Abstract
Mammalian genomes are partitioned into domains that replicate in a defined temporal order. These domains can replicate at similar times in all cell types (constitutive) or at cell type-specific times (developmental). Genome-wide chromatin conformation capture (Hi-C) has revealed sub-megabase topologically associating domains (TADs), which are the structural counterparts of replication domains. Hi-C also segregates inter-TAD contacts into defined 3D spatial compartments that align precisely to genome-wide replication timing profiles. Determinants of the replication-timing program are re-established during early G1 phase of each cell cycle and lost in G2 phase, but it is not known when TAD structure and inter-TAD contacts are re-established after their elimination during mitosis. Here, we use multiplexed 4C-seq to study dynamic changes in chromatin organization during early G1. We find that both establishment of TADs and their compartmentalization occur during early G1, within the same time frame as establishment of the replication-timing program. Once established, this 3D organization is preserved either after withdrawal into quiescence or for the remainder of interphase including G2 phase, implying 3D structure is not sufficient to maintain replication timing. Finally, we find that developmental domains are less well compartmentalized than constitutive domains and display chromatin properties that distinguish them from early and late constitutive domains. Overall, this study uncovers a strong connection between chromatin re-organization during G1, establishment of replication timing, and its developmental control.
Mammalian cells replicate their genomes in a defined temporal order (“replication-timing program”) that is cell type-specific and altered in many diseases (Rhind and Gilbert 2013). Almost half of the genome changes replication timing during development in units of 400–800 kb, designated replication domains (RDs) (Ryba et al. 2010). As a result, RDs can be categorized as replicating either at the same time in all cell types (constitutive RDs) or at different times in different cell types (developmental RDs) (Hiratani et al. 2008). The boundaries of both constitutive and developmental RDs are stable across cell types, but the subnuclear position, chromatin structure, and transcriptional state change coordinately with replication timing changes for developmental RDs. In general, early replication is correlated with transcriptional activity (Hatton et al. 1988; Hiratani et al. 2010; Ryba et al. 2010). Although RDs are regulated independently, tandem RDs often replicate at similar times, resulting in larger regions of similar replication timing that can fractionate or consolidate during differentiation depending on the distribution of constitutive and developmental domains (Hiratani et al. 2008).
The replication-timing program is re-established in each cell cycle at a point during early G1 phase termed the timing decision point (TDP). The TDP is coincident with the cytogenetically observed 3D repositioning and anchorage of chromatin domains to their final interphase positions in the newly formed nucleus (Dimitrova and Gilbert 1999; Walter et al. 2003; Thomson et al. 2004). The 3D organization of interphase chromatin in mammals has been linked to several genome-wide processes such as DNA replication, long-range gene regulation, and chromosomal translocations (Ryba et al. 2010; Moindrot et al. 2012; Zhang et al. 2012; Denholtz et al. 2013; Ay et al. 2014; Sima and Gilbert 2014). Applications of genome-wide chromatin conformation capture (Hi-C) have revealed organization of chromatin into sub-megabase topologically associating domains (TADs) that are preserved across cell types and compartmentalization of chromatin contacts between TADs (inter-TAD interactions) into larger scale folding states that are cell type-specific (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Nora et al. 2012). Strong correlations between replication timing and chromatin contacts in the interphase nucleus have revealed two important structure-function relationships. First, boundaries of TADs align to the boundaries of RDs, demonstrating that TADs are the units of replication timing control (Pope et al. 2014). Second, inter-TAD interactions segregate the genome into spatial compartments that precisely reflect the replication-timing program (Ryba et al. 2010; Yaffe et al. 2010). This interphase organization of chromatin contacts is dismantled during mitosis (Naumova et al. 2013), suggesting the hypothesis that TADs and inter-TAD contacts are re-established after each mitosis at the TDP. Here, we test this hypothesis using 4C-seq (4C) to examine the re-establishment of interphase chromatin organization and its relation to replication timing in a mouse mammary epithelial cell line (C127) in which the cell cycle regulation of replication timing has been extensively studied (Wu et al. 2006; Lu and Gilbert 2007; Lu et al. 2010; Splinter et al. 2012).
Results
Nuclear compartments are established during early G1
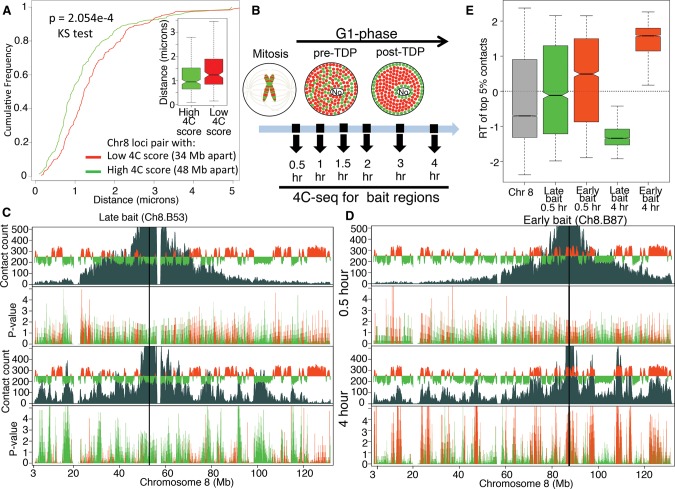
Employing replication timing profiles from 28 different mouse cell lines (replicationdomain.org), we identified 4C viewpoints (“baits”) across three mouse chromosomes (Chr 8, Chr 16, Chr 17) whose replication times are either constitutively early, constitutively late, or developmentally regulated (Supplemental Table 1; Supplemental Fig. 1). Cells were synchronized by mitotic shake-off after a brief nocodazole treatment and released into G1 phase for 0.5, 1, 1.5, 2, 3, and 4 h (Methods). Using this protocol, cells are synchronized precisely and homogeneously in metaphase and retain features of mitotic cells that are lost upon longer nocodazole blocks, including retention of Cyclin A and activation of APC ubiquitin ligase activity within minutes of release (Wu et al. 1997; Okuno et al. 2001). Within 1 h of release, the mitotic index dropped from 98% to 2%, demonstrating rapid and efficient reversibility of the block (Supplemental Fig. 2A). 4C profiles from each time point were highly reproducible across both biological and technical replicates (Supplemental Table 2; Supplemental Fig. 3). 4C profiles at 3 and 4 h were consistent with virtual 4C profiles generated using previously published Hi-C data from asynchronous mouse embryonic stem cells (ESCs) (Supplemental Fig. 3A,B) (Dixon et al. 2012). 4C contact frequencies at 3 and 4 h were also correlated with distances between locus pairs measured by 3D FISH, demonstrating correspondence to proximity in nuclei (Fig. 1A; Supplemental Fig. 4).
Figure 1.
Establishment of interphase chromatin compartments during early G1. (A) Validation of 4C by 3D FISH. Cumulative frequency curves and box plots of distances (in microns) between FISH signals from a pair of regions with low 4C contact count (red, n = 51) or high 4C contact count (green, n = 57). (B) Schematic diagram of spatial distribution of early (red) and late (green) RDs within the nucleus before and after the establishment of a replication-timing program at the TDP in mouse C127 cells. Time points chosen for 4C after release from mitosis are indicated with black arrows. (No) Nucleolus. (C,D) “Contact count”: smoothed 4C data (dark gray) overlaid with the replication timing (RT) profile (red/green is early/late replicating) for a late replicating bait Ch8.B53 (C) and an early replicating bait Ch8.B87 (D). Both baits were analyzed at 0.5 h (top) and 4 h (bottom) after mitosis. “P-value”: negative log10 of contact P-values (Methods) colored in red or green for early or late replicating regions, respectively. Between 0.5 and 4 h after mitosis, contacts between bait and other intra-chromosomal regions become more significant and focused to those with similar RT as the bait. (E) Distribution of replication timing (RT) values for the strongest (top 5% highest z-scores) contacts with either the early replicating (Ch8.B87, red) or the late replicating (Ch8.B53, green) bait. Positive values of RT indicate early replication, negative values of RT indicate late replication, and zero indicates middle replication. The higher the magnitude the earlier or later the region replicates. For reference, the RT distribution of the whole Chr 8 is shown in gray. Contacts at 0.5 h did not have statistical significance, whereas 4 h had significant contacts to regions with the same RT as the bait (Supplemental Fig. 7A).
4C data collected from cells in mitosis or at 0.5 h after mitosis showed a gradual drop-off of contact frequency with distance from the bait, which resembled the pattern seen in Hi-C data collected from human cells blocked in mitosis (Supplemental Fig. 5; Naumova et al. 2013). In contrast, by 4 h the 4C profiles had drastically changed, with contact frequencies dropping precipitously at sites aligned with the RD boundaries flanking the bait location (Fig. 1C,D). For both early and late replicating baits, the strongest contacts (top 5% highest z-scores) changed dramatically from being random with respect to replication timing to being clearly compartmentalized. Contacts with regions that replicated at times similar to the bait became strongly enriched, while regions that replicated at different times from the bait became strongly depleted (Fig. 1C–E).
TADs and inter-domain contacts are established coincident with the TDP
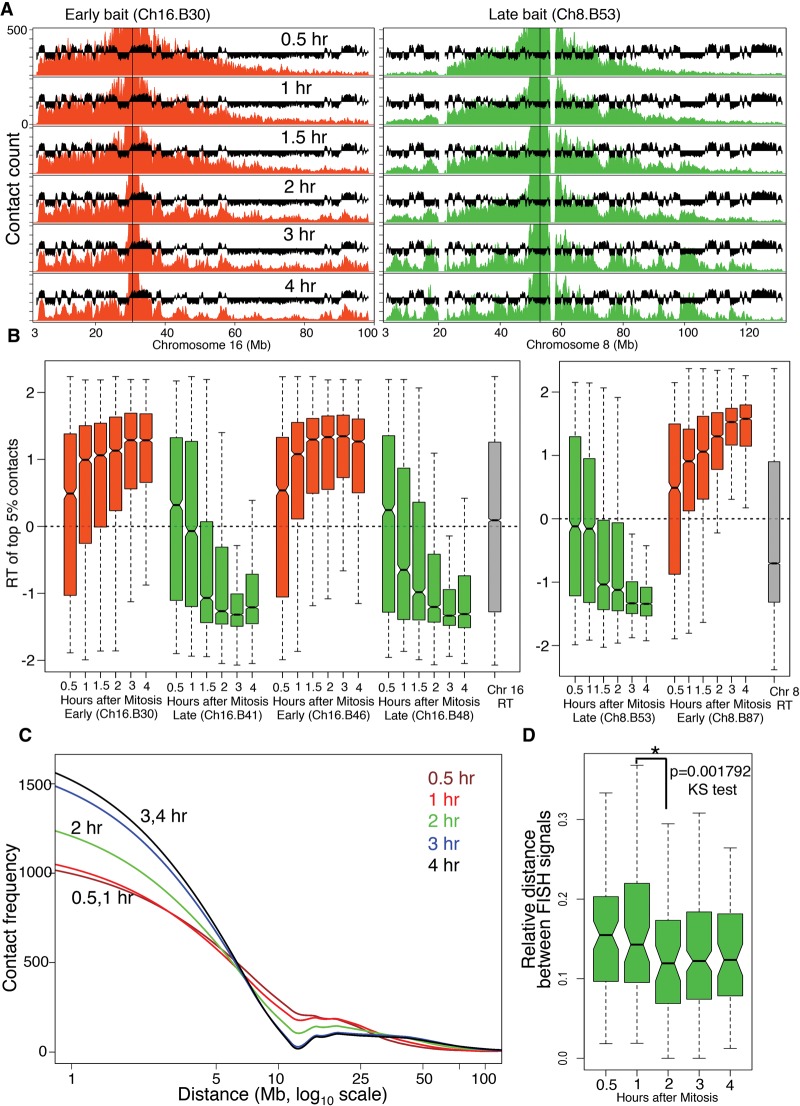
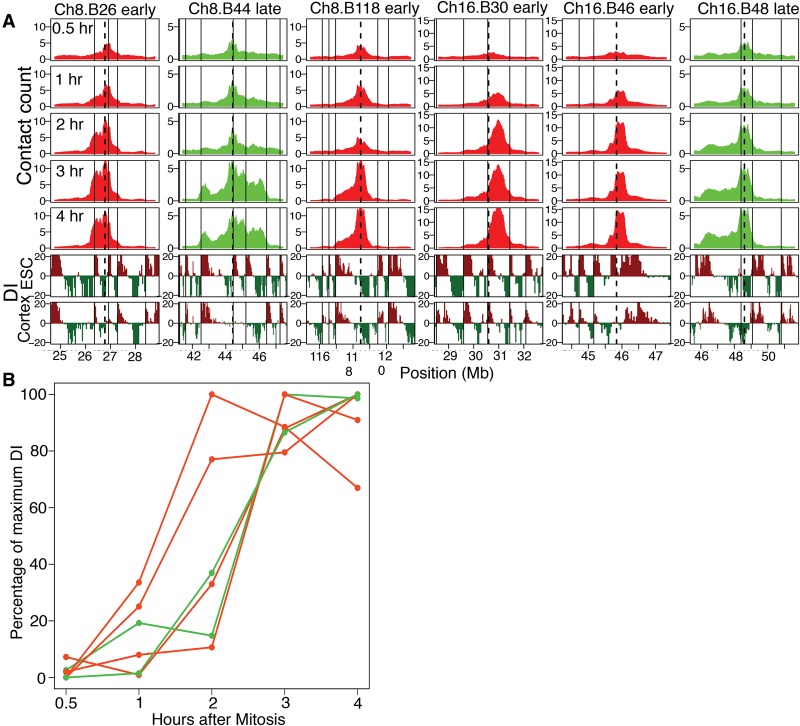
The TDP in C127 cells occurs 2–3 h after mitosis (Fig. 1B; Wu et al. 2006; Lu et al. 2010). To find the precise time point at which chromatin acquires interphase conformation, we compared 4C profiles between 12 baits positioned within three chromosomes across several G1-phase time points (Fig. 1B). The results revealed that the interphase organization of the chosen RDs is established coincident with the TDP (Fig. 2). Four lines of evidence support this folding behavior of the representative regions, independent of their replication timing. First, the temporal progression of spatial compartmentalization for domains harboring each bait was visually obvious without any computational analysis as an increase in the contact frequency with regions of similar replication timing as the bait. Compartmentalization of both early and late RDs was detectable as early as 1 h and reached completion by 3 h after mitosis (Fig. 2A; Supplemental Fig. 6A). This folding behavior is further revealed by quantification of spatial compartmentalization, determined by plotting the replication timing of the strongest contacts (top 5% highest z-scores) at each time point after mitosis (Fig. 2B; Supplemental Fig. 6B). Alternatively, applying a more rigorous test for quantifying significant contacts (Methods), which also corrects for multiple testing, revealed that significant contacts did not appear until the TDP (Supplemental Fig. 7A). As expected, inter-chromosomal contacts were rare and difficult to quantify. However, for those baits that had sufficient inter-chromosomal contact signal strength, significant contacts (Methods) also became compartmentalized after the TDP (Supplemental Fig. 7B). Second, the decay in contact frequency with increasing genomic distance from the bait for each time point showed significant differences between pre- and post-TDP profiles (Fig. 2C), reminiscent of Hi-C data collected during mitosis vs. interphase (Naumova et al. 2013). The differences in decay of contact frequency showed a transition from a mitosis-like organization observed at 30 min and 1h to an interphase-like configuration observed by 3 and 4 h (Fig. 2C). Third, we measured the 3D distance between two loci with similar replication timing on Chr 8 using fluorescent in-situ hybridization (FISH) at different time points after release from mitosis. Since FISH doesn't require a large number of cells, these experiments were performed in the complete absence of nocodazole. The results show that the relative 3D distance between the two loci decreases with time after release from mitosis and stabilizes by 2 h post-mitosis, consistent with the 4C data (Fig. 2D). Fourth, we examined the formation of TAD structures as measured by the appearance of periodic transitions in upstream or downstream contact bias (directionality bias) that disappear during mitosis (Dixon et al. 2012; Naumova et al. 2013). The kinetics of TAD formation during early G1 phase could be visualized by the appearance of a sharp drop in contact frequency at replication domain/TAD boundaries and also by the appearance of an asymmetric distribution of contact frequency to either side of the bait (directionality bias) (Fig. 3A). We chose six bait regions with visually identifiable directionality bias and quantified their directionality biases at each G1-phase time point (Methods), revealing a switch from negligible to strong directionality bias that suggests formation of TADs (Fig. 3B).
Figure 2.
Interphase chromatin compartments are established coincident with TDP. (A) Representative 4C contact count plots for baits positioned in early replicating (red) and late replicating (green) domains for the entire chromosome. 4C profiles are overlaid with C127 replication timing data (black). (B) Distribution of replication timing values for the strongest (top 5% highest z-scores) contacts with early replicating (red) and late replicating (green) baits across Chr 8 and Chr 16. For reference, the RT distributions of the whole chromosomes 8 and 16 are shown in gray. Contacts before the TDP did not have statistical significance, whereas significant contacts begin to appear coincident with the TDP and become more compartmentalized after the TDP (Supplemental Fig. 7A). (C) Loess (local polynomial regression fitting) fitted decay of 4C contact frequency with distance from the bait (decay curve) averaged for all baits from Chr 8 and Chr 16. (D) FISH-based measurement of 3D proximity normalized to the size of the nucleus (relative distance) (Methods) for two late loci (Supplemental Fig. 4) at different points after release from mitosis (nocodazole-free synchrony). There is a significant increase in 3D proximity (P < 0.001792) between before 1 h and after 2 h, which is coincident with the TDP. N = 91, 82, 57, 184, 67 for 30 min, 1, 2, 3 and 4 h, respectively. Mitosis is not included in the time course due to the inability to normalize to the state of condensation of mitotic chromatin (see Supplemental Methods).
Figure 3.
TADs are formed coincident with the TDP. (A) 4C contact counts shown for 1 to 3 Mb to either side of early/late baits. Below the contact count plots is the directionality index (DI) (Dixon et al. 2012) calculated from Hi-C in mouse ESC and cortex cell lines, shown to indicate the downstream (dark red) or upstream (dark green) bias. The vertical solid black line indicates TAD boundaries in mouse ESCs, and the dotted black line indicates the position of the bait. The kinetics of TAD formation can be visualized by the appearance of a sharp drop in contact frequency at RD/TAD boundaries. (B) Formation of TADs measured as an increase in directionality bias of the contacts with time, for baits positioned at constitutive early (red) and late (green) RDs (Ch8.B26, Ch8.B44, Ch8.B118, Ch16.B30, Ch16.B46, Ch16.B48). The y-axis shows the percentage of DI relative to the maximum DI for each bait.
Taken together, these data demonstrate that replication domains/TADs have distinct contact partners and establish both domain boundaries and inter-domain contacts within the same brief early G1 phase time frame as the establishment of a replication-timing program.
Post-TDP chromatin organization is preserved in G0 and G2
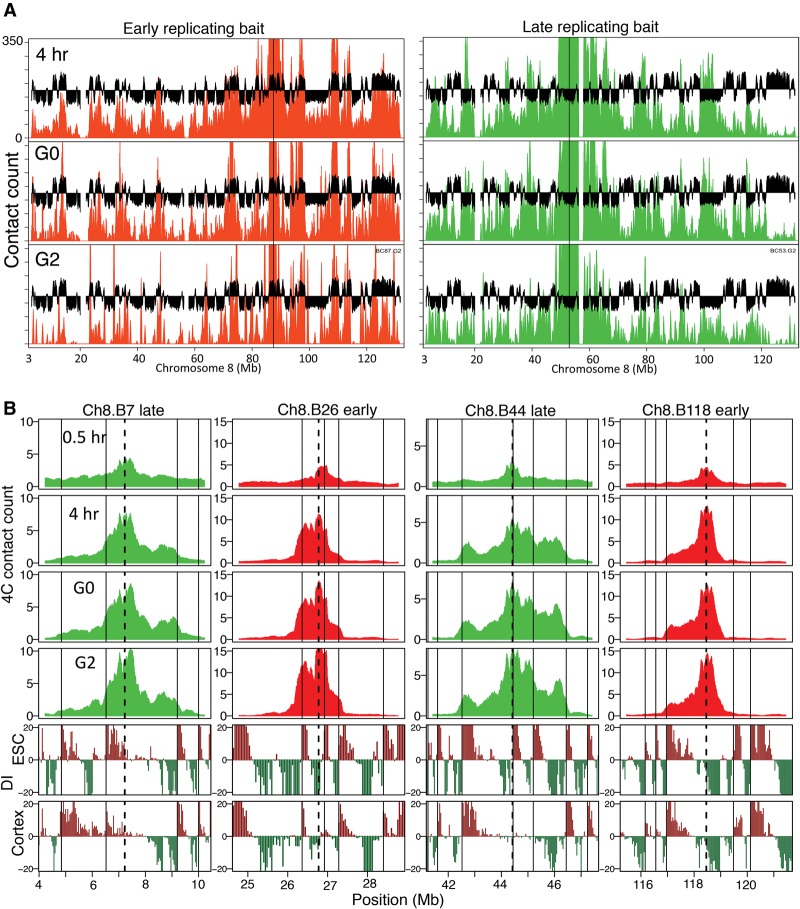
To further probe the relationship between chromatin contacts and replication timing, we examined quiescent and G2-phase cells, whose chromatin retains or has lost replication-timing determinants, respectively (Bridger et al. 2000; Lu et al. 2010). Cytogenetic studies in fibroblasts have demonstrated that chromatin undergoes dramatic organizational changes during entry into quiescence, including repositioning of chromosomes, distortion of heterochromatin organization, and changes in distribution of replication foci, while replication timing is retained (Bridger et al. 2000; Lu et al. 2010). In contrast, a recent 4C study found that chromatin contacts in resting human lymphoblasts were identical to those during interphase (Moindrot et al. 2012), suggesting that either lymphocytes do not undergo the cytogenetically visible organizational changes seen in fibroblasts during quiescence or that these dramatic organizational changes can occur while retaining interphase contacts. To distinguish between these two possibilities, we performed 4C in serum-starved C127 cells (Supplemental Fig. 2) using a previously published protocol where quiescence-associated chromosome re-organization has been observed (Lu et al. 2010). 4C profiles in quiescence were very similar to post-TDP cells and exhibited TAD organization and similar compartmentalization patterns predicted by the replication time of the bait region (Fig. 4A,B; Supplemental Fig. 8). These results demonstrate that interphase chromatin contacts and the replication-timing program are established together and preserved together even under conditions that lead to widespread cytogenetically detectable chromatin spatial re-organization.
Figure 4.
Chromatin organization in G0 (quiescence) and G2 are similar to post-TDP. (A) 4C contact counts for an early (red) and late (green) replicating region, displayed as in Figure 2A, comparing 4-h interphase time point to G0 and G2. (B) 4C contact counts shown for 1 to 3 Mb to either side of early/late baits shows conserved TADs and directionality bias (as in Fig. 3A) during G0 and G2 phase, similar to the 4-h time point.
In contrast to quiescence, G2 phase is a period where RT is lost but the interphase spatial organization of chromatin as measured cytogenetically remains unchanged (Lu et al. 2010). The complete lack of temporal specificity to replication when induced to initiate within G2 phase nuclei, similar to what is seen in the pre-TDP stage of G1 phase, led us to hypothesize that chromatin organization within G2 nuclei may have returned to the pre-TDP configuration. We prepared G2 phase C127 cells by releasing from mitosis for 15 h and including nocodazole during the last 3 h to block fast cycling cells from entering the next G1, followed by eliminating mitotic cells by shake-off prior to collection (Supplemental Fig. 2). Unexpectedly, the G2 4C profiles exhibited TAD organization and compartmentalization patterns similar to post-TDP (Fig. 4A,B; Supplemental Fig. 8). Hence, while interphase chromatin contacts in early G1 may be necessary to establish replication timing, those chromatin contacts are not sufficient to dictate the program within G2 phase nuclei.
Developmental domains exhibit decreased compartmentalization and distinct chromatin properties
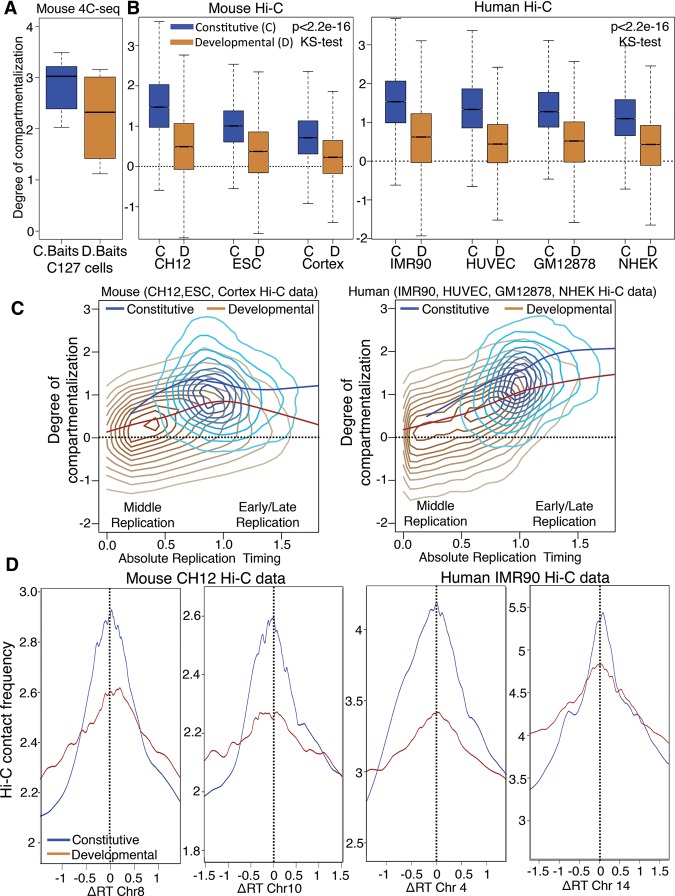
We observed that, even after achieving their interphase configuration, certain baits were less spatially compartmentalized than others (Supplemental Fig. 6A). Since only one out of seven constitutive baits vs. two out of five developmental baits were poorly compartmentalized (Supplemental Fig. 6A), we investigated whether structural plasticity (i.e., poor compartmentalization) might be related to developmental plasticity (tendency to change replication timing during development). Indeed, quantifying compartmentalization as the log ratio between contacts to the same compartment vs. contacts to the opposite compartment for each of our baits (degree of compartmentalization) (Fig. 5A; Methods) suggested that developmental domains were less compartmentalized. Applying this same definition to normalized Hi-C contact counts from three mouse cell lines and four human cell lines at 50-kb resolution (Dixon et al. 2012; Imakaev et al. 2012; Rao et al. 2014) validated, genome-wide, that developmental domains were indeed significantly less compartmentalized than constitutive domains (Fig. 5B). Although developmental domains can replicate as early or late as constitutive domains, they generally switch replication timing to or from mid-S phase (Hiratani et al. 2008, 2010). However, their reduced compartmentalization was not a consequence of frequent middle S phase replication time, as the degree of compartmentalization of developmental domains (brown line) was always lower than constitutive domains (blue line) for any given replication timing value (Fig. 5C). Finally, we compared the contact strength and the difference in replication timing value (ΔRT) for pairs of interacting loci using Hi-C data, revealing that constitutive domains have significantly stronger contacts to domains with similar replication timing (ΔRT = 0) than developmental domains (Fig. 5D).
Figure 5.
Developmental domains are less compartmentalized than constitutive domains. (A) Degree of compartmentalization (Methods) measured for constitutive baits (C baits), in blue, vs. developmental baits (D baits), in brown. The difference in degree of compartmentalization between C baits and D baits did not have statistical significance (P = 0.7374, KS test); therefore, we performed genome-wide comparison using Hi-C. (B) Degree of compartmentalization for constitutive (C) regions vs. developmental domains (D) (50-kb windows) across three mouse cells types and four human cell types using Hi-C data. The difference in degree of compartmentalization between constitutive domains and developmental domains showed high statistical significance (P < 2.2 × 10−16, KS test). (C) Density contour plot (lighter colors indicate less density) with local regression fitting (loess) for degree of compartmentalization vs. absolute replication timing value for developmental (brown) vs. constitutive (blue) regions. (D) Plot comparing frequency of Hi-C contacts vs. difference in replication timing value for pairs of 50-kb windows using Hi-C data. The top 50 percent of contacts from the Hi-C data was used for the analysis. Blue and brown lines show local regression fitting of the data for constitutive and developmental regions.
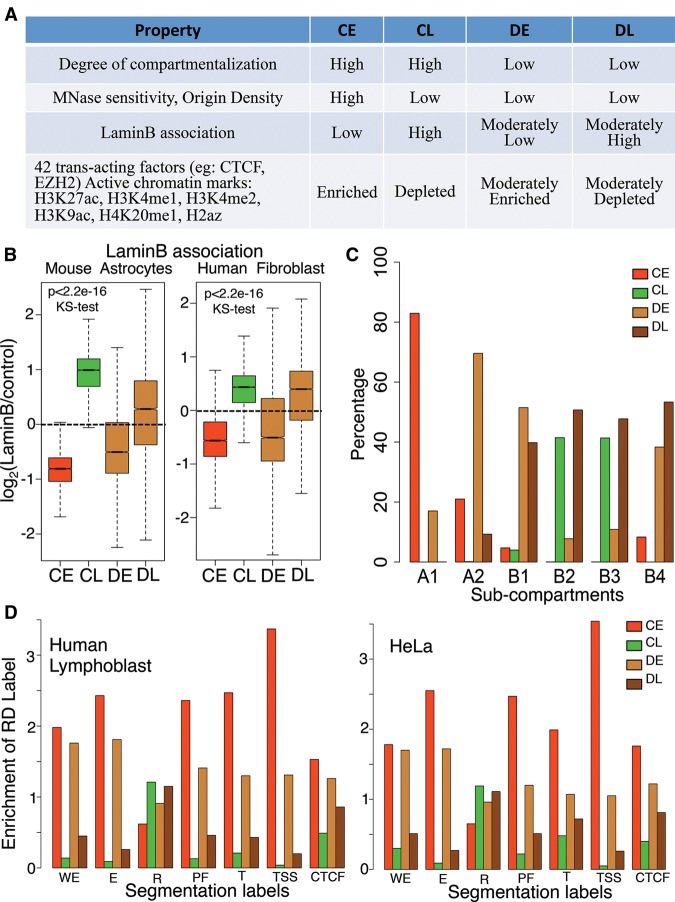
These results suggest that developmental RDs have fundamentally different chromatin organization principles. Indeed, we recently reported that developmental RDs display low nuclease sensitivity when either early or late replicating (Supplemental Fig. 9B), despite a global correlation of nuclease sensitivity to early replication (Takebayashi et al. 2012). To systematically analyze the differences between constitutive domains and developmental domains, constitutive RDs were further subclassified as constitutive early (CE) or constitutive late (CL) and similarly, for each given cell type, developmental RDs were subclassified as developmental early (DE) or developmental late (DL). We next compared several properties of chromatin, exploiting data from this report as well as other published data sets (Fig. 6A; Supplemental Table 3; Supplemental Fig. 9; Guelen et al. 2008; Peric-Hupkes et al. 2010; Bernstein et al. 2012; Besnard et al. 2012; Dixon et al. 2012; Takebayashi et al. 2012). Enrichment of various chromatin features was correlated to replication timing for constitutive domains, while developmental domains showed moderate enrichment that was less correlated to replication timing. For example, association with the nuclear lamina, a transcriptionally repressive, late replicating, nuclear compartment (Burke and Stewart 2013), was less constrained by replication timing for developmental vs. constitutive RDs (Fig. 6B). Reduced compartmentalization of developmental RDs may be related to an intermediate affinity of these domains for the lamina.
Figure 6.
Developmentally regulated regions have different organizational principles. (A) Table comparing chromatin features between constitutive early and late (CE, CL) and developmental RDs that are either early (DE) or late (DL) in the particular cell type analyzed for each respective chromatin feature (Supplemental Table 3). (B) Lamin B association for constitutive early/late and developmental early/late RDs. (C) Distribution of constitutive early/late and developmental early/late RDs within the six subcompartments defined by Hi-C data in GM12878 (Rao et al. 2014). (D) Segway/ChromHMM analysis of human lymphoblast (GM12878) and HeLa cells showing the enrichment of seven chromatin labels within constitutive early/late and developmental early/late RDs. (WE) Weak enhancer, (E) predicted enhancer, (R) predicted repressed regions, (PF) promoter flanking region, (T) predicted transcribed regions, (TSS) predicted promoter region including TSS, (CTCF) CTCF enriched element. Note that in all cases, the properties of developmental domains are less distinct and less dependent on their replication time than constitutive domains.
Recently, high-resolution Hi-C from human GM12878 was used to classify the genome into six subcompartments (two active [A1, A2] and four inactive [B1 to B4]) based on long-range chromatin interactions. Our classification of the genome into four categories by developmental regulation of replication parsed out predictably into these six subcompartments (Fig. 6C; Rao et al. 2014). Specifically A1, which replicates the earliest, comprised mostly constitutive early RDs, whereas A2, that has both active and repressive marks (H3K9me3), comprised mostly developmental early RDs. Subcompartment B1, which correlated with facultative heterochromatin, was almost exclusively composed of developmental early and late domains, whereas B2 and B3, which comprised mainly heterochromatin, were devoid of analyzed chromatin marks, and were enriched for constitutive late and developmental late RDs (Fig. 6C). B4, which consists of a small percentage of the genome and is heavily enriched for KRAB-ZNF genes, comprised almost exclusively developmental domains.
We next investigated a set of seven chromatin labels defined in human lymphoblasts and HeLa cells, via a combination of Segway and ChromHMM (Methods; Hoffman et al. 2013). All but one of these labels was more correlated to replication timing for constitutive vs. developmental domains (Fig. 6D). The one label that did not correlate was repressive marks, which have individually been shown to have a poor correlation to replication timing (Ryba et al. 2010). Furthermore, a recent segmentation method identified domains of cell type-specific vs. constitutive gene expression that were enriched for developmental vs. constitutive RDs, respectively (Libbrecht et al. 2015)¸ Finally, we individually examined 44 trans-acting factor binding sites and 11 chromatin marks. Consistent with the chromatin segmentation analysis, we observed intermediate enrichments and depletions for developmental RDs in 42 trans-acting factors and six chromatin marks (Fig. 6A; Supplemental Fig. 8). The remaining trans-acting factors and chromatin marks displayed either no enrichment (notably, the heterochromatin marks H3K9me3 and H3K27me3) or other patterns of enrichment between the domains (Supplemental Fig. 8). Together, these results reveal that developmental RDs are distinct from constitutive RDs both in the level of enrichment of various chromatin properties and in the correlation of those properties with replication timing, which may underlie their compartmentalization plasticity.
Discussion
In this study, we used 4C-seq at different time points after mitosis to study the re-organization of interphase chromatin, categorized with respect to temporal and developmental regulation of DNA replication. Our results demonstrate that TADs, as well as the inter-TAD contacts that create distinct subnuclear chromatin compartments, are formed coincident with the establishment of a replication-timing program. This organization is preserved when cells withdraw from the cell cycle into quiescence, a state in which replication-timing determinants are preserved despite cytogenetically visible re-organization of chromatin. These results provide compelling evidence supporting the replication domain model for replication-timing regulation and suggest that the 3D organization of chromatin is necessary to establish and maintain a temporal order of replication. However, we found that TADs and inter-TAD contacts are maintained during G2 phase when the determinants of replication timing are lost (Lu et al. 2010), demonstrating that 3D organization is not sufficient to maintain replication timing. Additional factors essential for replication timing, such as RIF1 (Cornacchia et al. 2012; Hayano et al. 2012), may become absent or relocalized during G2 phase. The formation of TADs and their segregation into distinct nuclear compartments may function as a scaffold to seed the assembly of compartments that are enriched or depleted for such factors (Gilbert 2001).
We also show that developmental and constitutive replication domains have distinct organization principles. In contrast to constitutive regions that interact almost exclusively with chromatin of similar replication time, the contacts of developmental regions tend to be less tightly constrained to chromatin of similar replication timing, suggesting a link between developmental and structural plasticity. Our results cannot distinguish whether developmental regions are more mobile, transiently visiting different compartments in the same cell, or are equally anchored as constitutive domains but reside in different compartments in different cells. Moreover, the enrichment of various chromatin features was considerably more correlated to replication time for constitutive vs. developmental regions. The lower degree of compartmentalization and intermediate enrichment of transcription factors and histone marks in developmental domains may underlie their ability to switch between active and inactive epigenetic states more easily during the course of development. Indeed, a comparison of constitutive and developmental domains between two cell types in Drosophila revealed similar findings (Lubelsky et al. 2014), and other studies have reported a similarity in nuclease sensitivity and origin density between developmental domains and constitutively late domains, regardless of their replication time (Besnard et al. 2012; Takebayashi et al. 2012).
In summary, our results show that 3D chromatin organization is established during early G1 phase coincident with the establishment of a replication-timing program at the TDP. 3D organization is not sufficient to maintain the replication-timing program in G2 phase, suggesting that it may create a scaffold to seed the assembly of essential, cell cycle-regulated factors. We also show that domains whose replication timing is regulated during development, which represent ∼50% of the genome (Hansen et al. 2010; Hiratani et al. 2010), comprise a novel subnuclear compartment defined by promiscuous nuclear compartmentalization and a chromatin composition that is poorly correlated to replication time. The evolutionary conservation of replication timing and its developmental regulation in all studied metazoans (Ryba et al. 2010; Yue et al. 2014) indicates that the novel properties of developmentally regulated RDs described here are a fundamental aspect of genome organization.
Methods
Cell culture and synchronization
C127 cells (ATCC) were cultured and synchronized in mitosis as previously described (Wu et al. 1997; Lu et al. 2010). Quiescent cells were prepared by serum starvation for 5 d as previously described (Lu et al. 2010). Cells were synchronized in G2 by releasing from mitosis for 15 h and including nocodazole during the last 3 h to block fast cycling cells from entering the next G1, followed by eliminating mitotic cells by shake-off prior to collection (Lu et al. 2010).
3D FISH
FISH experiments were performed as previously described (Takebayashi et al. 2012) with modifications described in Supplemental Methods. For time-course experiments, a significant increase in the radius of the nuclei was observed during the first few hours of G1; therefore, the distance between probes was normalized to the radius of the nucleus, to give “relative distance.”
Multiplex 4C-seq
4C-seq was performed and 4C reads were processed for statistical significance assignment as previously described (Splinter et al. 2012), with modifications described in Supplemental Methods. Directionality index (DI) was calculated as previously described (Dixon et al. 2012), with modifications described in Supplemental Methods.
Classification of human and mouse genomes into constitutive/developmental early and late regions
Twenty-eight mouse replication timing data sets and 21 human published replication timing data sets (replicationdomian.org) were used for the annotation of the human and mouse genome into constitutive/developmental early and late regions. The data sets were windowed into 40- or 50-kb bins. Then, the following criteria were used for the annotation of both human and mouse data sets. Windows with replication timing values >0.5 in all data sets were labeled constitutive early, and below −0.2 in all data sets were labeled constitutive late. Certain large late domains had replication-timing values higher than −0.5 in a few data sets. To prevent the removal of these large late domains from the CL category just because of these outliers, we set the threshold of CL lower than CE. Windows where the magnitude of replication-timing value is more than 0.5 in all data sets but with a sign change (positive to negative or vice versa) in at least one data set were labeled developmental. Developmental segments were further classified into developmental early or developmental late for a given cell line depending on whether the segment replicated early (RT > 0) or late (RT < 0) in that cell line.
Data access
The 4C-seq data generated for this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE66579. Replication-timing labels (constitutive early [CE], constitutive late [CL], or developmental [D]) for both human (hg19) and mouse (mm9) genomes are available as Supplemental Files (Supp-RT-human-50kb-bins.xlsx, Supp-RT-mouse-40kb-bins.xlsx).
Supplementary Material
Acknowledgments
We thank B. Robinson, J. Zimmerman, and C.T. Garcia for help with 3D FISH; B. Moindrot, M. Denholtz, and W. de Laat for advice on 4C; and M. Libbrecht and A. Kundaje for help with navigating ENCODE data. This research was supported by NIH Grants GM083337 and GM085354 to D.M.G. and HG00700 and AI106775 to W.S.N. V.D. is supported by the American Heart Association under Award Number 13PRE17060020.
Author contributions: V.D. and D.M.G. devised experiments; V.D. and J.S. performed the experiments; V.D., F.A., J.S., and D.L.V. analyzed data; V.D., F.A., J.S., W.S.N., and D.M.G. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.183699.114.
References
- Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert J-P, Noble WS, Le Roch KG. 2014. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res 24: 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, Lemaitre J-M. 2012. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19: 837–844. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol 10: 149–152. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. 2013. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol 14: 13–24. [DOI] [PubMed] [Google Scholar]
- Cornacchia D, Dileep V, Quivy J-P, Foti R, Tili F, Santarella-Mellwig R, Antony C, Almouzni G, Gilbert DM, Buonomo SBC. 2012. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31: 3678–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. 2013. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13: 602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. 2001. Nuclear position leaves its mark on replication timing. J Cell Biol 152: F11–F15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza M, Talhout W, Eussen B, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton KS, Dhar V, Brown EH, Iqbal MA, Stuart S, Didamo VT, Schildkraut CL. 1988. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol 8: 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H. 2012. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, Lyou Y, Townes TM, Schübeler D, Gilbert DM. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, et al. 2013. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 41: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord R, Naumova N, Goloborodko A, Lajoie B, Dekker J, Mirny L. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht MW, Ay F, Hoffman MM, Gilbert DM, Bilmes JA, Noble WS. 2015. Joint annotation of chromatin state and chromatin conformation reveals relationships among domain types and identifies domains of cell type-specific expression. Genome Res 25: 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum N, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie B, Sabo P, Dorschner M, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Gilbert DM. 2007. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 179: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li F, Murphy CS, Davidson MW, Gilbert DM. 2010. G2 phase chromatin lacks determinants of replication timing. J Cell Biol 189: 967–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. 2014. DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A. 2012. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res 40: 9470–9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E, Lajoie B, Schulz E, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum N, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J 20: 4263–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman S, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven R, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. 2013. DNA replication timing. Cold Spring Harb Perspect Med 3: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Gilbert DM. 2014. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev 25: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, van de Werken HJG, Klous P, de Laat W. 2012. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods 58: 221–230. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Dileep V, Ryba T, Dennis JH, Gilbert DM. 2012. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc Natl Acad Sci 109: 12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol 14: 166–172. [DOI] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Yu G, Gilbert DM. 1997. Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods 13: 313–324. [DOI] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM. 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174: 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. 2010. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho Y-J, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148: 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.