Abstract
Natural products produced by microorganisms and plants are a major resource of antibacterial and anticancer drugs as well as industrially useful compounds. However, the native producers often suffer from low productivity and titers. Here we summarize the recent applications of heterologous biosynthesis for the production of several important classes of natural products such as terpenoids, flavonoids, alkaloids, and polyketides. In addition, we will discuss the new tools and strategies at multi-scale levels including gene, pathway, genome and community levels for highly efficient heterologous biosynthesis of natural products.
Keywords: synthetic biology, chassis, heterologous biosynthesis, natural products, terpenoids, alkaloids, flavonoids, polyketides
1. Introduction
Molecules from natural sources, especially bacteria, fungi and plants, have proven to be a large reservoir for new pharmaceuticals, therapeutic agents and industrially useful compounds1. To date, natural products and their derivatives command a substantial market share in pharmaceutical industry in the past 30 years, comprising 61% of anticancer compounds and 49% of anti-infectives2. For example, erythromycin from Saccharopolyspora erythraea is an antibiotic, which has an antimicrobial spectrum similar to that of penicillin and can be used to treat various diseases3. Lovastatin (Merck's Mevacor) isolated from Aspergillus terreus is often used to reduce risk of cardiovascular disease, while paclitaxel extracted from the bark of the Pacific yew, Taxus brevifolia, is a mitotic inhibitor used in cancer chemotherapy. There are also important natural products discovered from other diverse sources, such as insects and marine sources. In the old days before 1940s, natural products of interest are isolated directly from plant sources (Figure 1a). However, bioactive compounds from natural sources are usually secondary metabolites of their native producers, thus in some cases, they exist at a relatively low level comparing with the production of primary metabolites. Therefore, in most cases, direct isolation and extraction is not an environmental friendly, sustainable and economical solution. Combinatorial chemistry was also introduced to synthesize and screen small molecules of medicinal importance (Figure 1b). Still, due to the structure complexity of natural products, chemical synthesis of these compounds is often impractical.
Figure 1.
Routes for heterologous biosynthesis of natural products. (A) Synthesis of natural products by traditional isolation from plants. (B) Synthesis and screening of small molecules by combinatorial chemistry. (C) Production of natural products by heterologous biosynthesis in microbes.
To solve these problems, researchers have engineered and optimized natural products biosynthetic pathways for decades. As genetic manipulation is either difficult or yet-to-be established for the majority of organisms, heterologous expression of a single gene, a cassette of genes, or an entire biosynthetic gene cluster in a genetically tractable host is a practical alternative route for identifying and engineering the corresponding natural product. In recent years, with the third revolution in biotechnology, which requires the convergence of life science, physical science and engineering disciplines, new solutions have been proposed for drug discovery, energy and food crisis4. The first revolution of biology was evidenced by the discovery of the double helix structure of DNA by James Watson and Francis Crick, while the second revolution began with the human genome project. Now, with the dawn of the post-genomics era came the third revolutionary change, which shifts the paradigm of the biology field from the understanding and engineering of microbial biosynthetic pathways towards the design and creation of novel metabolic circuits and biological systems. With the newly developed biosystems, the yield of target natural products could be increased noticeably in a heterologous host. For example, Keasling and his colleagues successfully redesigned the biosynthesis process of the artemisinin precursor, artemisinic acid, in heterologous hosts Escherichia coli and Saccharomyces cerevisiae5-7, which produced the artemisinic acid with a much higher productivity, titer and yield and now the process has been transferred to a pharmaceutical company for commercialization. In addition, with the versatility of synthetic biology approaches, new compounds could be discovered or generated by pathway refactoring or advanced combinatorial biosynthesis (Figure 1c)8, 9.
Microbes, especially E. coli and yeast, have proven to be useful organisms for heterologous biosynthesis of secondary metabolites for decades. They have been engineered to balance and optimize natural product biosynthetic pathways for cost-effective production of high value compounds, as well as to discover novel compounds10. Recent advances in synthetic biology and genome engineering facilitate the development of new tools for construction, controlling and optimization of metabolic pathways in microbes for heterologous biosynthesis.
Herein, we review the recent progresses made in heterologous biosynthesis of several different classes of important natural products, as well as how the newly developed synthetic biology tools could help future heterologous biosynthesis of valuable natural products. In addition, we will highlight some of the most recent advances in heterologous biosynthesis of natural product in the past five years.
2. Heterologous Biosynthesis of Major Classes of High Value Natural Products
Natural products are a very important source for the development of medicines. Many natural products and the derivatives have been developed as antibacterial, anticancer, antifungal, antiviral and antiparasitic drugs2. Butler et al. systematically summarized the development of drugs in clinical trials from natural products and the derivatives11. In this section, we will provide a comprehensive overview of the production of different classes of high-value compounds from microbes. In addition, we will discuss the recently developed metabolic engineering and synthetic biology approaches employed to engineer microorganisms for heterologous expression of natural product pathways, especially for higher production of practically important natural products.
2.1 Terpenoids
Terpenoids, also called isoprenoids, are the largest class of plant metabolites, consisting of more than 40,000 molecules12. The most famous plant terpenoids have been used as the major drugs to treat life-threatening diseases, such as the anticancer drug paclitaxel and the antimalarial drug artemisinin13. Some other terpenoids are supplemented with daily nutritional diets, such as lycopene and linalool. Terpenoids usually consist of different numbers of basic five-carbon isopentenyl diphosphate (IPP) units with a series of assembly, cyclization and group modification reactions14. Traditional methods for producing these compounds usually rely on multistep extraction from plants or organic chemical synthesis, both of which can result in low yield, high cost and sometimes severe pollutions to the environment.
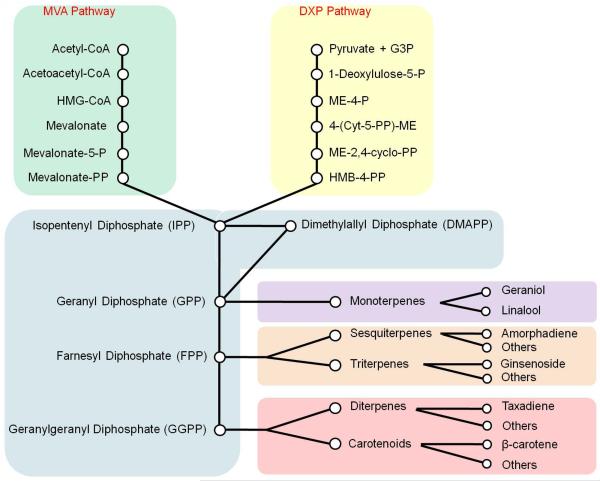
In microbes, such as E. coli and yeast, the terpenoids biosynthetic pathways typically work with very low fluxes. The important precursors for terpenoids, IPP and its isomer dimethylallyl pyrophosphate (DMAPP), are synthesized from acetyl-CoA through the DXP pathway in E. coli-like bacteria and the MVA pathway in yeast-like fungi. In yeast, some highly active IPP compounds can also be synthesized based on the basic C5 units. For example, geranyl pyrophosphate (GPP) is a C10 structure generated by condensing one molecule IPP and one molecule DMAPP. Following this reaction, an additional IPP molecule can be combined to make a C15 structure, farnesyl pyrophosphate (FPP), and further with another IPP to make a C20 structure, geranylgeranyl pyrophosphate (GGPP). In native yeast, ergosterol, as the final metabolite of the successive condensation reactions, is derived from C30 squalene which is synthesized by condensation of two FPP molecules (Figure 2). These reactions in bacteria and fungi enable the introduction of genes of plant origins to construct the heterologous terpenoids biosynthetic pathways. Monoterpenoids are synthesized from GPP; sesquiterpenoids from FPP; diterpenoids and carotenoids from GGPP; and triterpenoids from squalene.
Figure 2.
Schematic of various biosynthetic pathway of terpenoids. Two pathways, the MVA pathway and the DXP pathway, provide the precursors for the biosynthesis of terpenes, which are the backbones to synthesis of terpenoids, including monoterpenoids, sesquiterpenoids diterpenoids, triterpenoids and carotenoids.
2.1.1 Monoterpenoids
The most well-known monoterpenoids that have been heterologously synthesized in engineered yeast are linalool and geraniol. Although wild-type yeast does not synthesize these monoterpenes, the mutants with blocked FPP synthetase that excretes geraniol and linalool have been characterized previously15. However, the enzyme involved in GPP dephosphorylation has not been identified yet.
Linalool is a value-added fine chemical, and can be used to synthesize linalyl acetate, geraniol, linalyl methyl sulfide, vitamin A, vitamin K and other medical intermediates16. Linalool is synthesized via the condensation of IPP and DMAPP by the endogenous mevalonate pathway. Several cis or trans linalool synthases were identified from different origins17-19. The engineered industrial yeast strain with a linalool synthase from Clarkia breweri produced linalool with no by-products20. In addition, over-expression of the hmg1 gene in the engineered yeast strain enhanced the production of linalool to 133 +/− 25 μg/L21.
Geraniol, as a widely used floral fragrance, is a typical monoterpene, and can be further transformed into ester flavors for food flavors, fruit flavors, ginner flavors, and cinnamon flavors22. It is also used for clinical treatment of chronic bronchitis and improvement of pulmonary ventilation function22. Geraniol is usually obtained from myrcene by esterification, saponification and fractionation in current industrial production. However, geraniol can also be synthesized from linalool. The geraniol synthase (GES) was isolated from Ocimum basilicum and expressed in S. cerevisiae to produce geraniol23, 24. Furthermore, blocking of FPP synthase increased geraniol synthesis24.
2.1.2 Sesquiterpenoids
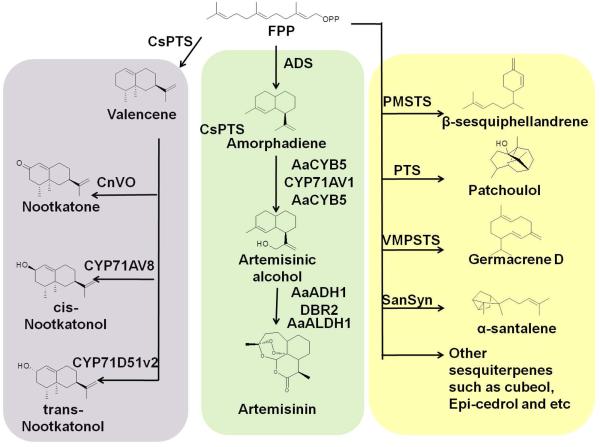
Sesquiterpenoids are the most diverse class of terpenoids, including industrially valuable products for fragrances, repellents, food colorants and high-value therapeutic molecules, such as epicedrol, artemisin, valencene, cubebol, patchoulol and santalene25-31. All these sesquiterpenoids are derived from a 15-carbon precursor, FPP. Different sesquiterpene synthases were used to catalyze the synthesis of different sesquiterpenes from FPP (Figure 3). β-sesquiphellandrene, with efficient anti-microbial and anti-oxidant activity, was synthesized by PMSTS (Persicaria minor sesquiterpene synthase)28, 29. Valencene, an odorant molecule, was synthesized by CsPTS (Citrus sinensis valencene synthase), and could be further oxidized to (+)-nootkatol, (−)-nootkatol or (−)-nootkatone by different kinds of P450 cytochrome oxidase (CYP71D51v2 from Nicotiana tabacum, CYP71AV8 from Cichorium intybus and CnVO from C. nootkatensis)31, 32. Patchoulol, the major constituent of patchouli oil, was synthesized by PTS (patchoulol synthase from Pogostemon cablin)25, 26. In addition, other valuable sesquiterpenes, including arteminisic acid, cubeol, germacrene D, β-caryophyllene, α-santalene, were recently produced in engineered microbes30, 33, 34. Importantly, the biosynthesis of artemisinin, a valuable and powerful antimalarial natural product extracted from Artemisia annua L, was identified and engineered. Firstly, FPP was converted to amorphadiene by ADS (amorphadiene synthase). Then amorphadiene was oxidized to artemisinic alcohol by CYP71AV1 (a kind of P450 cytochrome oxidase). It was then reduced by DBR2 (artemisinic aldhyde reductase) to dihydroartemisinic aldehyde and oxidized to dihydroartemisinic acid by ALDH1 (aldehyde dehydrogenase from A. annua). Finally, dihydroartemisinic acid was converted to artemisinin via an allylic hydroperoxide intermediate, and light and oxygen were required for this conversion5.
Figure 3.
Schematic of sesquiterpenoids biosynthesis. Farnesyl diphosphate (FPP), the 15-carbon precursor, is converted to different sesquiterpene by various sesquiterpenoid synthases (ADS, amorphadiene synthase; CsPTS, valencen synthase; PMSTS, Persicaria minor sesquiterpene synthase; PTS, patchoulol synthase; VMPSTS, Vanda mimi palmer sesquiterpene synthase; SanSyn, santalene synthase). Some of these sesquiterpenes can be further oxidized to their derivatives by various cytochrome P450 oxidases (CYP71AV8, CnVO, CYP71D51V2 and CYP71AV1). In the biosynthesis pathway of artemisinin, the well-known sesquiterpenoid, the artemisinic alcohol can be further converted to artemisinin or artemisinic acid by several enzymes (AaALDH1, artemisinic aldehyde dehydrogenase; ADH1, artemisinic alcohol dehydrogenase; DBR2, artemisinic aldehyde reductase; CPR1, reductase of CYP71AV1; CYB5, cytochrome b5 from A. annua).
Most of these sesquiterpenes have been successfully produced in engineered microbes such as E. coli, S. cerevisiae and other industrial microorganisms. To increase the production of these sesquiterpenes, the enzymes from plants or the flux of the whole pathway need to be modified. For example, in engineering patchoulol producing yeast, PMSTS was fused with ERG20 (FPP synthase) to optimize the local concentration and spatial organization of these two enzymes and to redirect the metabolic flux towards the product26. Combined with the repression of ERG9, the final patchoulol titer reached 23 mg/L26. In addition, higher FPP concentration and increased production of valencene was achieved by localizing the enzyme to the mitochondrion35. Another successful example of heterologous biosynthesis of sesquiterpene is the optimized biosynthesis of artemisinic acid6. The systematic optimization started with regulating the proportion of CYP71AV1 and AaCPR1, followed by overexpressing AaCYB5 (cytochrome b5 from A. annua) and native HEM1 to optimize the oxidation of amorphadiene which is the key step of the synthesis of artemisinic acid. At the same time, the native CTT1 (catalase) is also overexpressed to reduce the oxidative stress produced by CYP71AV1. AaADH1 (alcohol dehydrogenase from A. annua) and AaALDH1 are overexpressed to promote the conversion of artemisinic acid from artemisinic alcohol. The final production of artemisinic acid reached up to 25 g/L6.
2.1.3 Diterpenoids
Diterpenoids are widely distributed natural products with important physiological effects, such as paclitaxel against cancer, tanshinones against inflammation, and stevioside as sweetener13, 36, 37. However, the amount of the diterpenoids in the host plants is usually very low, let alone the slow growth rate of plants. For example, the paclitaxel content is only at 10-ppm level in the most productive species, Taxus brevifolia38. In addition, complex diterpenoids molecules are quite difficult for chemical synthesis de novo39. Therefore, heterologous expression of the pathways in microbes is very promising for diterpenoids production.
Tanshinones are the main lipophilic components of the Chinese medicinal herb danshen (Salvia miltiorrhiza). Modular pathway engineering (MOPE) was successfully developed at assemble and optimize the biosynthetic pathway of miltiradiene, the precursor to tanshinones, in S. cerevisiae40. The fusion of SmCPS and SmKSL and the fusion of GGPPS (GGPP synthase) and FPPS, together with overexpression of tHMGR led to significant improvement of miltiradiene production (365 mg/L) and reduced byproduct accumulation significantly40. Based on the increased production of miltiradiene, incorporation of genes coding for CYP76AH1 and phyto-CYP reductase genes led to heterologous production of ferruginol, the precursor to danshinones41.
Paclitaxel, as a diterpenoid, has been approved for treatment of breast, ovarian and lung cancers, as well as coronary artery disease42. Although some studies tried to figure out the biosynthetic pathway for paclitaxel in fungal endophytes and improve the paclitaxel production by fermentation43, a recent study indicated no paclitaxel pathway could be identified in fungal endophytes44. Due to the high clinical needs and high costs of production, researchers have been mining the key genes for paclitaxel biosynthesis for decades. Most of the biosynthetic reactions have been elucidated, and several important genes or enzymes have been characterized (Figure 4). Based on the elucidated partial pathway for paclitaxel production, the heterologous synthesis of paclitaxel precursors were investigated in different microbes. E. coli was firstly used as the host cell for the heterologous biosynthesis of taxadiene45. The heterologous biosynthesis of taxadiene in K-derived E. coli and B-derived E. coli indicated that the background of the chassis affected taxadiene synthesis46. A multivariate-modular approach increased titers of taxadiene to approximately 1 g/L (~15,000-fold increase) in an engineered E. coli strain47. Besides E. coli, S. cerevisiae was also used as the chassis to produce taxadiene48. Five enzymes catalyzing sequential transformations from IPP and DMAPP were heterologously expressed in S. cerevisiae, and taxadiene titer was improved to 8.7 ± 0.85 mg/L by overexpression of tHMGR and UPC2-149. Six different GGPPSs were analyzed using enzyme-substrate docking strategy, and GGPPS from Taxus baccata cuspidate showed the best performance in docking with substrate molecule FPP and led to the highest taxadiene production (72.8 mg/L) ever reported in yeast so far50.
Figure 4.
Schematic of the paclitaxel biosynthetic pathway. Paclitaxel is synthesized by the combination of the core taxane and the side chain. The core taxane structure is synthesized by the modification of the taxadiene, which is produced by the cyclization of GGPP. The solid arrows in the pink background mean the steps with known reactions and known enzymes. The solid arrows in the light blue background mean the steps with known reactions but unknown enzymes. The dotted arrows mean the steps with unknown reactions and known enzymes. FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate. TDC1, Taxadiene synthase; CYP725A2: taxane 13 alpha-hydroxylase; CYP725A1, taxane 10 beta-hydroxylase; TBT, taxoid 2a-O-benzoyl transferase; DBAT, 10-deacetylbaccatin III 10-O-acetyltransferase; PAM, phenylalanine aminomutase.
Although the paclitaxel precursors were produced with high efficiency by heterologous biosynthesis in different chassis, the heterologous biosynthesis of paclitaxel is still impossible due to the lack of information for several key biosynthetic steps. Therefore, some alternative bioconversion approaches were developed to produce paclitaxel. An Enterobacter sp. was found to convert 7-xylosyl-10-deacetyltaxol (7-XDT) to 10-deacetyltaxol (10-DT), which is the most abundant byproduct in Taxus. The highest conversion rate and yield of 10-DT from 7-XDT reached 92% and 764 mg/L, respectively51. A xylosidase from Enterobacter sp was discovered and heterologously expressed in yeast52. The engineered yeast could robustly convert 7-XDT into10-DT with over 85% conversion rate and the highest yield of 8.42 mg/mL52.
2.1.4 Triterpenoids
Triterpenoids are synthesized by condensation of six C5 IPP basic units through end-to-end coupling reactions. Triterpenoids are widely distributed in nature and can be divided into two main categories, pentacyclic triterpenoids (such as ginsenoside) and tetracyclic triterpenoids (such as amyrin-derivative terpenoids), according to the number of carbon rings contained in the compounds53.
The chemical compositions of the ginseng complex have extensive biological activities and unique pharmacological actions. Medical and pharmacological research proved that one of the main effective components of ginseng is a group of ginsenosides, which are considered to possess medical value as anti-tumor, antiviral and cholesterol-decreasing drugs and precursors. Genetic engineering of plants was tried to improve the ginsenoside production by overexpression of the key genes coding dammarane synthase and amyrin synthase. However, limited success was achieved54.
Three biosynthetic pathways for various ginsenosides were successfully constructed in S. cerevisiae55, and they are responsible for the alternative synthesis of protopanaxadiol, protopanaxatriol and oleanolic acid, the three basic aglycons of ginsenosides. The engineered S. cerevisiae was transformed with expression cassettes of β-amyrin synthase, oleanolic acid synthase, dammarenediol-II synthase, protopanaxadiol synthase, protopanaxatriol synthase and NADPH-cytochrome P450 reductase from different plants. Through additional overexpression of some endogenous genes, the final engineered strain produced 17.2 mg/L protopanaxadiol, 15.9 mg/L protopanaxatriol and 21.4 mg/L oleanolic acid.
The main tetracyclic triterpenoids successfully synthesized by engineered microbes are amyrin-derivative compounds, including α-amyrin type and β-amyrin type. Several key functional genes were introduced into yeast cells to produce various amyrin structures53, 56. The main product based on β-amyrin precursor is oleanolic acid55. Catharanthus roseus is an important medicinal plant and commercial source of monoterpenoid indole alkaloids, ursolic acid and oleanolic acid. The EST collection from C. roseus leaf epidermome was analyzed, and a cDNA (CrAS) encoding 2,3-oxidosqualenecyclase and a cDNA (CrAO) encoding amyrin C-28 oxidase were successfully discovered57. The functions of CrAS and CrAO were analyzed in S. cerevisiae. The co-expression of CrAO and CrAS in yeast led to ursolic- and oleanolic acids production. In addition, the cellsco-expressing CrAO and AtLUP1 from Arabidopsis thaliana produced betulinic acid. Another typical triterpenoid is ginsenosides compound K (CK)58. Among the several structures of ginsenosides, CK is the main functional component presenting bioactivities of anti-inflammation, hepatoprotection, antidiabetes and anti-cancer. The potential CK biosynthetic pathway was designed in yeast, including acytochrome P450 (CYP716A47) from Panax ginseng, a NADPH-cytochrome P450 reductase (ATR2-1) from Arabidopsis thaliana, and a Dammarenediol-II synthase (PgDDS) from P. ginseng, and the CK synthase was explored based on bioinformatics analysis. CK was synthesized heterologously for the first time in engineered microbes by two pathways, especially considering active CK could not be detected in original Panax plants. The final production of CK reached 1.4 mg/L.
2.1.5 Tetraterpenoids
Tetraterpenoids consist of eight isoprene units and are mainly involved in metabolic precursors and products in the biosynthesis of carotenoids with unique UV-Vis absorption spectra59. Carotenoids serve as precursors for vitamin A biosynthesis, but more importantly, they demonstrate coloring and antioxidant properties attributed to their structures. Therefore, their expanded production is appealing.
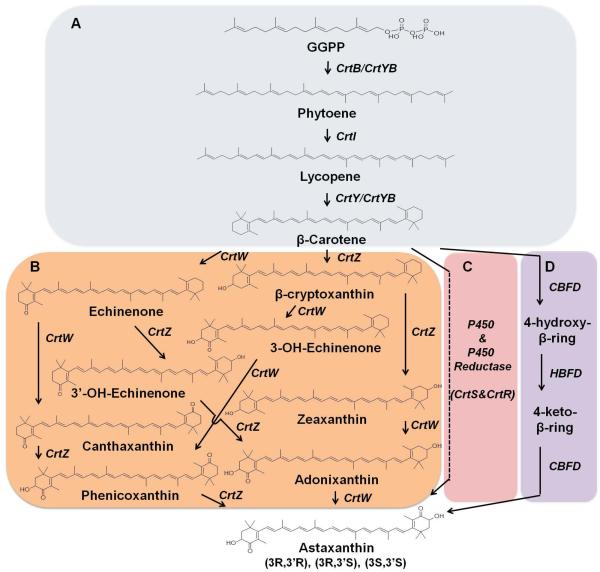
Many kinds of hosts can produce carotenoids, such as Agrobacterium aurantiacum, Blakeslea trispora, several Pantoea bacteria, Xanthophyllomyces dendrorhous and Haematococcus pluvialis. Commonly in carotenogenic bacteria and algae, biosynthesis of carotenoids starts from GGPP, which is synthesized by GGPP synthase (CrtE) either through direct condensation of 5-carbon precursor, IPP and DMAPP, or from 15-carbon precursor, FPP. A tail to tail condensation of two GGPP molecules catalysed by phytoene synthase (CrtB) leads to the formation of the primal carotenoid, phytoene, which is colorless. Then, through successive introduction of four double bonds in the phytoene skeleton by phytoene desaturase (CrtI), lycopene is synthesized. Using lycopene as substrate, lycopene β-cyclase (CrtY) catalyzes the formation of β-ionone rings at both ends to produce β-carotene. Different from some bacteria and algaes, the phytoene synthase and phytoene desaturase belong to a bifunctional enzyme in red yeast, X. dendrorhous, and other fungi. B. trispora, X. dendrorhous and H. pluvialis are well-known microorganisms capable of synthesizing astaxanthin. In both prokaryotes and eukaryotes, biosynthesis of astaxanthin is the results of ketolation at the 4 and 4’-position and hydroxylation at the 3 and 3’ positions of the β-ionone rings of β-carotene. In bacteria, ketolase (CrtW) and hydroxylase (CrtZ) are required for astaxanthin biosynthesis. However, due to their weak selectivity toward keto- and hydroxyl-substrate, quite a few intermediates accumulated when combining bacteria crtW and crtZ to produce astaxanthin60, 61. In red yeast X. dendrorhous, astaxanthin synthase (CrtS), a cytochrome P450 enzyme, in combination with its cytochrome P450 reductase (CrtR), are responsible for astaxanthin biosynthesis60. In plant Adonis aestivalis, the 4-position of β-ring of β-carotene is activated by 4-dehydrogenase-β-ring (CBFD), followed by further dehydrogenation to yield a carbonyl catalyzed by 4-hydroxy-β-ring 4-dehydrogenase (HBFD). And then, the 3-position is added by CBFD with a hydroxyl group, which leads to the synthesis of astaxanthin. This pathway was proved to work efficiently in E. coli (Figure 5)62.
Figure 5.
Schematic of the various biosynthetic pathways of carotenoids. (A) The common pathway for lycopene and β-carotene biosynthesis in bacteria and fungi. IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate; CrtE, GGPP synthase; CrtB, phytoene synthase; CrtI, phytoene desaturase; CrtY, lycopene β-cyclase; (B,C,D) The astaxanthin biosynthetic pathway in carotenogenic bacteria and algae (B), red yeast X. dendrorhous (C), and plant Adonis aestivalis (D). CrtZ, β-carotene hydroxylase; CrtW, β-carotene ketolase; CrtS, P450 cytochrome monooxygenase; CrtR, cytochrome P450 reductase; CBFD, carotenoid β-ring 4-dehydrogenase; HBFD, carotenoid 4-hydroxy-β-ring 4-dehydrogenase.
Heterologous biosynthesis of carotenoids was mostly carried out in two hosts, E. coli and baker’s yeast. In E. coli, the main products are lycopene and β-carotene. By combining the native MEP pathway and the heterologous MVA pathway, 1.35 g/L lycopene63 and 2.47 g/L β-carotene with a yield of 70 mg/g DCW64 was produced by fed-batch fermentation in E. coli. Most recently, 3.2 g/L β-carotene was achieved under flask cultivation employing a similar strategy65. With a targeted engineering strategy, a very high maximum productivity of 74.5 mg/L/h and up to 1.23 g/L of lycopene was produced in 100 L fed-batch fermentation66. ATP and NADPH recycling was engineered in E. coli to enhance terpenoids production67. Five alternative functional modules were designed for combinatorial metabolic engineering, including the heterologous synthesis module, MEP module, ATP module, PPP module, and TCA module, which led to an optimized strain capable of producing 2.1 g/L β-carotene with a yield of 60 mg/g DCW. In addition, expression tuning the expression level of the genes encoding α-ketoglutarate dehydrogenase, succinate dehydrogenase and transaldolase B increased NADPH and ATP supplies, leading to 3.52 g lycopene/L (50.6 mg/g DCW) in fed-batch fermentation68.
In yeast, carotenoids are the first class of terpenoids to be produced. In the early period, like the studies in E. coli, engineering efforts sourced carotenoid biosynthetic genes from several microorganisms, in particular the red yeast X. dendrorhous and the soil bacterium Pantoea ananatis, and achieved low-level production of target carotenoids from endogenous precursors69. Since then, researchers began to engineer yeast strains to accumulate more quantities of GGPP precursor for carotenoid production by either introducing a heterologous GGPP synthase gene crtE69, 70 or overexpressing the native gene bts160, 71. By using this combinatorial genetic engineering strategy, the expressions of several genes were optimized, leading to 5.9 mg/g DCW β-carotene and astaxanthin60, 69-71. Recently, a set of marker recyclable integrative plasmids (pMRI) was employed for decentralized assembly of the β-carotene biosynthetic pathways72. By using GAL inducible promoters, the pathway could be switched on at certain time, leading to production of 11 mg/g DCW of total carotenoids (72.57 mg/L) and 7.41 mg/g DCW of β-carotene. The engineered yeast strain exhibited high genetic stability after 20 generations of subculturing. After down-regulating the squalene synthase (ERG9), 1156 mg/L (20.79 mg/g DCW) of total carotenoids was achieved by high-cell density fermentation73. Interestingly, by exploiting the antioxidant properties of carotenoids, adaptive evolution was successfully applied to boost carotenoids production74. In another study, carotenoid was used as a phenotypic marker to screen the yeast deletion collection to reveal new genes with roles in enhancing terpenoid production75, which provides potential targets for subsequent metabolic engineering.
2.2 Flavonoids
Flavonoids are a large group of plant secondary metabolites containing at least one hydroxylated aromatic ring. Flavonoids, when consumed in human diet, could help to promote human health and prevent certain diseases, as it can prevent cancer by preventing cellular oxidation processes and stopping cell degradation and aging76. The fact that only a few of these compounds can be produced as pure products in small quantities from limited plant species makes flavonoids as attractive candidates to be synthesized in engineered microbes, such as E. coli and S. cerevisiae76-78.
S. cerevisiae is a suitable host to synthesize flavonoid compounds as its natural metabolism provides necessary amino acid precursors tyrosine and phenylalanine for downstream exogenous pathways. To date, more than 20 pathways from fungus and plant have been implanted into S. cerevisiae to produce different kinds of flavonoid products, including flavanones, flavones, isoflavones, and flavonols77, 78.
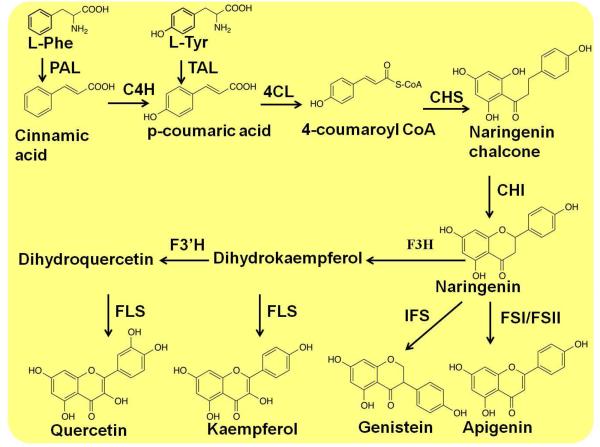
p-Coumaric acid is a common intermediate for almost all flavonoids biosynthesis. Phenylalanine ammonia lyase (PAL) first catalyzes the deamination of phenylalanine to cinnamic acid, and cytochrome P450 cinnamate-4-hydroxylase (C4H) acting with partner reductase (CPR) subsequently catalyzes the hydroxylation of cinnamic acid to p-coumaric acid. The enzyme from the fungus Rhodospridium toruloides was introduced into yeast to exhibit a tyrosine ammonia lyase (TAL) activity and thus was able to bypass C4H to produce p-coumaric acid from tyrosine79.
The synthesis of naringenin chalcone was first achieved in an engineered yeast, through co-expression of three different enzymes, including R. toruloides PAL, Arabidopsis thaliana 4-coumarate: CoA ligase (4CL), and Hypericum androsaeemum chalcone synthase (CHS)80. The following chalcone isomerase (CHI) could catalyze the cyclization of chalcones to flavanones (Figure 6)77, 81-83.
Figure 6.
Schematic of the various biosynthetic pathways of flavonoids. PAL, phenylalanine ammonia lyase; TAL, tyrosine ammonia lyase; 4CL, 4-coumarate:Co1-ligase; C4H, cinnamate-4-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; FSI, flavone synthase I; FSII, flavone synthase II; IFS, isoflavone synthase; TAL, tyrosine ammonia lyase.
Based on the biosynthesis of flavonones, the introduce of cytochrome P450 enzymes, such as isoflavone synthase (IFS), flavone synthase II (FSII) and flavone synthase I (FSI), can lead to the production of isoflavone genistein and the flavone apigenin77, 81, 82. Co-expression of flavanone 3b-hydroxylase (F3H), flavonoid 3-hydroxylase (F3’H), and flavonol synthase (FLS) achieved the synthesis of kaempferol and quercetin in yeast77. The production of some stilbenoids, such as resveratrol, has also been achieved in yeast77, 84.
2.3 Alkaloids
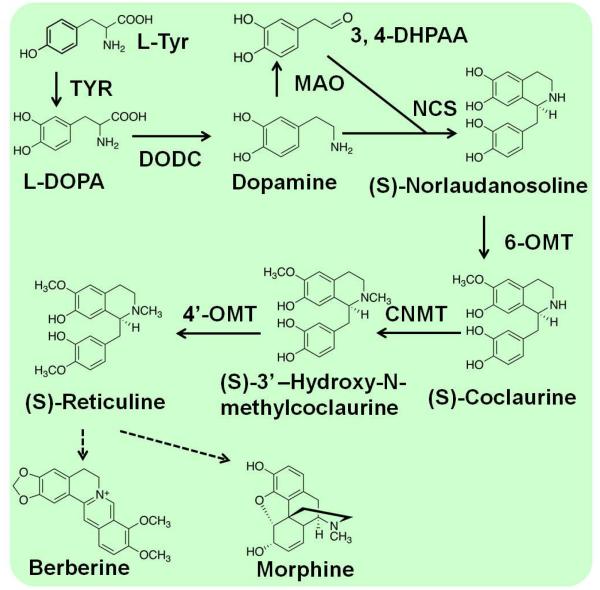
Alkaloids are a large class of nitrogen-containing compounds in plant secondary metabolites with low molecular weight2. Most alkaloids are derived from amines produced by the decarboxylation of amino acids, such as histidine, lysine, ornithine, tryptophan, and tyrosine. One of the largest groups of pharmaceutical alkaloids is benzylisoquinoline alkaloids (BIAs), including the antitussive codeine, the analgesic morphine, and the antibacterial drugs berberine. BIAs are widely used in human health and nutrition, at present they are mainly obtained by extraction from plants, which limited the diversity of BIAs structures available for drug discovery due to their low abundance. Many of the native plant hosts accumulate only a selected few BIA products. BIAs are produced through (S)-reticuline, which begins with the condensation of two L-tyrosine derivatives, 4-hydroxyphenylacetaldehyde (4HPAA) and dopamine (Figure 7). (S)-reticuline is a main branch-point metabolite in the biosynthesis of many kinds of BIAs, and it plays an impartment role in the development of novel antimalarial and anticancer drugs. A number of recent reports have successfully transplanted portions of (S)-reticuline biosynthesis pathways into microbial hosts, such as E. coli and S. cerevisiae85-89.
Figure 7.
Schematic of the various biosynthetic pathways of alkaloids. CNMT, coclaurine-N-methyltransferase; DODC, DOPA decarboxylase; MAO, monoamine; NCS, norcoclaurine; TYR, tyrosinase; 3,4-DHPAA, 3,4-dihydroxyphenylacetaldehyde; 6-OMT, norcoclaurine 6-O-methyltransferase; 4’-OMT, 3’-hydroxy-N- methylcoclaurine 4’-O-methyltransferase.
(S)-Reticuline and some other intermediates in the berberine and morphinan branches could be synthesized in yeast by overexpressing enzymes from three plant sources and humans. A glucocorticoid inducible promoter and in situ promoter titration were applied to tune enzyme expression levels and optimize the productivity85. Reticuline could also be produced from dopamine in E. coli by heterologous expression of biosynthetic enzymes (i.e., MAO, NCS, 6OMT, CNMT, and 4-OMT)86. About 7.2 mg/L magnoflorine and 8.3 mg/L scoulerine were produced from dopamine via reticuline by using different combination cultures of engineered E. coli and S. cerevisiae86. The productivity of reticuline could be improved by fine-tuning the possible rate-limiting step88. Notably, S. cerevisiae expressing CYP80G2 and CNMT converted reticuline to magnoflorine86. An alkaloid biosynthetic pathway from L-tyrosine cells was constructed in E. coli, which produced 46.0 mg/L of (S)-reticuline from glycerol87. Recently, a biosynthesis pathway for the production of dihydrosanguinarine and sanguinarine from (R,S)-norlaudanosoline was constructed with 10 genes in S. cerevisiae89. It represents the most complex plant alkaloid biosynthetic pathway ever reconstructed in yeast and proves the potential of the synthesis of ever more complex plant natural products in engineered microbes.
2.4 Polyketides
Polyketides are another diverse group of natural products with important applications in the pharmaceutical industry. They have been isolated and characterized from a variety of natural sources, including plants, fungi, and bacteria. They are of high interests as they often exhibit a broad spectrum of biological activities, which are attributed to their structural complexity and diversity. Those diverse and complex structures are produced by polyketide synthases (PKSs), which polymerize acyl-Coenzyme As (CoAs) in a fashion similar to fatty acid synthesis (Figure 8).
Figure 8.
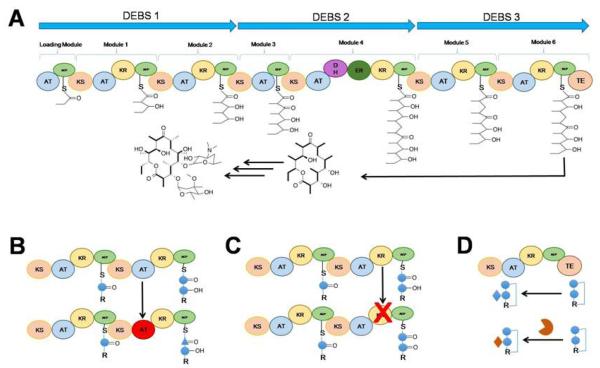
Schematic of the biosynthetic pathway of erythromycin. (A) the biosynthetic assembly line for the polyketide antibiotic erythromycin. AT: acyltrasferase; ACP: acyl carrier protein; KS: ketosynthase; KR:ketoreductase; DH:dehydratse ; ER:enoylreductase; TE: thioesterase. (B) Domain engineering; (C) Domain inactivation; (D) Engineering of tailoring reactions.
Erythromycin A is a potent polyketide antibiotic produced by the Gram-positive bacterium Saccharopolyspora erythraea90. It can be used to treat various diseases, such as respiratory infections, whooping cough, syphilis, Legionnaires’ disease, gastrointestinal infections, acne, as well as used as a substitution for penicillin. After the identification and characterization of the DEBS PKSs gene cluster responsible for biosynthesis of erythromycin, functional heterologous expression of these proteins and their mutants in S. coelicolor and E. coli were employed91, 92. This not only helped to reveal the biochemical basis for these highly controlled megasynthases, but also allowed the rational design of its biosynthetic products, and led to the generation of diverse polyketide libraries. Researchers have engineered the microbes for heterologous production of various erythromycin derivatives for further studies in the hope of improving their pharmacological properties. One notable example is the incorporation of unnatural starter units?to change the chain initiation process by replacing the loading domain AT with other AT domains, or incorporating synthetic oligoketide precursors into the downstream?DEBS pathway93-95. Besides the precursors, the extender unit can also be replaced in the erythromycin DEBS system96-98. Since the DEBS AT domains of each module are the primary gatekeepers for the incorporation of an extender unit into the polyketide chain, replacing AT domains with other ones that accept other extender units could result in the incorporation of other functional groups into the final polyketide products. Later on, combinatorial polyketide biosynthesis by de novo design and rearrangement of modular DEBS genes with other eight?PKS genes created 154 bimodular combinations and allowed the production of novel derivatives as well99. Recently, a new E. coli platform for erythromycin analogue production was established by the production of alternative final erythromycin compounds exhibiting bioactivity against multiple antibiotic-resistant Bacillus subtilis strains100. Both the native deoxysugar tailoring reactions, D-desosamine and L-mycarose deoxysugar pathways, were replaced with the alternative D-mycaminose and D-olivose pathways to produce new erythromycin analogues in E. coli.
Besides understanding the basic biochemistry and production of new derivatives, newly developed synthetic biology platforms offer new strategies for productivity enhancement. Random mutagenesis, recombinant DNA techniques, and process development were combined and applied to a S. coelicolor strain expressing the heterologous DEBS101. Fermentation achieved the production of 1.3 g/L of 15-methyl-6-dEB, and the productivity was increased by over 100% in a fermentation process. Furthermore, the introduction of an engineered mutase-epimerase pathway in E. coli enabled a 5-fold higher production of 6-dEB102, 103. In addition, after the carbon flux of the biosynthetic pathway was redirected and an extra copy of a key deoxysugar glycosyltransferase gene was introduced, a 7-fold increase in erythromycin A titer was achieved104. In those studies, E. coli-derived production of erythromycin was enabled by the introduction of the entire erythromycin pathway (20 genes in total) using separate expression plasmids. However, this may cause metabolic burden and plasmid instability. Recently, the E. coli erythromycin A production system was upgraded by altering the design of the expression plasmids needed for biosynthetic pathway introduction, and a 5-fold increased erythromycin A production was achieved105. On the other hand, in the native host, by the chromosome gene inactivation technique based on homologous recombination with linearized DNA fragments, one TetR family regulator SACE_7301 was identified to enhance erythromycin production in S. erythraea17.
Another recent example is for the biosynthesis of cholesterol-lowering drug pravastatin106 derived from the natural product compactin107. Pravastatin inhibit 3β-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase, which catalyzes the rate-limiting step in cholesterol biosynthesis. Pravastatin is obtained after stereoselective hydroxylation of compactin at the C-6 position and this is often achieved by a two-step production process in industry108. Novel expression vector has been used to carry a gene for cytochrome P450 monooxygenase, and 3.3-fold increased production of pravastatin has been achieved. Optimized batch culture yielded 14.3 g/l of pravastatin after 100 h109. Metabolic reprogramming of this antibiotics has been achieved by introducing both the compactin pathway and a new cytochrome P450 from Amycolatopsis orientalis (CYP105AS1) into the β-lactam–negative P. chrysogenum DS50662. More than 6 g/L pravastatin was obtained via further engineering of the CYP105AS1 enzyme at a pilot production scale110.
3. New tools for Heterologous Biosynthesis of Natural Products
Prior to synthetic biology, several genetic engineering and metabolic engineering strategies were developed for the optimization of heterologous pathway expression. For example, expression of single genes can be optimized by codon-optimization, promoter engineering, gene copy number, translation regulating sequence mutation, post-translational modification and protein engineering. While for multiple gene expressions, metabolic modelling is widely used to guide the coordination of multiple gene expression in the metabolic network, such as identifying the targets of limiting steps. However, the lack of the parameters during the heterologous gene expression, such as the physiological changes of the host and the property parameters of the heterologous proteins in the host environment111, limited the applications of metabolic modelling.
In the past few years, several powerful synthetic biology tools have been developed for the optimization of heterologous pathways in engineered microbial hosts. As engineered heterologous microbes offer an optimized and standardized platform or cell factory for natural product production, advances in their metabolism engineering would benefit the reconstruction of the whole cellular network and heterologous natural product biosynthesis pathways.
3.1 Engineering at the Parts Level
Besides the basic physical construction of a heterologous pathway, the achievement of maximal yield of target products relies on many other factors, including optimization of metabolic flux, reduction of intermediate toxicity, and tuning proper stress on the host cell. Some general strategies for the design of gene or protein parts are needed for optimal expression and organization of individual pathway enzymes. The expression of gene parts can be tuned by modifying gene copy numbers, transcriptional activity, or post-transcriptional processing. The individual proteins can be organized as fusion proteins or in artificial scaffolds for coupled activities in metabolic pathways. The natural and heterologous enzymes can also be organized inside and outside of certain organelles to enhance the product yields.
3.1.1 Tuning the Expression of Gene Parts
In engineered microbes, expressions of gene parts, especially heterologous gene parts, can be fine-tuned throughout the whole central dogma process, including gene replication, transcription, translation, and post-translation. The development of central dogma based engineering toolkits has attracted attention for decades since the discovery of the double helix structure of DNA and the lac operon112, 113. More recently, the rapid development of synthetic biology calls for much more powerful central dogma based engineering toolkits. These toolkits possess some common characteristics, such as standardization, universality, real-time controllability, reusability, and modularity. Several strategies for standardized gene parts storage and ligation have been established recently, such as Biobrick, Bglbrick and SEVA114-116. Gene parts have standard identification sites that can be digested by type I or type IIS restriction enzymes and be ligated to another part containing the same couple of sites. However, there is a paradox about standardization and universality of the gene parts. Several excellent tools to tune gene expression were developed by combination of standardized rules and universal manipulations (Figure 9). At the gene level, variant copies of genes on different plasmids showed obvious effects on the synthesis of target products117. At the transcriptional level, promoter strength, mutation of promoter functional sites, addition of the upstream activation sites, and inducible artificial promoters have been successfully used in optimization of multiple genes expression118-124. At the translational level, special sequence repeats, synthetic riboswitches, rational ribosome binding sites design tools, random translation starting sites, tuneable intergenic region options, and artificial codons have been used for fine-tuning of gene expression125-130. Most of these methods showed great potentials in tuning of multiple genes and optimization of multiple genetic modules. In addition, novel 4-base codons and unnatural amino acids were introduced in the translation process for novel and orthogonal polypeptides elongation and protein synthesis130. Most recently, two synthetic bases X and Y were added to genetic codons in addition to natural bases A, T, C and G131.
Figure 9.
Strategies to tune the gene expression levels in the natural product biosynthetic pathways. The expression of heterologous genes are regulated at the gene level (variation on copy numbers of genes, selection of plasmids), at the transcriptional level (promoter strength, mutation of promoter functional sites, addition of upstream activation sites, and inducible artificial promoters), and at the translational level (special sequence repeats, synthetic riboswitches, rational ribosome binding sites, random translation starting sites, tuneable intergenic region options, and artificial codons). The codon optimization of the heterologous genes is another efficient strategy to tune gene expression.
Based on these ideas of engineering the central dogma process, next generation of DNA synthesis technologies can be used to construct large libraries of genes with modified codons and varying expression levels. By taking advantage of computational design and metagenomic information, de novo DNA synthesis enables us to obtain artificial or natural genes for the required enzymes in an expanding range (Figure 9). For example, 89 methyl halide transferases found in metagenomic sequences from diverse organisms were synthesized and screened for higher enzymatic activities132. 154 and 1,468 reporter genes were constructed and characterized as libraries to study codon usage of genes133. Oligo pools were combined with multiplexed reporter assays for construction of more than 14,000 reporter constructs. The transcriptional and translational rates of these constructs were used to analyze the effects of N-terminal codon bias on protein expression levels134.
3.1.2 Making Artificial Protein Fusions
Natural metabolism is under tight regulations, such as positive or negative feedbacks. The heterologously expressed enzymes often suffer from flux imbalances because of the lacking of well-established regulatory mechanisms. In addition, the enzymes in the same metabolic pathway may be distributed at different regions inside the cell, thus diffusion or degradation of the intermediates occurs and the efficiency of the whole pathway would be decreased. Simply blocking the competitive pathways, in many cases, is not sufficient since it might be lethal or hard to implement. In fact, the high expression level of engineered pathways may compete with biomass production or other primary metabolite pathways for nucleotides and amino acids, which results in opposite to what is expected.
Fusion of proteins could bring active sites of enzymes into a closer proximity, which facilitates substrate channelling (Figure 10) and relieves the side-pathway competition. Protein fusion strategies have been used in the endogenous MVA pathway26. FPPS, encoded by ERG20, catalyses the condensation of 5-carbon units IPP and DMAPP to produce FPP, which is the general precursor for biosynthesis of sesquiterpenes and geranylgeranyl pyrophosphate (GGPP), and then GGPP can be converted into diterpenes and tetraterpenes. In natural growth conditions, most of the FPP is used for production of ergosterol, which is essential for the cellular maintenance. Therefore, the FPP is a main branching point for the synthesis of sesquiterpenoids and triterpenes (Figure 2). Fusing FPPS together with patchoulol synthase (PTS, encoded by Pogostemon cablin gene PatTps177) and GGPPS redirected the flux to the production of the sesquiterpene patchoulol and GGPPS respectively, and enhanced the production by 2-fold and 8-fold, respectively26, 135. Besides fusion of these two endogenous proteins, heterologous enzymes copalyl diphosphate synthase (SmCPS from Salvia miltiorrhiza) and kaurene synthase-like (SmKSL from Salvia miltiorrhiza) were fused for biosynthesis of tanshionones precursor miltiradiene in yeast40. Simply fusing FPPS with Bst1p made the production from trace to 1.0 mg/L. According to the coimmunoprecipitation experiments, they predicted SmKSL and SmCPS may form a complex in vivo, which inspired them to fuse these two enzymes for more efficient production. By comparison of fused SmCPS-SmKSL and SmKSL-SmCPS, the latter pattern was more effective by offering a 2.9-fold increase of miltiradiene to 3.1 mg/L40.
Figure 10.
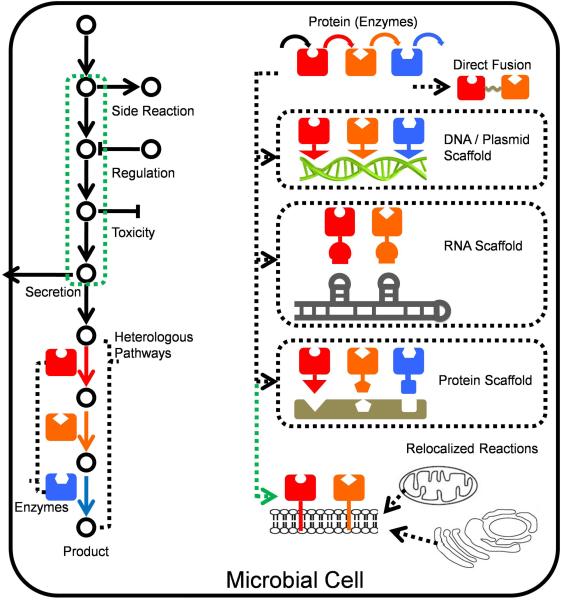
Strategies to enhance the pathway flux by modifying the distances of proteins and protein re-localization. In order to enhance heterologous metabolic flux, fusion of proteins with different linkers is efficient. DNA-, RNA- and protein-based scaffolds are used to assemble the heterologous enzymes to improve the sequential reactions. Re-localization of enzymes switches the subcellular position of heterologous proteins to enhance the production by either enhancing enzyme concentration or taking advantage of environmental differences. Colourful solid arrows represent the heterologous enzymes.
Construction of scaffolds is another commonly used strategy to couple several enzymes catalysing sequential reactions in the same pathway. This type of constructs will not affect other metabolic pathways significantly due to its controllability. There are different types of scaffolds, such as plasmid scaffolds, RNA scaffolds and protein scaffolds (Figure 10). Plasmid scaffolds possess the advantage of accommodating many interaction motifs and linkers of variable lengths without solubility issues, such as the linking of the glucose oxidase and horse radish peroxidase via a lysine residue to short DNA oligo nucleotides136. However, the disadvantage is that enzymes must be significantly modified with multiple zinc finger domains (usually 3–4 domains with a total addition of 90-120 amino acids)137. RNA scaffolds were designed and constructed using multidimensional RNA structures for the spatial organization of bacterial metabolism. The synthetic RNA modules were functionally discrete and formed 1D and 2D scaffolds in which the palindromic regions were disfavoured and the assembly order of RNA strands were controlled by insuring tile formation before polymerization. This RNA-based scaffolding system was applied to increase the production of hydrogen, and an increase of 48-fold over the unscaffolded, tagged enzymes in E. coli was observed138. Besides plasmid scaffolds and RNA scaffolds, protein scaffolds were constructed by co-localization of pathway enzymes to synthetic complexes using well-characterized protein–protein interaction domains and their specific ligands. This strategy is able to increase the effective concentrations of the metabolic intermediates around the enzymes.
The mevalonate producing pathway is consisted of three enzymes (acetoacetyl-CoA thiolase (AtoB) from E. coli, hydroxy-methylglutaryl-CoA synthase (HMGS) and HMGR from S. cerevisiae) in the engineered E. coli139. Difference in the expression levels of these 3 enzymes led to a “bottle-neck” effect and the accumulation of toxic intermediate HMG-CoA. Thus the synthetic protein scaffolds were applied to spatially recruit metabolic enzymes, and the 1:2:2 ratio produced 77-fold of mevalonate compared with the wild strain139. No harm to the growth of the strain was observed, no matter how the inducer concentration varied139. At the same time, increasing the copy number of the protein scaffolds led to enhanced productivity.
3.1.3 Re-localizing Proteins
Primarily for eukaryotic microbial hosts, many enzymes, such as cytochrome P450s, are thought to bind to the endoplasmic reticulum or mitochondrial membrane via the hydrophobic membrane binding domain at the N-terminus in their native hosts. Therefore, this membrane binding domain may be required for function, especially when these cytochrome P450s are expressed in heterologous hosts. Mature forms of these enzymes without the signal peptides are often used in heterologous expression. However, the cytochrome P450 with both N-terminal membrane insertion and C-terminal HA epitope-tag deletion exhibited the highest expression level than other forms with partial deletion140. Relatively upstream pathways are also effective targets for compartmentalization. S. cerevisiae was engineered to produce 2,3-butanediol (BDO) by the introduction of a cytosolic acetolactate synthase (cytoILV2)141. Acetyl-CoA level was improved in the cytosol by the combined disruption of competing pathways and introduction of heterologous biosynthetic pathways to supply more precursors for desired product biosynthesis in yeast142. n-Butanol production was improved to 3-fold through acetyl-CoA enhancement by introduction of heterologous acetyl-CoA biosynthetic pathways including pyruvate dehydrogenase (PDH), ATP-dependent citratelyase (ACL), and PDH-bypass142.
In contrast with expressing mitochondrial enzymes in cytosol, some enzyme and metabolic pathways were also expressed in mitochondria instead of in cytoplasm, because mitochondria have many potential advantages for metabolic engineering, such as the sequestration of diverse metabolites, containing intermediates of many central metabolic pathways, and the environment in the mitochondrial matrix is highly different from that in the cytoplasm143. Mitochondrion was also used as an alternative location for heterologous expression of metabolic engineering instead of cytosol to enhance the flux to FPP35. CsTPS1 (valencene synthase from Citrus sinensis) was located into mitochondria by the fusion with mitochondrial targeting signal peptide of COX4, which brought a 3-fold rise in valencene titers compared with cytosol CsTPS1. The expression of mitochondrion-targeted FPPS (mtFPPS) further increased the production by 40%. Addition of another copy of cytosolic CsTPS1 further increased the production to 1.5 mg/L, which was an 8-fold enhancement. This strategy was also performed on amorph-4,11-dienesynthase (ADS), and mtADS was constructed, which strongly enhanced the amorphadiene production to 20 mg/L. Accordingly, for upstream pathways, Avalos et al. engineered yeast mitochondria to produce isobutanol, isopentanol and 2-methyl-1-butanol through Ehrlich degradation pathway, which originally occurs in cytoplasm, while the upstream pathway is confined to mitochondria, which creates a substantial bottleneck in the transportation143. They developed a standard, flexible set of vectors to target identical pathways to different subcellular parts. Targeting Ll-kivd (α-ketoacid decarboxylase from Lactococcus lactis) and Sc-adh7 (alcohol dehydrogenase from S. cerevisiae) to mitochondria increased the titer of isobutanol approximately 220% to 486 mg/L143. The results showed that the compartmentalization the pathway into mitochondria achieved higher local enzyme concentrations and increased the availability of intermediates and released the restriction on transportation.
3.2 Engineering at the Pathway Level
Similar to organic synthesis of a target chemical, the heterologous production of a target product in engineered microbe hosts is also affected by common factors such as substrate supply, reaction efficiency, reaction thermodynamics and kinetics, yield, productivity, toxicity, and cost-efficiency. Although some operation units are different for alternative products and can be optimized case by case, some universal pathway engineering strategies would facilitate the improvement of these biochemical reactions. The successful production of artemisinic acid synthesis in a heterologous host offered us a good example of how to optimize cellular metabolic pathways for terpenoid production, and these strategies could be applied to other biochemical molecules as well.
3.2.1 Mining of Heterologous Pathways
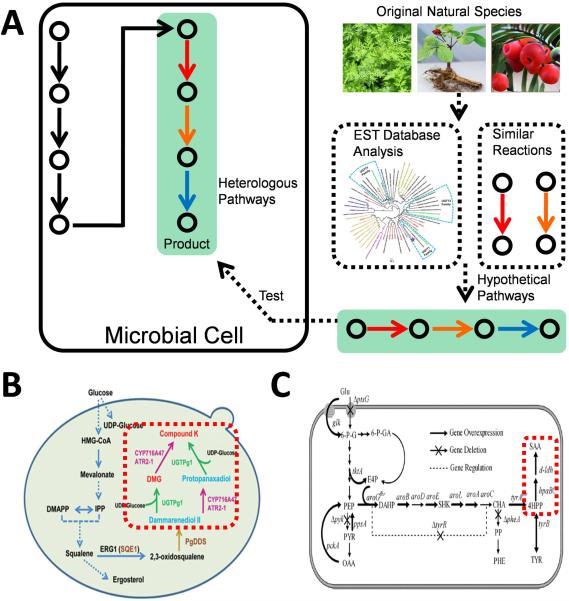
A typical synthetic biology approach for heterologous biosynthesis of target products begins with the exploration of genes and pathways encoding functional enzymes. However, there are still many pathways remaining to be explored, and the typical strategy the ones studied is to express them heterologously, either to make more use of the known pathway or decipher the cryptic pathway (Figure 11). The first trial to introduce a whole pathway heterologously was done for terpenoid production144. Targeted amorpha-4,11-diene, the sesquiterpene olefin precursor to artemisinin, as the final product, they introduced the mevalonate terpenoid pathway from S. cerevisiae into E. coli and the concentration reached 24 μg caryophyllene equivalent/mL, which approved the functionalization of fungi pathway in the cytosol of bacteria. For cryptic pathway exploration, besides the analysis of systems biology, synthetic biology also takes in practice the construction of transcription units of any possible ORFs based on bioinformatics analysis, as in the example of the mining of the gene encoding the CK synthase (Figure 11)58. CK, as the main functional component of ginsenoside, possesses bioactivities of anti-inflammation, hepatoprotection, antidiabetes and anti-cancer145. The potential CK biosynthetic pathway was designed in yeast, including a cytochrome P450 (CYP716A47) from Panax ginseng, a NADPH-cytochrome P450 reductase (ATR2-1) from Arabidopsis thaliana, and a Dammarenediol-II synthase (PgDDS) from Panax ginseng, with other involved enzymes originated from S. cerevisiae. In this study, a proprietary cDNA database including 479 689 assembled cDNA contigs was established based on 9 Panax EST datasets available from the NCBI GenBank. 16 ORFs were amplified and expressed in S. cerevisiae, through which way, the enzyme coding CK synthase was obtained from natural resources and primarily verified and the strain produced 1.4 mg/L of CK58.
Figure 11.
Exploration and construction of novel natural product biosynthetic pathways. (A) Heterologous expression of uncharacterized genes or proteins with potential functions to catalyze the specific reaction is applied to explore new pathways for natural product biosynthesis. (B) Heterologous expression of uncharacterized genes to explore the biosynthetic pathway for ginsenoside CK. (C) Heterologous expression of protein with similar activity to construct a novel biosynthetic pathway for Salvianic acid A. Colourful solid arrows represent the heterologous enzymes. Adapted by permission from Macmillan Publishers Ltd: [Cell Research] (ref. 58), copyright (2014). Reprinted from Metabolic Engineering, 19, Yao YF, Wang CS, Qiao J, Zhao GR., Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway, Pages 79-87, Copyright (2013), with permission from Elsevier.
Sometimes a novel pathway for product synthesis can also be designed. For example, an unprecedented artificial salvianic acid A (SAA) biosynthesis pathway was developed in E. coli146. Two enzymes D-lactate dehydrogenase (D-LDH) from Lactobacillus pentosus and hydroxylase complex encoded by hpaBC from E. coli, were introduced into E. coli to convert 4-hydroxyphenylpyruvate (4HPP) into SAA. D-LDH and its variants were selected to catalyze the chiral reduction of ketone group in 4HPP according to their performance in transformation of phenylpyruvate into phenyllactic acid (Figure 11). Co-expression of d-ldh and its mutants and hpaBC together with optimizations on the upstream pathway resulted in 7.1 g/L of SAA with a yield of 0.47 mol/mol glucose146.
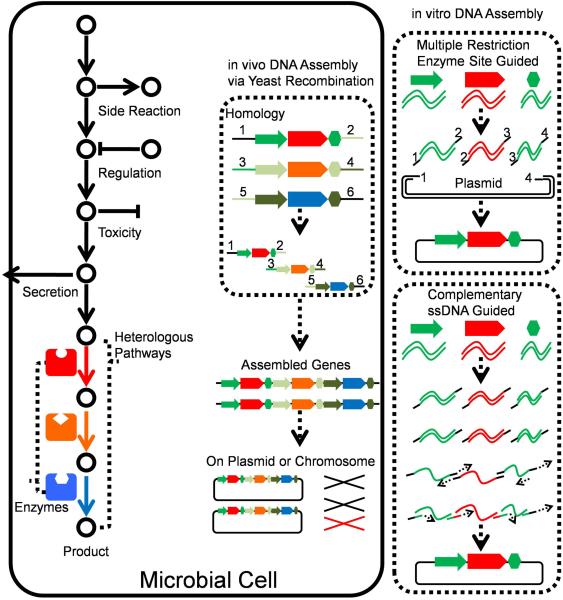
3.2.2 DNA Assembly-Assisted Pathway Construction
To synthesize a desired product in heterologous organisms, the introduced biosynthetic pathways and upstream metabolic pathways are required to be considered together as the supply of precursors affects the ultimate yield of the final product. In recent years, the studies on synthetic biology also expand to strategies of engineering central carbon metabolisms, which can be summarized as combinatorial engineering (Figure 12). Most of these strategies were designed rationally; however, some could also be utilized to build up a randomized library in the pre-design frame. In some strategies, variation in expression levels of individual genes were introduced, and those alternative expressions were combined during DNA assembly process (Figure 12). One prominent example is the Customized Optimization of Metabolic Pathways by Combinatorial Transcriptional Engineering (COMPACTER) method in which assembly of promoter mutants and pathway genes resulted in the combination of different gene expression levels in a target pathway147. A more recent work utilized Gibson assembly method to introduce coding sequences after the initial codon ATG for expanding expression range of the genes coding violacein synthesizing enzymes, causing variant combinations of multi-gene transcriptional levels128.
Figure 12.
Strategies to optimize natural product biosynthetic pathways assisted by DNA assembly methods. A heterologous pathway can be optimized by changing different promoters to regulate the expression of different genes. DNA assembly in vivo and in vitro can be used to introduce variations either inside the genes or in regulation parts (promoters or regulators) to build a library, which can be used to select the desired strains with the heterologous pathway. Colourful solid arrows represent the heterologous enzymes.
3.2.3 Modular Pathway Engineering for Metabolic Precursor Synthesis
To produce high-level products, simple modification of the expression levels of genes in the target pathway is the most rational method for the pathway optimization. These methods could be classified into 3 categories (Figure 13): (1) increasing the supplement of the substrate, (2) increasing the whole flux of the desired biosynthetic pathway, and (3) decreasing or eliminating the flux of the branch pathway. Here, we take the optimization of the MVA pathway as an example to demonstrate how to optimize a biosynthesis process at the pathway level.
Figure 13.
Strategies of modular pathway engineering for metabolic precursor synthesis. Strategies to increase metabolic precursors include: (1) increasing the supply of the substrate, such as adding new biosynthetic pathway of precursor; (2) increasing the whole flux of the desired biosynthetic pathway, such as improving the gene expression for precursor synthesis; (3) decreasing or eliminating the flux of the branch pathways, such as deleting the branch pathways. These strategies can be achieved by manipulating the copy number of genes, changing promoters and introducing either heterologous upstream or heterologous downstream pathways. Colourful solid arrows represent the heterologous enzymes. Red arrow with two tailors means the down regulation.
Acetyl coenzyme A (acetyl-CoA) is a central metabolite in carbon and energy metabolism. The mevalonate pathway in S. cerevisiae is initiated from acetyl-CoA, a key precursor to a wide range of valuable secondary metabolites148. Moreover, acetyl-CoA supply in the mitochondrial matrix is essential for cell growth because it is used to replenish the mitochondrial tricarboxylic acid (TCA) cycle with C2 units and produce ATP149. Optimizing the supply of acetyl-CoA becomes a logical approach to increase the production of the target compounds. Chen et al. enhanced the supply of acetyl-CoA in the cytoplasm through a combined push-pull-block strategy. The push part included the over-expression of the endogenous alcohol dehydrogenase (ADH2) and NADP-dependent aldehyde dehydrogenase (ALD6), and a codon-optimized acetyl-CoA synthase variant from Salmonella enterica (ACSSEL641P). The pulling of acetyl-CoA towards the products of interest was enhanced by the over-expression of ERG10, encoding acetyl-CoA C-acetyltransferase, that catalyzes the conversion of acetyl-CoA to acetoacetyl-CoA. The block part of the strategy decreased reduction of acetyl-CoA and was achieved through removing two key enzymes involved in the consumption of acetyl-CoA. One is the peroxisomal citrate synthase and the other is cytosolic malate synthase. This strategy improved the production of α-santalene by 4-fold150. Lian et al. redirected the glycolytic flux towards acetyl-CoA by inactivating ADH1 and ADH4 for ethanol formation and GPD1 and GPD2 for glycerol production, resulting in 4-fold improvement in n-butanol production. Subsequent introduction of heterologous acetyl-CoA biosynthetic pathways, including pyruvate dehydrogenase (PDH), ATP-dependent citrate lyase (ACL), and PDH-bypass, further increased n-butanol production. Among them recombinant PDHs localized in the cytosol (cytoPDHs) was most efficient and increased n-butanol production by additional 3-fold142.
To increase the overall flux of the MVA pathway, enhancing the reaction rate of the limiting step (HMG-CoA to mevalonate) was considered. tHMG1, the truncated HMG1 which only contains the catalytic domain of HMG1, is commonly used in engineering of the MVA pathway in yeast. tHMG1 was often overexpressed by strong promoter and has many copies in the cell7, 33, 151. On the other hand, UPC2-1, a modified global transcriptional regulator of ergosterol biosynthesis was often used to up-regulate the expression level of the entire MVA pathway7, 33, 151.
For production of terpenes, the native biosynthesis pathway of ergosterol always needs to be down-regulated. The most useful method is to reduce the expression level of essential gene ERG9 by different weaker promoters such as pMET, pCTR3, and pHXT16, 7. Knocking out the lppA and STC genes reduced the production of fansesol while the important precursor FPP was accumulated33.
Optimization of the MVA pathway needs the combination of different strategies. A very successful example is the biosynthesis of amorphadiene, which can be synthesized from FPP by amorphadiene synthase encode by ADS. Westfall et al. overexpressed all genes responsible for the MVA pathway from ERG10 to ERG20 under the GAL promoter and integrated all these genes into genome. Among them, the key enzyme tHMG1 had three copies to enhance the limiting-step of the MVA pathway. ERG9 was down-regulated by substitution of the native promoter of ERG9 by pMET. During fermentation, ethanol was the sole carbon source for production, which was helpful for supplying more acetyl-CoA for biosynthesis. The accumulated precursor FPP, could be converted into amorphadiene efficiently by highly expressed ADS. Subsequent fermentation optimization led to 40 g/L products, which was the highest production compared to previous reports7.
3.2.4 Biosensor-directed Real-time Control and Evolution
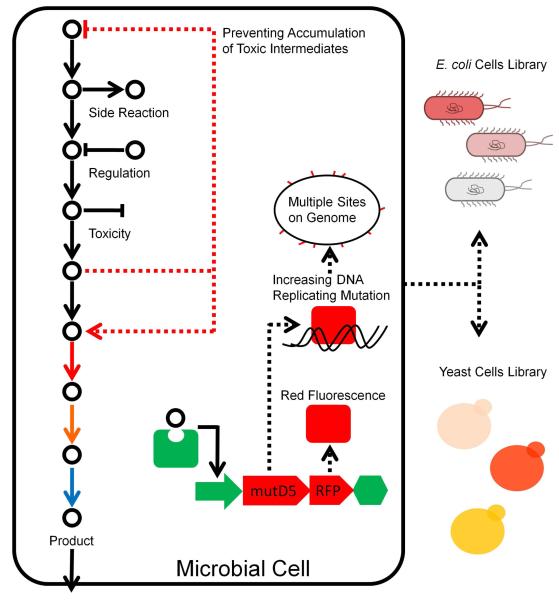
In recent years, development of intracellular small molecule screening methods led to generation of cellular self-modification systems with the small molecules functioning as signal molecules (Figure 14). Some of the small molecules are produced by the cell, such as N-acylhomoserine lactones (AHL) and N-butyrylhomoserine lactone (BHL), which could be synthesized by special enzymes in cells and diffused to coherent cells to regulate multi-cell activities through gene circuits. While some other similar molecules could only be added from outside to tune genes’ transcription and translation processes, such as isopropyl thiogalactoside, arabinose, anhydrotetracycline, and theophylline. Light is also used as a signal to regulate gene expression and change cellular activity152-154. However, most of these earlier systems could only be operated in a pre-determined way or via serendipity. Recent advances offered us some biosensor-directed real-time metabolic control methods (Figure 14). It is often difficult to accumulate the products in engineered cells due to the toxicity of target intermediates. Metabolite response promoters were explored and utilized in metabolic self-regulation155. The whole-genome transcript arrays were employed to identify promoters that responded to these intermediates. The promoters could control expression of certain genes in a real-time manner, causing enhanced amorphadiene production by 2-fold over traditional constructions in E. coli. This approach could also be extended to other toxic intermediates for dynamic regulation155. A similar metabolite biosensor system was constructed to achieve the feedback regulated evolution of phenotype (FREP) based on production of target products156. In the designed adaptive control system, the genomic mutation rate would be increased by expression of mutD5 to generate diversity in the population, and be decreased only when the concentration of target metabolites came to required amounts. The synthetic transcription factors were utilized to construct artificial biosensors to respond for metabolites without natural sensors. The FREP was verified by evolving increased tyrosine and terpenoid production.
Figure 14.
Biosensor-directed real-time control and evolution. The promoters inhibited by the target products are used as the biosensors to control the expression of DNA mutator proteins, such as mutD5. This design can be used to generate a mutant library. Meanwhile, the same promoter also control the expression of a fluorescent protein, which is used as the reporter for phenotype selection. Colourful solid arrows represent the heterologous enzymes. Adapted by permission from Macmillan Publishers Ltd: [Nature Communication] (ref. 156), copyright (2013).
3.3 Engineering at the Genome Level
Genome engineering approaches can be grouped into three categories: genome editing, transcriptome engineering, and genome synthesis157. Genome editing precisely or combinatorially modifies the target genome at multiple loci while transcriptome engineering essentially targets regulatory elements by mutating endogenous regulators or introducing artificial ones. Genome synthesis involves hierarchical assembly of short chemically synthesized DNA fragments into microbial genomes. Because of the large size of a genome, genome engineering is typically coupled with high throughput screening technologies. In the past few years, genome engineering has been increasingly used for heterologous biosynthesis of natural products.
Based on the allelic replacement on the lagging strand by the oligos during replication, multiplex automated genome engineering (MAGE) was developed in E. coli, and applied to improve the biosynthesis of lycopene by simultaneously modifying 24 genes in the corresponding biosynthetic pathway158. This method was recently extended to S. cerevisiae159. By applying the same principle, multiplex iterative plasmid engineering (MIPE) was developed to engineer the heterologous metabolic pathway in the plasmid, and a clone with 2.67-fold improved production of riboflavin was achieved in less than a week160. Another related approach, RNA interference (RNAi)-assisted genome evolution (RAGE) could continuously improve target trait(s) by accumulating multiplex beneficial genetic modifications in an evolving yeast genome161.
Modular artificial nucleases have been increasingly used for genome editing. The zinc-finger domain, which typically recognizes 3-nucleotide DNA motifs, was the first to be exploited. Sequence-specificity was further increased by engineering zinc-finger nucleases (ZFNs) in a way that requires their heterodimerization through the FokI domain for efficient cleavage162. The modular approach was taken a step further with the discovery of the TAL effector (TALE) DNA-binding modules of Xanthomonas bacteria, and their simple DNA recognition code163. However, to target unique sequences in a eukaryotic genome, long TAL arrays need to be assembled in order, because each TAL repeat targets a single nucleotide. More recently, clustered regularly interspaced short palindromic repeats (CRISPR) have been developed as a general method for genome engineering including activating transcription/translation, silencing transcription/translation, up/down regulating gene expression, and modifying multiple sites simultaneously, each accompanied with permanent changes of genome sequence. Compared to ZFNs and TALENs, the CRISPR technology has radically improved the accessibility of gene targeting due to its straightforward approach for customizing sequence specificity via target-specific guide RNAs. Its targeting efficiency is comparable with the best efficiency achieved using TALENs in a wide range of animals and plants164. Recently, it was shown that the endonuclease domains of the Cas9 protein can be mutated and co-expressed with a guide RNA to specifically interfere with transcriptional elongation and work like RNAi. This system was named CRISPR interference (CRISPRi), which can efficiently repress expression of a target gene with no detectable off-target effects165. In addition, dCas9 was used with existing optogenetic or chemical-induced proximity systems to dynamically analyze recruitment of a broad range of chromatin modifiers to a specific DNA sequence, both for customized activation and repression166. CRISPR/Cas transcription factors (CRISPR-TFs) have the potential to be integrated for the tunable modulation of gene networks167. A rapid, efficient, and potentially scalable strategy based on CRISPR/Cas system was recently developed to generate multiple gene disruptions simultaneously in S. cerevisiae168.
Genome synthesis with novel design is the extreme of genome editing for natural product synthesis. So far, genome synthesis based on the natural genome sequence has succeeded in several virus and bacteria, such as Mycoplasma genitalium, Mycoplasma mycoides and Phaeodactylum tricornutum169-171. However, due to the lack of understanding of the biology, the de novo design of a genome has not been attempted. On the other hand, an initiative to redesigning the genome of S. cerevisiae named SC 2.0 was launched recently. Numerous loxP sites were introduced into the redesigned genome and the SCRaMbLE (synthetic chromosome rearrangement and modification by loxP-mediated evolution) was developed by combining loxP sites with the inducible Cre recombinase (Figure 15)172. The SCRaMbLE system in Sc2.0 is a very useful tool for minimization of the yeast genome and generation of a library of mutant genomes at a large scale173-175. Such mutant genomes may offer the potential chassis for heterologous biosynthesis of different natural products.
Figure 15.
The SCRaMbLE system based on genome synthesis. Based on the Cre-LoxP system, the SCRaMbLE (synthetic chromosome rearrangement and modification by loxP-mediated evolution) is able to generate a mutant library by gene deletion, gene reversion, gene insertion, and gene translocation at a genome scale. Colourful solid arrows represent the heterologous enzymes. Reproduced with permission from ref. 175, copyright 2014 John Wiley and Sons.
3.4 Engineering at the Community level
Many complex natural products are synthesized by very long pathways such as paclitaxel. It is often a challenge for one cell to express such long heterologous pathways due to the heavy metabolic burden associated with expression of so many heterologous enzymes and production burden176, 177. Most of the microbial consortium engineering focus on the lignocelluloses bioconversion to produce chemicals and fuels132, 178. The progress on the fundamental studies and the applications of artificial consortium was reviewed recently179. More recently, engineering of the microbial consortium has been applied to biosynthesis of complex natural products177. Taxadiene, as the scaffold molecule of paclitaxel, can be currently produced at much higher titer in the engineered E. coli compared to engineered yeast47, 50. However, heterologous expression of cytochrome P450s, the downstream enzymes for taxadiene transformation, is more suitable in yeast than in E. coli47, because yeast possesses the advanced protein expression machinery and abundant intracellular membranes. Therefore, metabolic pathway for taxane biosynthesis was distributed into the consortium of E. coli and S. cerevisiae, and this synthetic consortium produced 33 mg/L oxygenated taxanes (Figure 16)177. Using the similar strategy, biosynthesis of ferruginol were also achieved in the artificial consortium of E. coli with the heterologous pathway of miltiradiene synthesis and S. cerevisiae with an cytochrome P450177. The concept of distributing the long metabolic pathways into consortium would significantly enhance the heterologous biosynthesis of natural products (Figure 16).
Figure 16.
Engineering of the microbial consortium to improve heterologous biosynthesis of complex natural products. (A) Schematic of the distribution of the long synthetic pathway for complex natural products into different strains of the consortium. (B) The artificial consortium with engineering E. coli and S. cerevisiae produce oxygenated taxanes using xylose and ethanol. Colourful solid arrows represent the heterologous enzymes. Adapted by permission from Macmillan Publishers Ltd: [Nature Biotechnology] (ref. 177), copyright (2015).
4. Conclusion and Future Prospects
Natural products have been and continue to be the source and inspiration for a substantial fraction of human therapeutics. The successfully commercialized production of several drugs or drug candidates shed lights on the future heterologous biosynthesis of important natural products. With the emergence of more recently genomics- and bioinformatics-guided synthetic biology approaches, alternative strategies have been developed for better heterologous expression of natural products biosynthesis pathways. In the near future, better designed parts, modules or biosystems could be created to facilitate the heterologous biosynthesis of natural products. Also, the new tools developed in the synthetic biology field, such as CRISPR/Cas system or RNAi system would be engineered for manipulation of biosynthetic pathways via gene activation/inactivation. Moreover, these techniques not only can help improving the production of value-added compounds, but also can lead to flourish in natural product discovery field, including genomics-driven discovery combined with genetic manipulations in heterologous hosts. In addition, the separation technology coupled with specific artificial systems will be developed to further enhance the heterologous biosynthesis of natural products.
E. coli, Streptomyces species, yeast and Aspergillus species are often used as heterologous hosts for expression of a single gene, a cassette of genes, or an entire biosynthetic gene cluster for identifying the corresponding natural product. Genes or pathways from bacteria were often heterologously expressed in E. coli or Streptomyces species, resulting in the synthesis of novel compounds52, 62-74. To facilitate the discovery of novel compounds in a bacteria heterologous host, several strategies have already been tested, such as activation of cryptic pathways via promoter replacement, generating genome-minimized heterologous hosts with clean background and combinatorial biosynthesis for new derivatives. Meanwhile, S. cerevisiae and Aspergillus species have often been used as heterologous hosts expressing genes from fungi180-190. Several strategies for activating silent gene clusters in fungal genomes have previously been developed and were recently reviewed elsewhere183, 190. As powerful analytical techniques (MS, NMR, and software for comparative data analysis) continue to evolve, we will continue to explore novel natural products from ‘silent’ natural product biosynthesis pathways by enabling detection and characterization of compounds present in minute quantities.
With the advent of new technologies for manipulation both of biosynthetic gene clusters and heterologous hosts, it is foreseeable that improved production of known compounds would be achieved; novel bioactive compounds with pharmaceutical or industrial importance could be discovered or generated and a greater understanding of the molecular complexities of natural products biosynthesis would be guaranteed.
Supplementary Material
Acknowledgements
This work was financially supported by the Ministry of Science and Technology of China (863 program: 2012AA02A701, 973 program: 2013CB733600), and the National Natural Science Foundation of China (major project: 21390203). HZ would like to acknowledge the financial support by National Institutes of Health (GM077596).
References
- 1.Luo Y, Cobb RE, Zhao H. Curr. Opin. Biotechnol. 2014;30:230–237. doi: 10.1016/j.copbio.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vannucchi V. Riv. Crit. Clin. Med. 1952;52:128–136. [PubMed] [Google Scholar]
- 4.Way JC, Collins JJ, Keasling JD, Silver PA. Cell. 2014;157:151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 5.Paddon CJ, Keasling JD. Nat. Rev. Microbiol. 2014;12:355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 6.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 7.Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E111–118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Nat. Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao Z, Luo Y, Zhao H. Mol. Biosyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quin MB, Schmidt-Dannert C. Curr. Opin. Biotechnol. 2014;29:55–61. doi: 10.1016/j.copbio.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler MS, Robertson AA, Cooper MA. Nat. Prod. Rep. 2014;31:1612–1661. doi: 10.1039/c4np00064a. [DOI] [PubMed] [Google Scholar]
- 12.Bohlmann J, Keeling CI. Plant J. 2008;54:656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 13.Pulido P, Perello C, Rodriguez-Concepcion M. Mol. Plant. 2012;5:964–967. doi: 10.1093/mp/sss088. [DOI] [PubMed] [Google Scholar]
- 14.Aharoni A, Jongsma MA, Bouwmeester HJ. Trends Plant Sci. 2005;10:594–602. doi: 10.1016/j.tplants.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Chambon C, Ladeveze V, Servouse M, Blanchard L, Javelot C, Vladescu B, Karst F. Lipids. 1991;26:633–636. doi: 10.1007/BF02536428. [DOI] [PubMed] [Google Scholar]
- 16.Chang MY, Shen YL. Molecules. 2014;19:6694–6706. doi: 10.3390/molecules19056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landmann C, Fink B, Festner M, Dregus M, Engel KH, Schwab W. Arch. Biochem. Biophys. 2007;465:417–429. doi: 10.1016/j.abb.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N. Plant J. 2008;55:224–239. doi: 10.1111/j.1365-313X.2008.03496.x. [DOI] [PubMed] [Google Scholar]
- 19.Dudareva N, Cseke L, Blanc VM, Pichersky E. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero O, Ramon D, Orejas M. Metab. Eng. 2008;10:78–86. doi: 10.1016/j.ymben.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Rico J, Pardo E, Orejas M. Appl. Environ. Microbiol. 2010;76:6449–6454. doi: 10.1128/AEM.02987-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, Viljoen AM. S. Afr. J. Bot. 2010;76:643–651. [Google Scholar]
- 23.Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. Plant Physiol. 2004;134:370–379. doi: 10.1104/pp.103.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oswald M, Fischer M, Dirninger N, Karst F. FEMS Yeast Res. 2007;7:413–421. doi: 10.1111/j.1567-1364.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- 25.Deguerry F, Pastore L, Wu S, Clark A, Chappell J, Schalk M. Arch. Biochem. Biophys. 2006;454:123–136. doi: 10.1016/j.abb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH. Appl. Environ. Microbiol. 2011;77:1033–1040. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson BE, Hart-Wells EA, Matsuda SP. Org. Lett. 2003;5:1629–1632. doi: 10.1021/ol034231x. [DOI] [PubMed] [Google Scholar]
- 28.Song AA, Abdullah JO, Abdullah MP, Shafee N, Othman R, Tan EF, Noor NM, Raha AR. PLoS One. 2012;7:e52444. doi: 10.1371/journal.pone.0052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song AA, Abdullah JO, Abdullah MP, Shafee N, Othman R, Noor NM, Rahim RA. FEMS Microbiol. Lett. 2014;355:177–184. doi: 10.1111/1574-6968.12469. [DOI] [PubMed] [Google Scholar]
- 30.Asadollahi MA, Maury J, Moller K, Nielsen KF, Schalk M, Clark A, Nielsen J. Biotechnol. Bioeng. 2008;99:666–677. doi: 10.1002/bit.21581. [DOI] [PubMed] [Google Scholar]
- 31.Asadollahi MA, Maury J, Schalk M, Clark A, Nielsen J. Biotechnol. Bioeng. 2010;106:86–96. doi: 10.1002/bit.22668. [DOI] [PubMed] [Google Scholar]
- 32.Song AA, Abdullah JO, Abdullah MP, Shafee N, Rahim RA. Int. J. Mol. Sci. 2012;13:1582–1597. doi: 10.3390/ijms13021582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scalcinati G, Partow S, Siewers V, Schalk M, Daviet L, Nielsen J. Microb. Cell. Fact. 2012;11:117. doi: 10.1186/1475-2859-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ignea C, Trikka FA, Kourtzelis I, Argiriou A, Kanellis AK, Kampranis SC, Makris AM. Microb. Cell. Fact. 2012;11:162. doi: 10.1186/1475-2859-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, Ovadis M, Abeliovich H, Vainstein A. Metab. Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Han JY, Fan JY, Horie Y, Miura S, Cui DH, Ishii H, Hibi T, Tsuneki H, Kimura I. Pharmacol. Ther. 2008;117:280–295. doi: 10.1016/j.pharmthera.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Geuns JM. Phytochemistry. 2003;64:913–921. doi: 10.1016/s0031-9422(03)00426-6. [DOI] [PubMed] [Google Scholar]
- 38.Mirjalili MH, Farzaneh M, Bonfill M, Rezadoost H, Ghassempour A. FEMS Microbiol. Lett. 2012;328:122–129. doi: 10.1111/j.1574-6968.2011.02488.x. [DOI] [PubMed] [Google Scholar]
- 39.Busch T, Kirschning A. Nat. Prod. Rep. 2008;25:318–341. doi: 10.1039/b705652b. [DOI] [PubMed] [Google Scholar]
- 40.Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G, Huang L, Zhao ZK. J. Am. Chem. Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 41.Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, Zhao ZK, Huang L. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jordan MA, Wilson L. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 43.Kusari S, Singh S, Jayabaskaran C. Trends Biotechnol. 32:304–311. doi: 10.1016/j.tibtech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Heinig U, Scholz S, Jennewein S. Fungal Diversity. 2013;60:161–170. [Google Scholar]
- 45.Huang Q, Roessner CA, Croteau R, Scott AI. Bioorg. Med. Chem. 2001;9:2237–2242. doi: 10.1016/s0968-0896(01)00072-4. [DOI] [PubMed] [Google Scholar]
- 46.Boghigian BA, Salas D, Ajikumar PK, Stephanopoulos G, Pfeifer BA. Appl. Microbiol. Biotechnol. 2012;93:1651–1661. doi: 10.1007/s00253-011-3528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB. Biotechnol. Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- 49.Engels B, Dahm P, Jennewein S. Metab.Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Ding MZ, Yan HF, Li LF, Zhai F, Shang LQ, Yin Z, Yuan YJ. PLoS One. 2014;9:e109348. doi: 10.1371/journal.pone.0109348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, Wang TT, Li JH, Zou JH, Chen YQ, Dai JG. J. Mol. Catal. B-Enzym. 2011;68:250–255. [Google Scholar]
- 52.Cheng HL, Zhao RY, Chen TJ, Yu WB, Wang F, Cheng KD, Zhu P. Mol. Cell. Proteomics. 2013;12:2236–2248. doi: 10.1074/mcp.M113.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips DR, Rasbery JM, Bartel B, Matsuda SPT. Curr. Opin. Plant Biol. 2006;9:305–314. doi: 10.1016/j.pbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Bulgakov VP, Khodakovskaya MV, Labetskaya NV, Chernoded GK, Zhuravlev YN. Phytochemistry. 1998;49:1929–1934. [Google Scholar]
- 55.Dai ZB, Wang BB, Liu Y, Shi MY, Wang D, Zhang XN, Liu T, Huang LQ, Zhang XL. Sci. Rep. 2014:4. [Google Scholar]
- 56.Huang LL, Li J, Ye HC, Li CF, Wang H, Liu BY, Zhang YS. Planta. 2012;236:1571–1581. doi: 10.1007/s00425-012-1712-0. [DOI] [PubMed] [Google Scholar]
- 57.Huang L, Li J, Ye H, Li C, Wang H, Liu B, Zhang Y. Planta. 2012;236:1571–1581. doi: 10.1007/s00425-012-1712-0. [DOI] [PubMed] [Google Scholar]
- 58.Yan X, Fan Y, Wei W, Wang PP, Liu QF, Wei YJ, Zhang L, Zhao GP, Yue JM, Zhou ZH. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu F, Yuan QP, Dong HR. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006;838:44–49. doi: 10.1016/j.jchromb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 60.Ukibe K, Hashida K, Yoshida N, Takagi H. Appl. Environ. Microbiol. 2009;75:7205–7211. doi: 10.1128/AEM.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemuth K, Steuer K, Albermann C. Microb. Cell. Fact. 2011;10:29. doi: 10.1186/1475-2859-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cunningham FX, Jr., Gantt E. Plant Cell. 2011;23:3055–3069. doi: 10.1105/tpc.111.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YS, Lee JH, Kim NH, Yeom SJ, Kim SW, Oh DK. Appl. Microbiol. Biotechnol. 2011;90:489–497. doi: 10.1007/s00253-011-3091-z. [DOI] [PubMed] [Google Scholar]
- 64.Nam HK, Choi JG, Lee JH, Kim SW, Oh DK. Biotechnol. Lett. 2013;35:265–271. doi: 10.1007/s10529-012-1072-7. [DOI] [PubMed] [Google Scholar]
- 65.Yang J, Guo L. Microb. Cell. Fact. 2014;13:160. doi: 10.1186/s12934-014-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu F, Lu L, Fu S, Zhong X, Hu M, Deng Z, Liu T. Process Biochem. 2015;50:341–346. [Google Scholar]
- 67.Zhao J, Li QY, Sun T, Zhu XN, Xu HT, Tang JL, Zhang XL, Ma YH. Metab. Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Sun T, Miao L, Li Q, Dai G, Lu F, Liu T, Zhang X, Ma Y. Biotechnol. Lett. 2014;36:1515–1522. doi: 10.1007/s10529-014-1543-0. [DOI] [PubMed] [Google Scholar]
- 69.Yamano S, Ishii T, Nakagawa M, Ikenaga H, Misawa N. Biosci. Biotechnol. Biochem. 1994;58:1112–1114. doi: 10.1271/bbb.58.1112. [DOI] [PubMed] [Google Scholar]
- 70.Miura Y, Kondo K, Saito T, Shimada H, Fraser PD, Misawa N. Appl. Environ. Microbiol. 1998;64:1226–1229. doi: 10.1128/aem.64.4.1226-1229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, van den Berg JA, van Ooyen AJJ. Appl. Environ. Microbiol. 2007;73:4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie WP, Liu M, Lv XM, Lu WQ, Gu JL, Yu HW. Biotechnol. Bioeng. 2014;111:125–133. doi: 10.1002/bit.25002. [DOI] [PubMed] [Google Scholar]
- 73.Xie W, Ye L, Lv X, Xu H, Yu H. Metab. Eng. 2014;28C:8–18. doi: 10.1016/j.ymben.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Reyes LH, Gomez JM, Kao KC. Metab. Eng. 2014;21:26–33. doi: 10.1016/j.ymben.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 75.Ozaydin B, Burd H, Lee TS, Keasling JD. Metab. Eng. 2013;15:174–183. doi: 10.1016/j.ymben.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 76.Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N. Biotechnol. J. 2007;2:1235–1249. doi: 10.1002/biot.200700184. [DOI] [PubMed] [Google Scholar]
- 77.Trantas E, Panopoulos N, Ververidis F. Metab. Eng. 2009;11:355–366. doi: 10.1016/j.ymben.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Siddiqui MS, Thodey K, Trenchard I, Smolke CD. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- 79.Vannelli T, Wei Qi W, Sweigard J, Gatenby AA, Sariaslani FS. Metab. Eng. 2007;9:142–151. doi: 10.1016/j.ymben.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Jiang H, Wood KV, Morgan JA. Appl. Environ. Microbiol. 2005;71:2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leonard E, Yan YJ, Lim KH, Koffas MAG. Appl. Environ. Microbiol. 2005;71:8241–8248. doi: 10.1128/AEM.71.12.8241-8248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ralston L, Subramanian S, Matsuno M, Yu O. Plant Physiol. 2005;137:1375–1388. doi: 10.1104/pp.104.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan YJ, Kohli A, Koffas MAG. Appl. Environ. Microbiol. 2005;71:5610–5613. doi: 10.1128/AEM.71.9.5610-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y, Yu O. Metab. Eng. 2011;13:455–463. doi: 10.1016/j.ymben.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 85.Hawkins KM, Smolke CD. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Minami H, Kim JS, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakagawa A, Minami H, Kim JS, Koyanagi T, Katayama T, Sato F, Kumagai H. Nat. Commun. 2011:2. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim JS, Nakagawa A, Yamazaki Y, Matsumura E, Koyanagi T, Minami H, Katayama T, Sato F, Kumagai H. Biosci., Biotechnol., Biochem. 2013;77:2166–2168. doi: 10.1271/bbb.130552. [DOI] [PubMed] [Google Scholar]
- 89.Fossati E, Ekins A, Narcross L, Zhu Y, Falgueyret JP, Beaudoin GA, Facchini PJ, Martin VJ. Nat. Commun. 2014;5:3283. doi: 10.1038/ncomms4283. [DOI] [PubMed] [Google Scholar]
- 90.Rawlings BJ. Nat. Prod. Rep. 2001;18:231–281. doi: 10.1039/b100191o. [DOI] [PubMed] [Google Scholar]
- 91.Kao CM, Katz L, Khosla C. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 92.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 93.Marsden AFA, Wilkinson B, Cortes J, Dunster NJ, Staunton J, Leadlay PF. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 94.Cane DE, Kudo F, Kinoshita K, Khosla C. Chem. Biol. 2002;9:131–142. doi: 10.1016/s1074-5521(02)00089-3. [DOI] [PubMed] [Google Scholar]
- 95.Liou GF, Lau J, Cane DE, Khosla C. Biochemistry. 2003;42:200–207. doi: 10.1021/bi0268100. [DOI] [PubMed] [Google Scholar]
- 96.Hutchinson CR. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3336–3338. doi: 10.1073/pnas.96.7.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato Y, Bai L, Xue Q, Revill WP, Yu TW, Floss HG. J. Am. Chem. Soc. 2002;124:5268–5269. doi: 10.1021/ja0127483. [DOI] [PubMed] [Google Scholar]
- 98.Hans M, Hornung A, Dziarnowski A, Cane DE, Khosla C. J. Am. Chem. Soc. 2003;125:5366–5374. doi: 10.1021/ja029539i. [DOI] [PubMed] [Google Scholar]
- 99.Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Nat. Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 100.Jiang M, Zhang HR, Park SH, Li Y, Pfeifer BA. Metab. Eng. 2013;20:92–100. doi: 10.1016/j.ymben.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 101.Desai RP, Leaf T, Hu ZH, Hutchinson CR, Hong A, Byng G, Galazzo J, Licari P. Biotechnol. Prog. 2004;20:38–43. doi: 10.1021/bp034171u. [DOI] [PubMed] [Google Scholar]
- 102.Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. Biochemistry. 2002;41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- 103.Murli S, Kennedy J, Dayem LC, Carney JR, Kealey JT. J. Ind. Microbiol. Biotechnol. 2003;30:500–509. doi: 10.1007/s10295-003-0073-x. [DOI] [PubMed] [Google Scholar]
- 104.Zhang HR, Skalina K, Jiang M, Pfeifer BA. Biotechnol. Prog. 2012;28:292–296. doi: 10.1002/btpr.702. [DOI] [PubMed] [Google Scholar]
- 105.Jiang M, Fang L, Pfeifer BA. Biotechnol. Prog. 2013;29:862–869. doi: 10.1002/btpr.1759. [DOI] [PubMed] [Google Scholar]
- 106.Serizawa N. Biotechnol Annu Rev. 1996;2:373–389. doi: 10.1016/s1387-2656(08)70017-6. [DOI] [PubMed] [Google Scholar]
- 107.Chen CH, Hu HY, Cho YC, Hsu WH. Curr. Microbiol. 2006;53:108–112. doi: 10.1007/s00284-005-0276-7. [DOI] [PubMed] [Google Scholar]
- 108.Matsuoka T, Miyakoshi S, Tanzawa K, Nakahara K, Hosobuchi M, Serizawa N. Eur. J. Biochem. 1989;184:707–713. doi: 10.1111/j.1432-1033.1989.tb15070.x. [DOI] [PubMed] [Google Scholar]
- 109.Fujii Y, Norihisa K, Fujii T, Aritoku Y, Kagawa Y, Sallam KI, Johdo O, Arisawa A, Tamura T. Biochem. Biophys. Res. Commun. 2011;404:511–516. doi: 10.1016/j.bbrc.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 110.McLean KJ, Hans M, Meijrink B, van Scheppingen WB, Vollebregt A, Tee KL, van der Laan JM, Leys D, Munro AW, van den Berg MA. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2847–2852. doi: 10.1073/pnas.1419028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Glick BR. Biotechnol. Adv. 1995;13:247–261. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- 112.Watson JD, Crick FH. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 113.Monod J, Jacob F. Cold Spring Harb. Symp. Quant. Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 114.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. J. Biol. Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. J. Biol. Eng. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Silva-Rocha R, Martinez-Garcia E, Calles B, Chavarria M, Arce-Rodriguez A, de Las Heras A, Paez-Espino AD, Durante-Rodriguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. Nucleic Acids Res. 2013;41:D666–675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jones KL, Kim SW, Keasling JD. Metab. Eng. 2000;2:328–338. doi: 10.1006/mben.2000.0161. [DOI] [PubMed] [Google Scholar]
- 118.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ellis T, Wang X, Collins JJ. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 121.Cox RS, 3rd, Surette MG, Elowitz MB. Mol. Syst. Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kinkhabwala A, Guet CC. PLoS One. 2008;3:e2030. doi: 10.1371/journal.pone.0002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Blazeck J, Garg R, Reed B, Alper HS. Biotechnol. Bioeng. 2012;109:2884–2895. doi: 10.1002/bit.24552. [DOI] [PubMed] [Google Scholar]
- 124.Liang J, Ning JC, Zhao H. Nucleic Acids Res. 2013;41:e54. doi: 10.1093/nar/gks1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Egbert RG, Klavins E. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16817–16822. doi: 10.1073/pnas.1205693109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kotter P, Weigand JE, Meyer B, Entian KD, Suess B. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Salis HM, Mirsky EA, Voigt CA. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 130.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 131.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr., Romesberg FE. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bayer TS, Widmaier DM, Temme K, Mirsky EA, Santi DV, Voigt CA. J. Am. Chem. Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 133.Kudla G, Murray AW, Tollervey D, Plotkin JB. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Goodman DB, Church GM, Kosuri S. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 135.Ohto C, Muramatsu M, Obata S, Sakuradani E, Shimizu S. Appl. Microbiol. Biotechnol. 2010;87:1327–1334. doi: 10.1007/s00253-010-2571-x. [DOI] [PubMed] [Google Scholar]
- 136.Wilner OI, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. Nat. Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 137.Collins CH, Yokobayashi Y, Umeno D, Arnold FH. Curr. Opin. Biotechnol. 2003;14:371–378. doi: 10.1016/s0958-1669(03)00091-0. [DOI] [PubMed] [Google Scholar]
- 138.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 139.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Nat. Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 140.Jennewein S, Wildung MR, Chau M, Walker K, Croteau R. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9149–9154. doi: 10.1073/pnas.0403009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lian J, Chao R, Zhao H. Metab. Eng. 2014;23:92–99. doi: 10.1016/j.ymben.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 142.Lian J, Si T, Nair NU, Zhao H. Metab. Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 143.Avalos JL, Fink GR, Stephanopoulos G. Nat. Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 145.Shibata S. J. Korean Med. Sci. 2001;16(Suppl):S28–37. doi: 10.3346/jkms.2001.16.S.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yao YF, Wang CS, Qiao JJ, Zhao GR. Metab. Eng. 2013;19:79–87. doi: 10.1016/j.ymben.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 147.Du J, Yuan Y, Si T, Lian J, Zhao H. Nucleic Acids Res. 2012;40:e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grabinska K, Palamarczyk G. FEMS Yeast Res. 2002;2:259–265. doi: 10.1016/S1567-1356(02)00110-1. [DOI] [PubMed] [Google Scholar]
- 149.Strijbis K, Distel B. Eukaryot. Cell. 2010;9:1809–1815. doi: 10.1128/EC.00172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. Metab. Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 151.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 152.Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang X, Chen X, Yang Y. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 154.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 155.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD. Nat. Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 156.Chou HH, Keasling JD. Nat. Commun. 2013;4:2595. doi: 10.1038/ncomms3595. [DOI] [PubMed] [Google Scholar]
- 157.Si T, Xiao H, Zhao H. Biotechnol. Adv. 2014 doi: 10.1016/j.biotechadv.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.DiCarlo JE, Conley AJ, Penttila M, Jantti J, Wang HH, Church GM. ACS Synth. Biol. 2013;2:741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Y, Gu Q, Lin Z, Wang Z, Chen T, Zhao X. ACS Synth. Biol. 2013;2:651–661. doi: 10.1021/sb400051t. [DOI] [PubMed] [Google Scholar]
- 161.Si T, Luo Y, Bao Z, Zhao H. ACS Synth. Biol. 2014 doi: 10.1021/sb500074a. [DOI] [PubMed] [Google Scholar]
- 162.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 163.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 164.Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant Methods. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Farzadfard F, Perli SD, Lu TK. ACS Synth. Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H. ACS Synth. Biol. doi: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- 169.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, 3rd, Smith HO. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 170.Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA, Smith HO, Venter JC, Glass JI. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 171.Karas BJ, Molparia B, Jablanovic J, Hermann WJ, Lin YC, Dupont CL, Tagwerker C, Yonemoto IT, Noskov VN, Chuang RY, Allen AE, Glass JI, Hutchison CA, Smith HO, Venter JC, Weyman PD. J. Biol. Eng. 2013:7. doi: 10.1186/1754-1611-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai J, Lindstrom DL, Boeke AC, Gottschling DE, Chandrasegaran S, Bader JS, Boeke JD. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, Cai Y, Zeller K, Agmon N, Han JS, Hadjithomas M, Tullman J, Caravelli K, Cirelli K, Guo Z, London V, Yeluru A, Murugan S, Kandavelou K, Agier N, Fischer G, Yang K, Martin JA, Bilgel M, Bohutski P, Boulier KM, Capaldo BJ, Chang J, Charoen K, Choi WJ, Deng P, DiCarlo JE, Doong J, Dunn J, Feinberg JI, Fernandez C, Floria CE, Gladowski D, Hadidi P, Ishizuka I, Jabbari J, Lau CY, Lee PA, Li S, Lin D, Linder ME, Ling J, Liu J, Liu J, London M, Ma H, Mao J, McDade JE, McMillan A, Moore AM, Oh WC, Ouyang Y, Patel R, Paul M, Paulsen LC, Qiu J, Rhee A, Rubashkin MG, Soh IY, Sotuyo NE, Srinivas V, Suarez A, Wong A, Wong R, Xie WR, Xu Y, Yu AT, Koszul R, Bader JS, Boeke JD, Chandrasegaran S. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dymond J, Boeke J. Bioeng. Bugs. 2012;3:168–171. doi: 10.4161/bbug.19543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jovicevic D, Blount BA, Ellis T. Bioessays. 2014;36:855–860. doi: 10.1002/bies.201400086. [DOI] [PubMed] [Google Scholar]
- 176.Brenner K, You L, Arnold FH. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 177.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Nat. Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Song H, Ding MZ, Jia XQ, Ma Q, Yuan YJ. Chem. Soc. Rev. 2014;43:6954–6981. doi: 10.1039/c4cs00114a. [DOI] [PubMed] [Google Scholar]
- 180.Yeh HH, Chang SL, Chiang YM, Bruno KS, Oakley BR, Wu TK, Wang CCC. Org. Lett. 2013;15:756–759. doi: 10.1021/ol303328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yin WB, Chooi YH, Smith AR, Cacho RA, Hu YC, White TC, Tang Y. ACS Synth. Biol. 2013;2:629–634. doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sanchez JF, Somoza AD, Keller NP, Wang CCC. Nat. Prod. Rep. 2012;29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wiemann P, Keller NP. J. Ind. Microbiol. Biotechnol. 2014;41:301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 184.Ishiuchi K, Nakazawa T, Ookuma T, Sugimoto S, Sato M, Tsunematsu Y, Ishikawa N, Noguchi H, Hotta K, Moriya H, Watanabe K. ChemBioChem. 2012;13:846–854. doi: 10.1002/cbic.201100798. [DOI] [PubMed] [Google Scholar]
- 185.Punya J, Tachaleat A, Wattanachaisaereekul S, Haritakun R, Boonlarppradab C, Cheevadhanarak S. Fungal Genet. Biol. 2013;50:55–62. doi: 10.1016/j.fgb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 186.Wasil Z, Pahirulzaman KAK, Butts C, Simpson TJ, Lazarus CM, Cox RJ. Chemical Science. 2013;4:3845–3856. [Google Scholar]
- 187.Xu W, Cai X, Jung ME, Tang Y. J. Am. Chem. Soc. 2010;132:13604–13607. doi: 10.1021/ja107084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang M, Beissner M, Zhao HM. Chem. Biol. 2014;21:257–263. doi: 10.1016/j.chembiol.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CC, Oakley BR. J. Am. Chem. Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Scharf DH, Brakhage AA. J. Biotechnol. 2013;163:179–183. doi: 10.1016/j.jbiotec.2012.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.