GM, Noggle SA, Weiler DA, Mitalipova MM, Condie BG. 2003. Directed neuronal differentiation of human embryonic stem cells. BMC Neurosci 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TC, Noggle SA, Palmarini GM, Weiler DA, Lyons IG, Pensa KA, Meedeniya ACB, Davidson BP, Lambert NA, Condie BG. 2004. Differentiation of human embryonic stem cells to dopaminergic neurons in serum-free suspension culture. Stem Cells 22: 1218–1238. [DOI] [PubMed] [Google Scholar]
- Schulz TC, Young HY, Agulnick AD, Babin MJ, Baetge EE, Bang AG, Bhoumik A, Cepa I, Cesario RM, Haakmeester C, et al. 2012. A scalable system for production of functional pancreatic progenitors from human embryonic stem cells. PLoS One 7: e37004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schübeler D. 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. limma: linear models for microarray data In Bioinformatics and computational biology solutions using R and bioconductor (ed. Gentleman R, et al. ), pp. 397–420. Springer, New York. [Google Scholar]
- Stoffregen EP, Donley N, Stauffer D, Smith L, Thayer MJ. 2011. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum Mol Genet 20: 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi SI, Dileep V, Ryba T, Dennis JH, Gilbert DM. 2012a. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc Natl Acad Sci 109: 12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi SI, Ryba T, Gilbert DM. 2012b. Developmental control of replication timing defines a new breed of chromosomal domains with a novel mechanism of chromatin unfolding. Nucleus 3: 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A. 2015. Transcription factor binding dynamics during human ES cell differentiation. Nature 518: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB. 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466. [DOI] [PubMed] [Google Scholar]
- Weddington N, Stuy A, Hiratani I, Ryba T, Yokochi T, Gilbert DM. 2008. ReplicationDomain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics 9: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. 2004. Replication timing of the human genome. Hum Mol Genet 13: 191–202. [DOI] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. 2010. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. 2002. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol Cell Biol 22: 4876–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.