Abstract
Recent studies have convincingly linked gut microbiota to traits relevant to atherosclerosis, such as insulin resistance, dyslipidemia and inflammation, and have revealed novel disease pathways involving microbe-derived metabolites. These results have important implications for understanding how environmental and genetic factors act together to influence cardiovascular disease (CVD) risk. Thus, dietary constituents are not only absorbed and metabolized by the host but they also perturb the gut microbiota, which in turn influence host metabolism and inflammation. It also appears that host genetics helps to shape the gut microbiota community. Here, we discuss challenges in understanding these interactions and the role they play in CVD.
Keywords: Host-gut microbiota interactions, Mucosal immune system, Trimethylamine-N-oxide, Germ-free mice, Polyphenolic metabolites, Systems biology approach
Introduction
Studies of the role of microbes in common diseases have exploded in recent years, due in part to technical advances such as the use of next-generation sequencing to profile samples. Such studies have now linked gut microbiota to ulcers, colitis, obesity, diabetes, cancer, fatty liver, autism, kidney stones, cardiovascular disease (CVD) and other disorders (reviewed in [1–3]). Among the most convincing of these associations are those for atherosclerosis and its risk factors, with experimental evidence of causal interactions in insulin resistance, bile acid metabolism, inflammation, and obesity. In addition, studies of certain microbiota derived metabolites have recently revealed novel pathways affecting atherosclerosis and cholesterol metabolism. A particularly exciting aspect of this work is the possibility for the development of new therapeutic interventions as well as diagnosis.
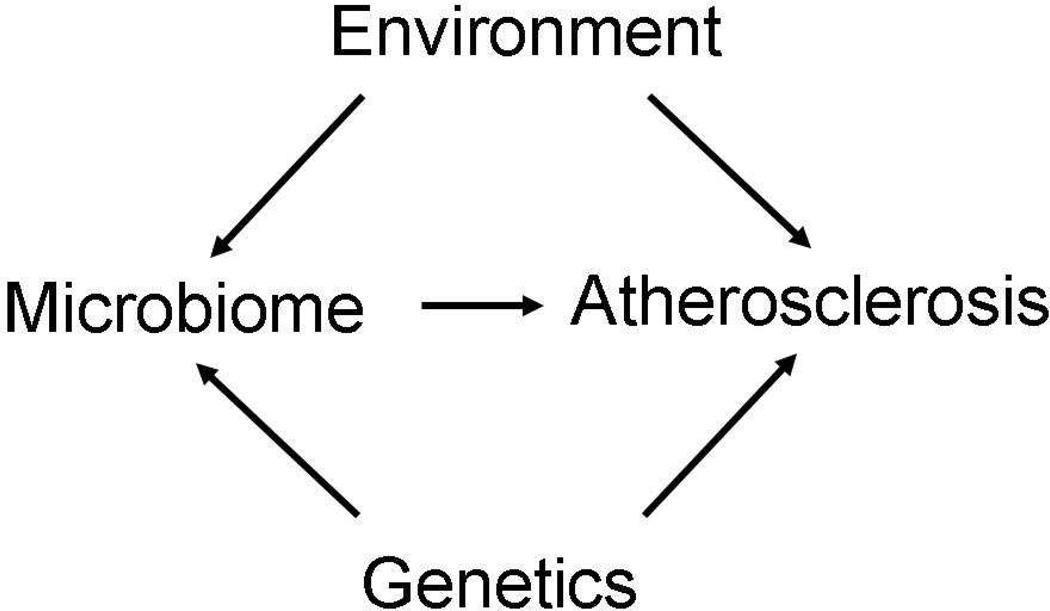
Our group, among others, has been interested in dissecting genetic and environmental interactions in atherosclerosis, and these recent findings introduce an additional key component, namely, the microbiota, that must be considered. Thus, in analyzing how a particular diet or genetic background influences the disease process, one must not only consider direct effects but also how the diet or genetic background perturbs the microbiome (Fig. 1). The great complexity of the gut microbiome and its sensitivity to environmental factors make such analyses challenging.
Figure 1. Genetic and environmental interactions in atherosclerosis: Role of the gut microbiota.
Environmental factors, particularly the diet, and host genetics can contribute to atherosclerosis and other CVD either directly or by perturbing microbiota.
Our review is divided into three parts. First, we briefly summarize some of the tools for studying microbiota, with an emphasis on gut microbiota. Second, we review the literature implicating microbes in atherosclerosis and its risk factors. And, third, we discuss approaches to unraveling the complex interactions between gut microbiota composition, diet, and genetics.
Studying Microbiota-Host Interactions
Human skin and mucosal surfaces and the gut are colonized by an untold number of microorganisms, comprised of bacteria, archaea, viruses, fungi, and other protozoans, collectively known as the microbiota (their genes are known as the microbiome). The human gut, for example, is colonized by about 100 trillion bacteria including at least 1000 distinct species [4]. The bacteria tend to associate with one another and the host in a mutualistic/commensal or opportunistic/parasitic manner. In the case of gut microbiota, it is clear that a symbiotic relationship has evolved. The host provides the microbiota with nutrients while the microbiota make possible the digestion of complex carbohydrates, balance immune and metabolic functions, and provide protection against opportunistic pathogens [4, 5]. The compositions of these microbial communities vary widely in the population due to differences in environmental factors and host genetic factors and, as discussed below, these differences appear to contribute to CVD.
The microbiota of an individual are “seeded” at birth through maternal physical contact, becoming more diverse within weeks and eventually stabilizing in adolescents or young adults. The gut microbiota play an important role in intestinal development and in shaping the immune system. Birthing, breastfeeding and weaning methods significantly impact the gut microbiota profiles of infants. Infants delivered by Caesarean section have bacterial composition more similar to maternal skin, unlike vaginally delivered infants, who have more Lactobacillus [3, 6]. Through development the predominantly aerobic bacteria are replaced by anaerobes, until approximately 2–3 years of age, where the profile more closely resembles that of an adult [7].
A very small fraction of gut microbiota have been cultured or studied in any detail. Most are obligate anaerobes rapidly killed by exposure to air and they live as communities highly in interdependent on other species. The distribution of microbes throughout the gut is highly structured and specialized in biological functions, varying considerably between different parts of gastrointestinal tract. In studies of mice the most commonly used sites are the cecum and different parts of small intestine (duodenum, jejunum and ileum), whereas in human studies stool samples are usually used.
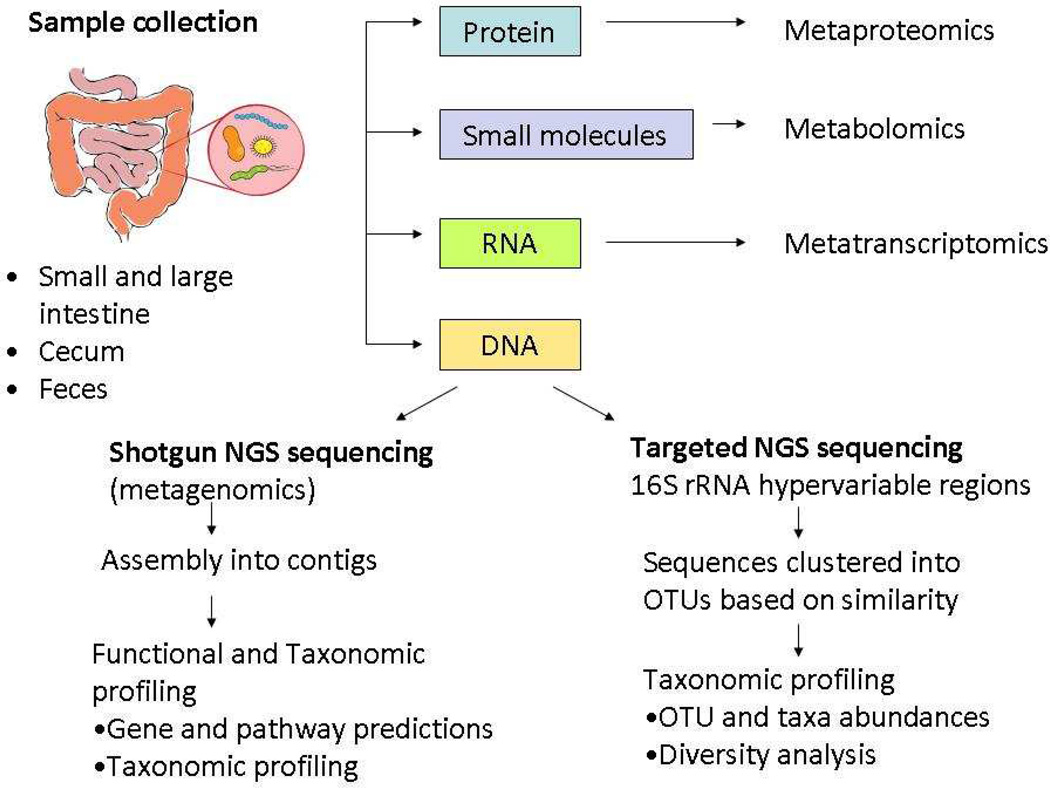
Because it is challenging to culture gut microbiota, culture-independent genomic methods have greatly stimulated this area of research (Fig. 2). Particularly important has been the use of next-generation sequencing for taxonomic profiling of samples, together with the development of bioinformatics techniques. Taxonomic profiling has revealed enormous interindividual variability due to factors such as diet [8] and other environmental exposures and factors such as age [7], sex [9], and host genotype [10–13]. Shotgun metagenomic sequencing, in which total DNA is extracted and sequenced, has the advantage of providing data on functional potential present in a given sample. Despite the considerable variability of gut microbiota composition, metagenomic shotgun sequencing has shown that these diverse communities share a core set of gene functions [14]. In concert with sequencing methods, new approaches for assessing the functionality of bacteria have been developed, such as screening for metabolites derived from microbiota [15].
Figure 2. Overview of culture-independent methods for microbial analysis.
High-throughput analysis of gastrointestinal tract microbiota via sequencing of DNA using targeted or shotgun sequencing or using other “omics” approaches. NGS, next generation sequencing; OTU, operational taxonomic units.
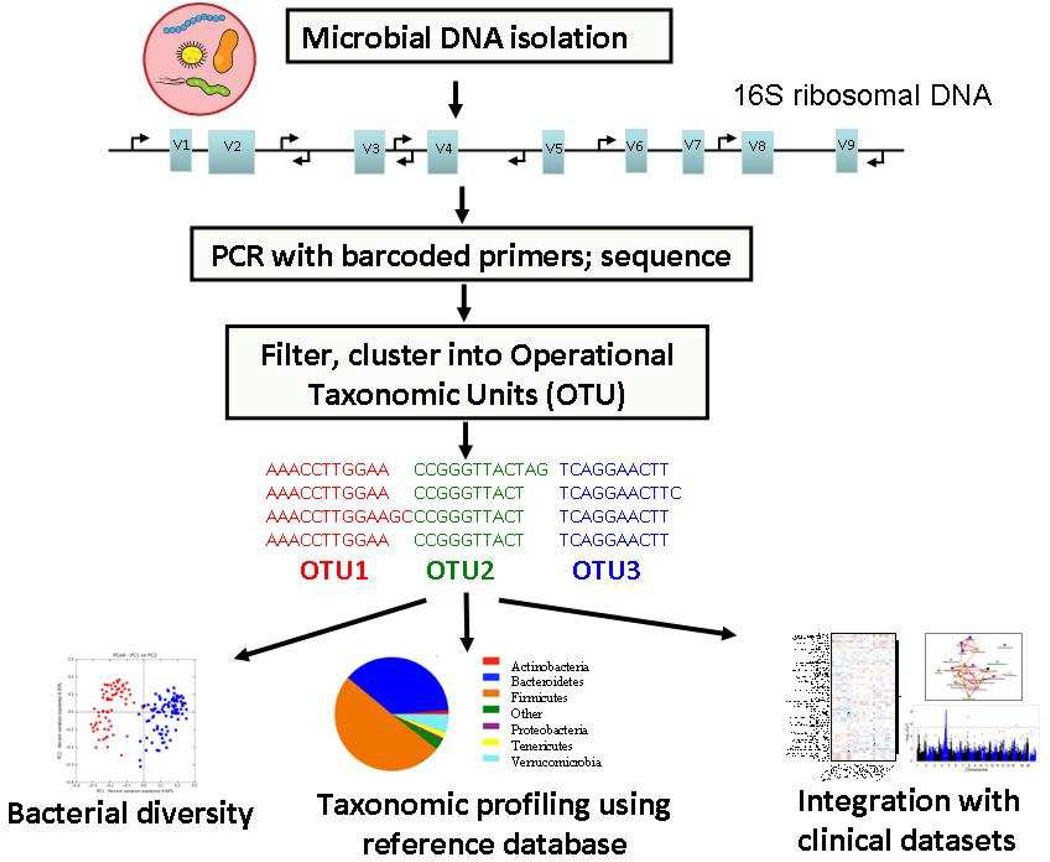
The most common approach to taxonomic profiling of gut microbiota is to amplify and then sequence hypervariable regions of bacterial 16S rRNA genes followed by clustering of the sequences into Operational Taxonomic Units (OTUs) (Fig. 3). Several analytic tools have been developed for analyzing next generation of DNA sequence data from targeted 16S and metagenomic analysis, such as QIIME [16], mothur [17] and VAMPS [18]. These enable an accurate downstream analysis of organism distribution, which can then be correlated with disease status or other phenotypes of interest.
Figure 3. Schematic representation of pipeline for profiling gut microbiota communities using 16S rRNA sequencing.
First, DNA is extracted from samples, followed by amplification of hypervariable region from 16S rRNA gene using specific barcoded primers. After next-generation sequencing (NGS), the raw input sequences are assigned to samples according to barcodes, quality filtered, clustered according to sequence identity into operational taxonomic units (OTUs), compared against a reference database to establish taxonomic identity. Once taxon information are obtained, various “pipelines” enable analysis describing community structure, diversity or biologically important associations.
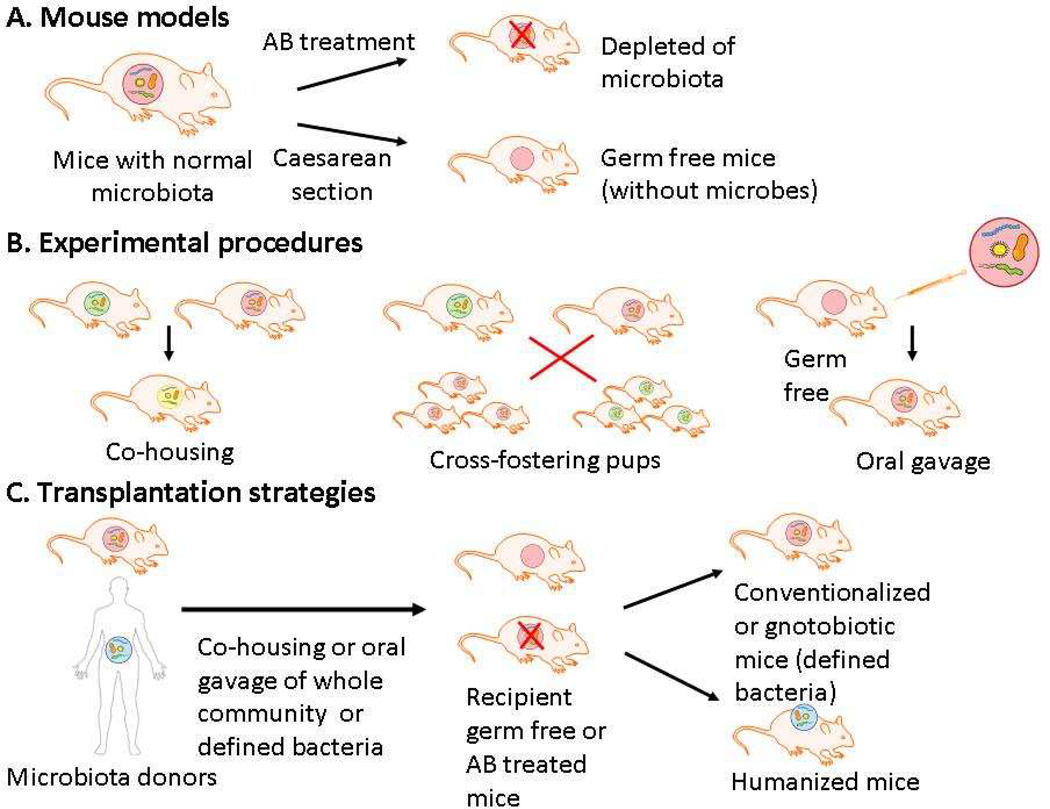
Mice harbor microbes that roughly resemble those found in humans and are the most widely used organism for experimental studies (reviewed in [19]). Mouse models allow perturbations in gut microbiota to be studied in highly controlled environments and thus are useful in assessing causality of the complex microbiota-host interactions and in developing mechanistic hypothesis. Figure 4 summarizes different experimental procedures with mice that are commonly used for studying gut microbiota. Germ-free mice or mice previously treated with antibiotics are often used for transplantation with human microbiota (humanized mice) or other microbes (conventionalized or gnotobiotic mice). Germ free mice are raised in the absence of any resident microorganisms, representing a defined assay system for studying the impact on the host of colonization of the gut by microbiota members. In addition, mice can be cross-fostered following Caesarian delivery, and since they are coprophagic, simply co-housing two strains allows transfer of gut microbiota.
Figure 4. Use of mice for analyses of the role of commensal microbes.
Schematic representation of different mouse models (A), experimental procedures (B) and transplantation strategies (C) for studying gut microbiota communities in mice.
An exciting aspect of studies of the gut microbiome in disease is the possibility of manipulating the microbiota composition through alterations in diet and the use of probiotics and prebiotics. “Probiotics” refers to the administration of live dietary supplements such as Lactobacilli or Bifido bacteria in yogurt. These can, for example, help alleviate symptoms of lactose intolerance [20]. Prebiotics are nondigestible food ingredients such as oligomers of fructose (fructans) and fibers such as Psyllium husk that can be utilized by selective microbiota. Such prebiotics have been shown to affect the absorption of minerals and the levels of plasma lipids [21]. Administration of fructans in mice on a high fat diet has been shown to decrease fat storage in white adipose and liver, improve glycemic control, and decrease systemic inflammation [22]. A high fat diet has been shown to downregulate the intestinal expression of certain antimicrobial peptides, such as Reg3g, and prebiotic treatment was shown to reverse this, providing one possible explanation for the effect on the microbiome composition [23].
The complexities of investigating the role of gut microbiota in disease are illustrated by two studies of atherosclerosis in germ-free mice. In an early study, Wright and colleagues [24] compared atherosclerosis development in germ-free versus conventional apolipoprotein E null (Apoe−/−) mice maintained on a Western-type high cholesterol, high fat diet. No differences in atherosclerosis development were observed in a large number of mice. Recently, however, Stepankova and colleagues [25] observed increased atherosclerosis in germ-free Apoe−/− mice consuming a standard diet when compared with conventional Apoe−/− mice, but did not see any differences between the two groups fed a Western diet. Their data indicate an overall protective effect of gut microbiota and suggest that this may be obscured by a high fat diet. Since gut microbiota compositions vary considerably between vivaria, even when the mice are maintained on similar diets, comparisons of studies in different labs are problematic. With respect to diets it is remarkable how even very small differences in dietary components can dramatically affect the microbiota composition (for example, refs. [26, 27]). Also, since microbiota appear to affect atherosclerosis by a number of distinct pathways (see below), some of which are pro-atherogenic and others anti-atherogenic, it is quite possible that these cancel each other in such comparisons as germ-free versus conventional mice. Complexities such as these can lead to problems of interpretation and to overinterpretation, as discussed by Hanage [28].
Studies Implicating Microbiota in Atherosclerosis
Chronic infection and atherosclerosis
The focus of this review is on “commensal” gut microbiota but we briefly discuss here evidence that pathogenic bacteria or viruses can promote CVD. Such evidence was first presented over a century ago based on studies in rabbits showing pathogen enhancement of fatty-streak lesions [29] and, since then, the concept has received support from numerous epidemiological and experimental studies [30–33]. Among the pathogens that have been implicated are Helicobacter pylori, Chlamydia pneumoniae, Porphyromonas gingivalis, and Aggregatibacter actinomycetemcomitans, hepatitis A virus, and various herpesviridae such as cytomegaloviruses. For example, the initial study implicating Chlamydia in atherosclerosis was the finding in the Helsinki Heart study that patients with CVD were more likely to exhibit antibodies to the bacteria [32]. A variety of subsequent experimental studies in model organisms supported a causal relationship [34]. Thus it was shown that C. pneumoniae can infect vascular cells and that such infections promote foam cell formation, leukocyte recruitment, smooth muscle cell proliferation, and lesion progression in mouse and rat models [35–39].
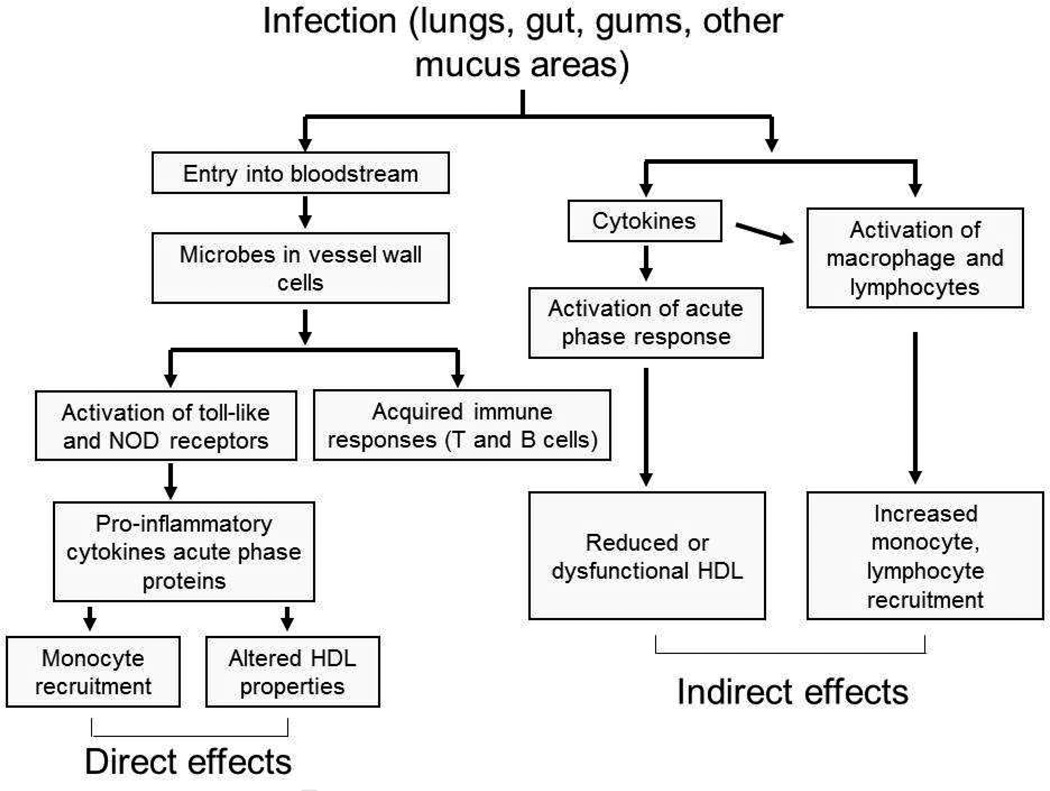
Chronic infections could contribute to atherosclerosis directly by invading vascular cells or leukocytes or indirectly by contributing to inflammation from distal sites such as the lungs or the gums [40] (Fig. 5). A variety of studies in humans have localized pathogens or their products to atheroma [41, 42], and in vitro studies have shown that infection of human endothelial cells or monocytes with pathogens such as C. pneumoniae can upregulate secretion of cytokines such as MCP-1, IL-8, IL-6 and TNF-α [43, 44]. Some recent data indicate that cholesterol crystals can activate the NLRP3 inflammasome [45–47], resulting in the release of cathepsins and an increase in mitochondrial reactive oxygen species. It is plausible that this could be aggravated by microbial products [45]. On the other hand, it is likely that infections can have systemic effects that promote atherogenesis. For example, proinflammatory cytokines such as IL-6 and serum amyloid A have been associated with atherosclerosis [48]. Campbell and colleagues [49] have recently shown that a single intranasal inoculation of C. pneumoniae in mice resulted in increases in plasma levels of a variety of cytokines as well as a decrease in the levels of the protective HDL protein paraoxonase. These investigators also showed that this was accompanied by an increase in the frequency of intraplaque hemorrhage in older Apoe−/− mice.
Figure 5. Chronic infection and atherosclerosis.
Microbial infections can influence atherosclerosis either directly, by entering the bloodstream and the vessel wall, or indirectly, by stimulating the production of cytokines or aspects of the immune system.
Such studies prompted a series of clinical trials with antibiotic interventions, including a number of large, randomized prospective studies of stable CVD (WIZARD, ACES, CLARICOR), acute coronary syndrome (PROVE-IT-TIMI), or peripheral artery disease (PROVIDENCE-1). None of the larger studies (thousands of participants) demonstrated any benefits of antibiotic treatment and in some studies treatment increased cardiovascular mortality (reviewed in [50]).
While these results have challenged the infection hypothesis, they clearly do not disprove it. Campbell and Rosenfeld [50] have noted several points that could explain the findings: 1. “End stage disease” may not be modifiable; 2. The pathogens may not be susceptible to the chosen antibiotics; 3. The events being measured (in some studies) such as plaque rupture may be independent of plaque progression; 4. Some microbes such as Chlamydia undergo developmental cycles giving rise to metabolically inactive forms that are not susceptible to antibiotics. Given the recent studies of the importance of gut microbiota in CVD traits, it is also possible that the antibiotic treatment negatively affected protective bacteria in the gut.
Since the development of next generation sequencing methods for determining the composition of microbiota a number of association studies have been reported for human oral, gut, and plaque microbiota [51]. For example, in one study, the microbial diversity of atherosclerotic endarterectomy plaques (N=35) was evaluated by sequence analysis of 16s ribosomal DNA. About one third contained bacterial DNA and 23 different species/phylotypes were detected, with the periodontal pathogen Aggregatibacter Actinomycetemcomitans being detected in 20% of samples [52].
Given the population association studies and supportive experimental studies in animal models, it is likely that chronic infection does contribute to CVD, despite the negative antibiotic trials. It would seem that further investigations as well as consideration of more specific therapeutic approaches are warranted.
Gut microbiota and atherosclerosis-related traits
It has long been known that gut bacteria are required for certain biochemical transformations such as the secondary modification of bile acids and the production of vitamin K and certain B vitamins. Early studies with germ-free rats showed that, in addition, gut microbiota contribute importantly to harvesting energy, as conventionally raised animals required 30% less caloric intake for maintenance of body weight than the germ-free counterparts [53]. Subsequent studies showed that bacteria play a role in the utilization of starch and various plant polysaccharides as well as several host-derived glycans such as chondriotin sulfate, mucin and heparin [21]. More recent studies showed that the gut microbiota of genetically obese mice differed in composition from their lean littermates in the relative abundances of the most abundant phyla, Bacteroidetes and Firmicutes, with the obese mice having increased capacity to harvest energy from the diet [54]. Furthermore, colonization of germ-free mice with human microbiota from obese individuals resulted in increased body fat as compared to microbiota from lean individuals [55, 56]. Over the past decade, a large body of work has supported a role for gut microbiota in many metabolic traits and in inflammation, and experimental studies, mostly in mice, have now provided substantial mechanistic information (reviewed in [15, 22]). Recently, some novel microbiota-derived metabolites have been shown to contribute to atherosclerosis and cholesterol metabolism. These are discussed in turn below
Gut microbiota and Short Chain Fatty Acids (SCFAs)
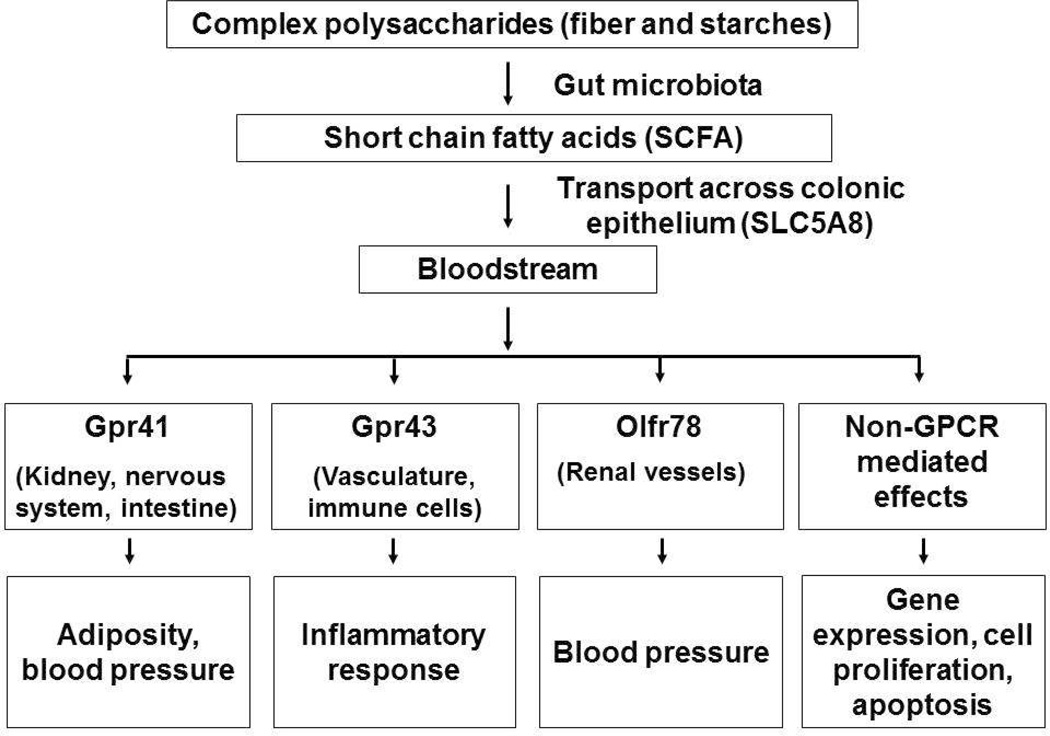
The primary products of bacterial hydrolysis are monosaccharides which are fermented to various SCFAs such as acetate, butyrate, and proprionate (reviewed in [21]). The SCFAs are transported across the mucosal epithelium by specific monocarboxylate transporters and they then act as regulatory molecules or substrates [57] (Fig. 6). The sodium-coupled monocarboxylase transporter SLC5A8 appears to be primarily responsible for SCFAs transport, although there is evidence for other transporters as well. SLC5A8 is expressed in the apical membrane of the colon and it transports SCFAs and Na+ with a 1:3 stoichiometry [58]. Certain SCFAs are ligands for the G-coupled protein receptors, GPR43 and 41. These receptors are widely expressed and have a role in the regulation of energy metabolism, immunity and blood pressure [22]. For example, activation of GRP43 on adipocytes by acetate and propionate results in the inhibition of lipolysis [59, 60]. Recently, an olfactory receptor (Olfr78) in kidney was shown to respond to SCFAs and influence renin secretion and blood pressure [61]. The different receptors, their sites of expression, and the traits affected are presented in Figure 6.
Figure 6. Microbiota derived Short Chain Fatty Acids (SCFAs) and atherosclerosis.
Microbiota can degrade and metabolize various complex carbohydrates, complementing the limited saccharolytic diversity encoded by the human genome. This results in the production of certain SCFAs which enter the circulation and activate receptors in a variety of all types to affect traits relevant to atherosclerosis.
Gut microbiota and the immune system
Gut microbiota interact with elements of both the adaptive and innate immune systems, contributing importantly to the development of the immune system and the maintenance of immune homeostasis (reviewed in [4, 5, 62]. Particularly important in mediating the pro-inflammatory effects of microbiota are the toll-like receptors (TLRs) and NOD-like receptors (NLRs) that recognize bacterial products such as lipopolysaccharides and peptidoglycans. Studies with mouse models have implicated several innate immune receptors, including TLR4, TLR2, NOD1, and NOD2, in various metabolic syndrome traits including obesity, insulin resistance, hepatic steatosis, and inflammation [62]. Similar studies with mice have implicated TLR4, TLR3, and TLR2 directly in atherosclerosis [63]. A series of studies have also linked inflammasome sensing to inflammation, obesity, and hepatic steatosis [64–66]. Inflammasomes are protein complexes which recruit and activate caspases, triggering the formation of cytokines IL-1β and IL-18 from their precursors. Inflammasome signaling can be initiated by a range of elements, including pathogenic infections, macromolecules, cytokines or reactive oxygen species [64, 67–69]. SCFAs can also significantly impact the immune system, in part by activating T-regulatory cells to suppress inflammation [70]. The immune system, in turn, shapes the composition of the gut microbiota through secretion of antibacterial proteins such as defensins, IgA, and Rog3g [4, 5]
Over-nutrition appears to contribute to low grade inflammation, one of the characteristics of metabolic syndrome [71, 72]. This may be mediated in part by nutrient components such as palmitate that can activate certain TLRs and NLRs [62]. The diet not only affects the gut microbiome [73] but can also affect the structure of the gut mucosa. Increased intestinal permeability and inflammation has been reported in patients with heart failure [74] and mice fed “Western” diets [75]. Such changes in barrier function would likely promote systemic inflammation by increasing the release of bacterial products into the circulation [76].
Gut microbiota and bile acid metabolism
Gut microbiota have long been known to contribute to the metabolism of bile acids, a class of lipids that are secreted into the intestine to solubilize lipids for absorption (Fig. 7). Bile acids are commonly divided into two categories, primary and secondary. The primary bile acids are synthesized by oxidation of cholesterol in the liver, whereas secondary bile acids are produced from primary bile acids through the action of gut microbiota (Fig. 7). The precise functions of the primary (chenodeoxycholic acid and cholic acid) and secondary (deoxycholic acid) bile acids are still poorly understood. Bile acids are of central importance in lipid metabolism since they are the major route for excretion of cholesterol. Following secretion into the intestine, about 95% of bile acids are reabsorbed for recycling (the enterohepatic circulation), with about 5% being lost. In addition to functioning in lipid solubilization, they signal through several receptors, including the farnesoid X receptor (FXR) and the bile acid responsive G-protein coupled receptor (TGR5). Such signaling influences not only lipid metabolism but also a wide range of physiologic processes [77]. Bile acids are also likely to shape the gut microbiome through their detergent and signaling properties [78–80].
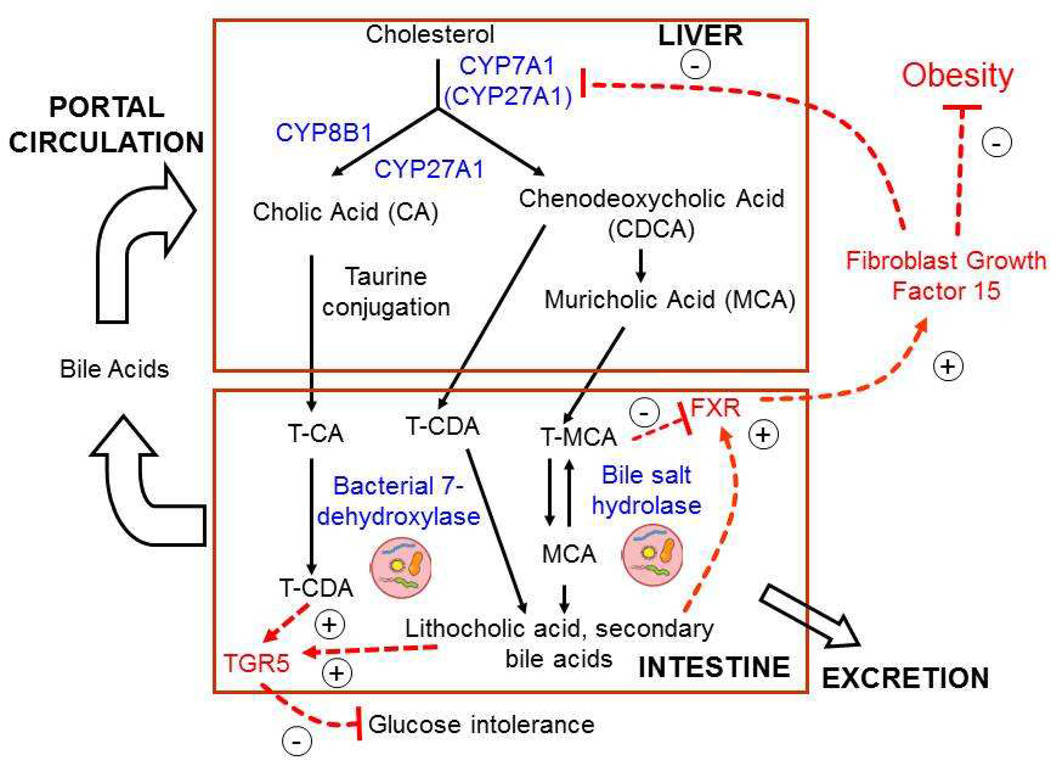
Figure 7. Bile acid metabolism, microbiota, and atherosclerosis risk factors.
The classic rate limiting bile acid pathway is catalyzed by CYP7A1, which converts cholesterol to 7-hydroxycholesterol. This is then converted to an intermediate by CYP8B1 and CYP27A1 to cholic acid (CA) and chenodeoxycholic acid (CDCA). In the alternative pathway, CYP27A1 converts cholesterol to 27-hydroxycholesterol, which is then converted to CDCA. In mouse liver, most CDCA is converted to muricholic acids (MCA), while in humans MCA is only found in trace amounts. Newly synthesized bile acids are conjugated to glycine or taurine and then secreted across the apical membrane of hepatocytes for storage in the gall bladder. In the intestine bacterial 7-dehydroxylase converts CA to CDA. CDCA is converted to either MCA or CDA, and unabsorbed CDCA can be metabolized by bacteria to lithocholic acid. The bacterial bile salt hydrolase (BSH), produced by Lactobaccili and other bacteria, disconjugates taurine-MCA prior to conversion to secondary bile acids. The FXR receptor plays a major role in regulating the negative feedback production of bile acids. FXR is highly expressed in intestine, and activation by bile acids results in increased expression of fibroblast growth factor 15 (FGF15), which then acts as an endocrine hormone to inhibit CYP7A1 expression. TGR5, on the other hand, increases secretion of the gut incretin GLP-1 [133], which enhances insulin production by β cells of the pancreas, lowering glucose levels [134, 135]. Mice lacking TGR5 showed blunted glycemic control [136].
Recent studies with mice have revealed surprising large effects of the gut microbiome on bile acids and also on systemic lipid metabolism and adiposity [81–83]. One important mechanism involves microbial bile salt hydrolase (BSH), involved in the deconjugation of tauro-muricholic acid (T-MCA) (Fig. 7). Thus, germ-free or antibiotic treated mice have elevated levels of T-MCA in the intestine and decreased FXR signaling [82, 84]. Recent studies showed that the T-MCA acts as an inhibitor of FXR, and that the downstream effects of the microbiome on systemic lipid metabolism and adiposity are mediated in part by FXR [84, 85]. The importance of BSH was further investigated using expression of the cloned bacterial enzyme in the gastrointestinal tract [86]. The BSH gene from two strains of Lactobacillus was cloned into the Escherichia coli strain K-12 that lacks the enzyme and germ-free mice were then monocolonized with the bacteria. As compared to BSH negative E. coli, mice colonized with the BSH expressing strains exhibited a dramatic reduction in T-MCA, increased FXR signaling in the intestine, and reduced CYP7A1 expression in the liver. Treatment of conventionally raised mice with the BSH-expressing strains.resulted in very significant reductions in weight gain on a high fat diet (46%), serum LDL-cholesterol (66%), and serum triglycerides (37%), relative to BSH-negative E. coli.
Several studies have implicated gut microbiota in weight loss following bariatric surgery [80, 87, 88] and a recent study by Ryan and colleagues [89], suggests that the benefits of bariatric surgery result in part from altered bile acid metabolism. It has been assumed that such surgeries, including vertical sleeve gastrectomy (VSG) in which about 80% of the stomach is removed, lead to weight loss and other benefits as a result of the ability of the stomach to accommodate a meal. Ryan and colleagues examined the effects of VSG surgery in mice with diet-induced obesity. They found remarkable effects on gene expression in intestinal regions as well as liver with enrichment for lipid and bile acid metabolism and pathways associated with gut microbiota, such as the glutathione metabolism and interferon signaling. To investigate a potential causal relationship between bile acid metabolism and obesity/diabetes, they investigated mice deficient in FXR. They discovered that, as compared to the wild type VSG mice, the FXR knockout VSG mice ate about 15% more calories, showed no improvement in insulin resistance measures, and failed to maintain extended weight loss. Associated with the surgery were changes in gut microbiota composition, and these were dependent in part on the expression of FXR. Several candidate taxonomic classes, including members of the genus Roseburia, were identified as microbes likely to be involved. Whether there is a causal relationship between gut microbiota and the response to VSG is still not certain (experiments with germ-free mice should address this). These findings suggest possible alternatives to the invasive and expensive bariatric surgery.
Gut microbiota and polyphenolic metabolites
Wang and colleagues [90] recently reported a novel mechanism regulating plasma lipids that involves diet, microbiota, and a microRNA (mRNA). It was previously shown that ingestion of anthocyanins, pigmented polyphenols found in fruits, vegetables, and red wine, protect against atherosclerosis, but because anthocyanins are poorly absorbed the mechanism has been unclear. These investigators identified a readily absorbable metabolite of anthocyanins, protocatechuic acid (PCA), and showed that it is a gut microbiota product. At physiologic concentrations, PCA, but not anthocyanin, reduced atherosclerosis in the Apoe−/− mouse model and promoted cholesterol efflux from macrophages by induction of ABCA1 and ABCG1. The mechanism of the induction was mediated by repression of macrophage microRNA 10b (miRNA-10b) which binds to 3’-untranslated regions on both ABCA1 and ABCG1. It is not yet known how PCA represses miRNA10b.
Gut microbiota and methylamine metabolism
Hazen and colleagues discovered that trimethylamine N-oxide (TMAO), derived from dietary choline and carnitine through the action of gut microbiota, is a major risk factor for atherosclerosis [27, 91, 92]. These investigators carried out an unbiased screen of plasma metabolites in a small case-control population to identify three metabolites of phosphatidylcholine that were associated with atherosclerosis: choline, betaine, and TMAO, with TMAO being the most strongly associated. This association was then validated in two additional studies involving about 2000 subjects presenting with cardiovascular disease phenotypes [91]. Subsequent validation involving over 4,000 subjects undergoing elective coronary angiography with a 3 year follow-up indicated that TMAO predicted a 2.5-fold increased risk of a major cardiac events independent of traditional cardiovascular risk factors (Fig. 8A) [93].
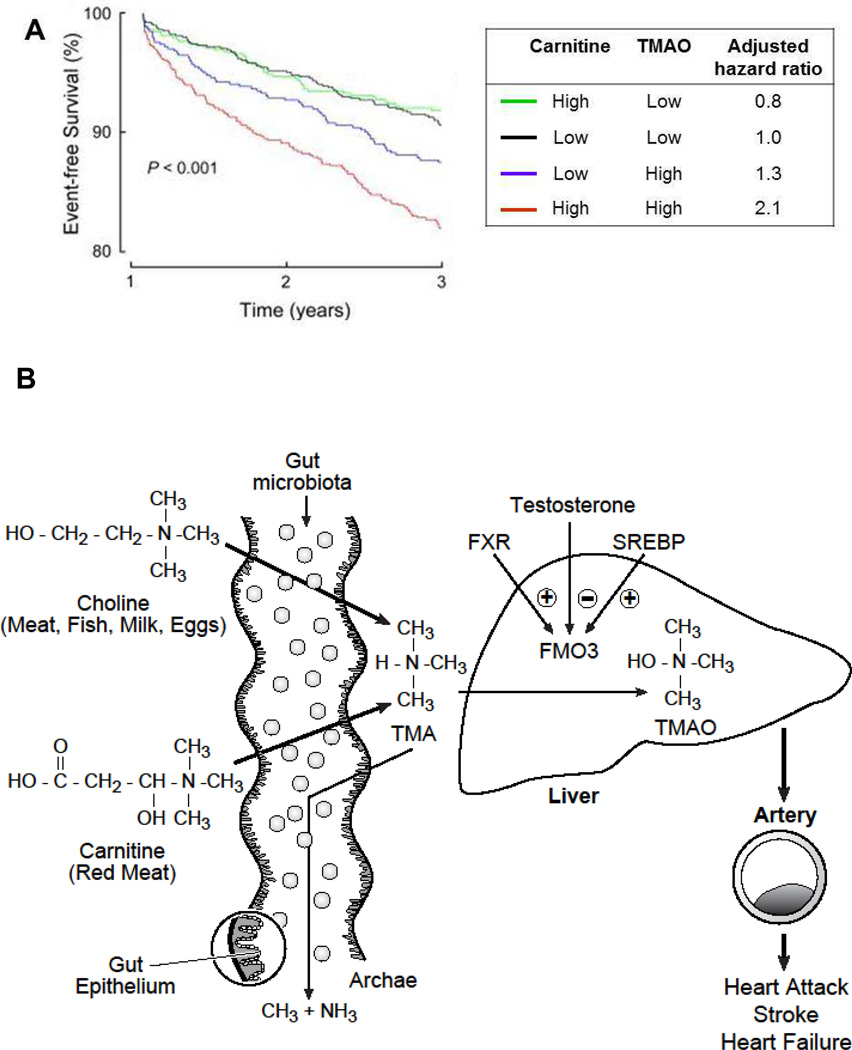
Figure 8. TMAO and atherosclerosis.
A) Survival curves showing that individuals with high plasma levels of plasma TMAO but not carnitine have a two-fold elevated risk of adverse cardiovascular events over a span of 3 years. Note that carnitine is associated with CVD events only if TMAO levels are high (Figure taken from Koeth et al., 2013, Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis, 19:576–85). B) Pathways for metabolism of TMAO. The primary sources of TMAO are dietary choline and carnitine. Choline is present in most foods as phosphatidylcholine (PC), a primary constituent of microbiomes which carnitine is particularly abundant in red meat. TMAO is also present at high levels in certain saltwater fish. In the gut, choline and carnitine are metabolized by distinct bacteria containing TMA lyases specific for choline (cutC and cutD) [137, 138]. In experimental studies in mice, most of the carnitine was first metabolized to γ- butyrobetaine and then to TMA by different bacterial species with differing distributions in the gut [26]. The TMA then enters the circulation where it is oxidized in part to TMAO by flavin monoxygenase (FMOs), primarily FMO3 in the liver [102]. TMA can also metabolize to methane by certain gut methanogenic archaea such as Methanomassiliicoccus luminyensis by reduction with H2.
The pathways for production of TMAO are summarized in Figure 8B. Dietary choline and carnitine are metabolized in the gut to trimethylamine (TMA) by several bacterial enzymes. The TMA then enters the circulation where it is transported to the liver and oxidized by flavin monooxygenases (FMOs), primarily FMO3, to TMAO. Antibiotic studies in both mice [91] and humans [93] confirmed an obligatory role of gut microbiota in the production of TMA and TMAO from phosphatidylcholine.
Based on studies in animal models, TMAO appears to be causal for atherosclerosis. In particular, feeding choline [91] or carnitine [27] to hyperlipidemic (Apoe−/−) mice increased TMAO levels and the development of atherosclerosis while prior antibiotic treatment blocked both the increase in TMAO levels and atherosclerosis [91]. Also, suppression of FMO3 in Apoe−/− mice using antisense oligonucleotides reduced both TMAO levels and atherosclerosis, although FMO3 suppression also had effects on lipid metabolism that may have confounded the interpretation [94]. TMAO administration to hyperlipidemic Apoe−/− mice increased foam cell formation, a hallmark of atherosclerosis, as well as atherosclerotic lesion development [27, 91]. TMAO has also been shown to exacerbate glucose tolerance and adipose tissue inflammation in mice fed a high fat diet [95]. A recent study revealed a strong association between TMAO levels and chronic kidney disease in a human population [96]. Certain rare individuals exhibit genetic or acquired deficiencies of FMO3 [97, 98] but it is unknown whether they have an altered incidence of CVD. It is as yet unclear how TMAO affects cholesterol metabolism or other pro-atherogenic pathways. TMA activates a G-protein coupled receptor, termed Trace Amine-Associated Receptor 5, TAAR5 [99, 100], but as yet, there is no known receptor for TMAO.
Undoubtedly, dietary differences account for a large part of variation in TMAO levels in human populations. The study of carnitine and TMAO by Koeth and colleagues [27] provides a striking example. Carnitine is found in highest concentrations in red meat and so these investigators examined a cohort of 51 omnivores and 26 vegetarians or vegans. Whereas the omnivores produced a large amount of TMAO following ingestion of a steak or labeled carnitine, TMAO production was barely detectable in the vegetarians, suggesting a shift in the microbes colonizing the gut in omnivores as compared to vegetarians. A recent human genome-wide association study of several thousand participants failed to identify any loci significantly associated with TMAO levels [101]. This does not rule out genetic factors but it suggests that environmental factors such as diet are likely to predominate in the variation of TMAO levels observed in human population studies.
The factors contributing to variations in TMAO levels have been studied in mice, where diet and other environmental factors can be more tightly controlled than in humans. Surveys of different inbred strains of mice have revealed consistent differences in circulating plasma TMAO levels [102, 103]. As in human populations, susceptibility to atherosclerosis segregated with TMAO levels in genetic crosses and associated with atherosclerosis across different strains. Genetic variations in the expression of FMO3 explained only a small fraction of the variation in TMAO levels [102]. A more important factor appeared to be differences among strains in gut microbiota composition. Recently, it was shown that transplantation of gut microbiota from a strain with high TMAO levels into a strain with low TMAO levels (following antibiotic treatment of the recipient) resulted in elevated TMAO and increased atherosclerosis [103]. Studies in mice have identified groups of bacteria associated with high TMA/TMAO production in response to choline or carnitine administration [27, 91] (Table 1). A recent study suggested that multiple bacterial species occupying different sections of the gut are involved in the formation of TMA from carnitine [26].
Table 1.
Analyses of gut microbiota associated with TMA/TMAO levels in mice*
| Taxonomy Level |
Taxa | Measured metabolite |
Correlation coefficient |
FDR2 |
|---|---|---|---|---|
| Carnitine diet | ||||
| Family | Lachnospiraceae | γBB | −0.513 | 0.001 |
| Genus | Prevotella | γBB | 0.605 | 0.029 |
| Genus | Parasutterella | γBB | 0.545 | 0.059 |
| Genus | Bacteroides | γBB | 0.503 | 0.095 |
| Genus | Anaeroplasma | TMAO | 0.677 | 0.016 |
| Genus | Porphomonadaceae | TMAO | −0.581 | 0.052 |
| Genus | Akkermansia muciniphila | TMAO | 0.519 | 0.081 |
| Family | Lachnospiracea | TMAO | 0.779 | 0.084 |
| Genus | Unknown Ruminococcaceae | TMAO | 0.725 | 0.084 |
| Choline diet | ||||
| Genus | Prevotella | TMAO | 0.44 | 0.001 |
| Family | Erysipelotrichaceae | TMAO | 0.40 | 0.01 |
| Order | RF39 | TMAO | 0.49 | 0.04 |
| γBB diet | ||||
| Genus | Akkermansia muciniphila | TMAO | 0.634 | 0.009 |
Apart from dietary modifications, several strategies for modulating TMAO levels have been proposed [92, 104] (Fig. 8B). For example, treatments of rats with meldonium, an aza-analogue of γ- butyrobetaine re, reduced TMA/TMAO production [105]. Studies in mice have shown that inhibition of FMO3 reduced TMAO levels while over-expression increased TMAO levels [94, 102]. FMO3 expression is regulated by bile acids and hormones and thus there is the potential for modulating the enzyme [102]. Conceivably, diet, probiotics, or prebiotics could be used to reduce levels of TMAO-producing bacteria or increase levels of TMA degrading bacteria. For example, some methanogenic archaea such as Methanomassiliicoccus have been shown capable of reducing methylamines such as TMA and thus introduction of these into subjects could potentially deplete TMA and reduce TMAO [106].
Dissecting the Role of Microbiota in CVD: The Road Ahead
Microbiota and atherosclerosis: The road ahead
Although based largely on studies in experimental organisms, the evidence that gut microbiota contribute to atherosclerosis susceptibility is convincing (Text Box 1). One clear lesson for atherosclerosis researchers is that differences in microbiota may well influence the results of in vivo studies. For example, we have noted striking differences in gut microbiota composition in mice maintained in different vivaria, even if fed identical diets (unpublished). Below, we discuss the factors contributing to variations of gut microbiota composition in populations. We then outline approaches to better understand gut microbiota interactions in atherosclerosis in experimental organisms and in human populations. Finally, we discuss possible therapeutic approaches.
Text Box 1. Microbiota and Atherosclerosis – A Summary.
Both epidemiologic and experimental evidence strongly support the possibility that chronic infection promotes atherosclerosis, despite the negative results of clinical trials with antibiotic treatment. Antibiotic treatment would affect commensal gut bacteria as well as any pathologic infection, complicating interpretation of the trials.
Gut bacteria metabolize complex carbohydrates to short chain fatty acids, which stimulate various G-protein coupled receptors to influence adiposity, lipid metabolism, immune functions and blood pressure.
Gut microbiota can promote systemic inflammation by activating the innate immune system, thereby influencing metabolic syndrome traits, hepatic steatosis and atherosclerosis.
Gut microbiota clearly impact bile acid metabolism and recent studies in mice have revealed an important role for bacterial bile salt hydrolase, involved in the deconjugation of taurine-muricholic acid, in FXR signaling and diet induced obesity. FXR signaling is also implicated in the responses to bariatric surgery.
Certain polyphenolic metabolites protect against hypercholesterolemia and atherosclerosis. Gut bacteria enzymatically modify these to increase their absorption in the intestine.
TMAO, a metabolite produced from dietary choline and carnitine through the action of gut microbiota, is strongly associated with atherosclerosis in human populations. Experimental studies in mice strongly support a causal relationship, possibly involving effects on cholesterol metabolism or inflammation of the vessel wall.
Factors influencing gut microbiota composition
Gut microbiota composition varies greatly in natural populations from invertebrates to humans. The major phyla in most stool samples from subjects are Bacteroidetes and Firmicutes, but these commonly vary 5-fold or more in abundance between subjects. Less abundant phyla such as Actinobacteria and Proteobacteria show even greater variation. Nevertheless, in adult individuals, the composition tends to remain relatively stable over time [107]. Different strains of experimental animals such as mice also show very substantial differences in gut microbiota composition, even if maintained on the same diets. This variation undoubtedly contributes to disease susceptibility, and an important challenge is to understand the nature of this variation. At least three factors are likely to be important: maternal transmission, diet, and genetic background. Other factors such as medication (particularly antibiotics), sex, aging, and disease undoubtedly also contribute.
The maternal environment provides the original inoculum of microbiota. Over the first few years of life, the composition of the gut microbiota becomes increasingly diverse, with additional maternal impact from breast feeding. The effects of the maternal environment have been studied experimentally in animals, and it is clear that they can last over multiple generations [10, 55]. Thus, when mice of one strain are fostered by mothers of a different strain, their microbiota will resemble that of the foster mothers and their pups will in turn continue to exhibit, in part, the microbiota composition of the foster mother.
Diet has been considered the primary factor driving gut microbiota composition [73, 108–110]. A dramatic example of the impact of diet is the decreased metabolism of carnitine in vegans as compared to omnivores. The complexity of the community is illustrated by the fact that carnitine is catabolized to TMA by two different groups of bacteria with distinct intestinal locations [26]. This emphasizes the specialized nature of members of the microbial community in terms of substrate preference and the intense competition for niche space. Although the effects of various diets on atherosclerosis have been studied in detail over the past decades, little is as yet known about how such diets affect gut microbiota.
Recent evidence indicates that host genetics also plays a role in shaping the gut microbiota, particularly when environmental differences are minimized, as in laboratory mice in a vivarium. The evidence for this originally came from mice carrying mutations affecting intestinal mucosa or immune responses [10, 111–116]. For example, TLR2 deficient mice are predisposed to develop obesity and insulin resistance and show increased Firmicutes levels. The phenotype can be transferred through microbiota transplantation to wild type mice and reversed with antibiotics [2, 77]. Also, our group has carried out analyses of gut microbiota in a large number of inbred mouse strains that have been separated for hundreds of generations from their shared ancestors and determined, based on the fraction of shared genetic material, that the heritability of gut microbiota is substantial (unpublished). Recently, studies of monozygotic and dizygotic twins provided evidence that genetic background contributes to the abundance of certain gut microbiota in humans as well [12]. The mechanisms mediating such effects are largely unknown but genetic differences in immune functions are strong candidates.
In all probability, gene-by-environment interactions are also likely to be important. For example, dietary changes may influence the gut microbiota directly by providing new substrates, or they may act indirectly, affecting the expression of factors such as Reg3γ, defensins and immunoglobulins. If the latter, host genetic variation could alter the response to the dietary change.
Dissecting host-microbiota interactions
As discussed above, there has been great progress in understanding the mechanisms by which microbiota contribute to disease and a long-term goal will be to induce a “healthy” microbiome. Although studies of probiotics and prebiotics have shown that it is possible to perturb the gut microbiome, translational efforts will depend on a better understanding of the nature of host-microbiota interactions. Initially this can be best addressed in experimental organisms, where dietary and other environmental factors can be controlled and tissues can be collected. A variety of experimental organisms, including Zebrafish, mice, rats, pigs, and even flies, have been studied, each with advantages and disadvantages [117]
One of the problems in studying gut microbiota is that they exist as communities, making it difficult to test the effects of individual species. Thus, studies in which individual cultured bacteria have been introduced into germ-free mice to test their metabolic consequences have had limited success. Another problem is that there are likely to be significant genetic and environmental interactions. For example, the results of a survey of the effects of dietary components on the microbiome will depend on the genetic background of the organism being studied [118].
Clearly, studies in which individual host genes have been mutated in mice have been informative in understanding how the host contributes to microbiota composition (reviewed in [13]). But, such studies depend on candidate genes and undoubtedly will miss many important pathways. Therefore, genetic mapping using linkage or genome-wide association in mice should be informative. Whether similar studies are feasible in humans, given the diversity of diets and environmental variables that occur in human populations, is unclear.
Gut microbiota appear to influence host metabolism and disease in large part by producing metabolites that enter the host circulation [119]. Some examples discussed above are the short chain fatty acids, TMAO, and polyphenols. Thus, broad metabolomic studies capable of identifying novel bacterial metabolites are likely to be productive. To examine the consequences of such metabolites, global host gene expression screens or analysis of epigenetic marks in relevant tissues could be utilized.
The 16S rRNA gene profiling of microbiota that has been employed in most studies thus far provides a very crude picture of the microbiota composition, and whole-genome sequencing of the microbiota as well as the host would provide additional power. It is becoming increasingly clear that bacteria often inhabit specific locations in the gut and on mucosal surfaces (the intestinal surface area in humans is ~200m2), and analysis of specific regions of the intestine (rather than simply studying fecal and cecal samples) should improve resolution.
In addition to in vivo studies in experimental organisms, in vitro models using bioreactors in which specific bacteria are cultured may be informative for addressing specific questions. Some novel approaches, such as a microfluidics-based Gut-on-a-Chip model have been proposed [117, 120].
Systems-based approaches in human populations
While reductionistic approaches in experimental organisms are rapidly advancing our understanding of host-gut microbiota interactions, their application in humans will be limited by genetic and environmental heterogeneity and the difficulty of experimental studies. Systems-based approaches, on the other hand, would seem to be well suited for this purpose, since they make use of the large inter-individual variation in microbiota to understand the molecular and clinical relationships. Such “systems” approaches were developed in the field of engineering but in the last decades have been increasingly applied to biologic problems. They have four basic steps: 1. Identify the parts (microbiota, host tissues); 2. Characterize the parts (function); 3. Identify the interactions of the parts (data integration); and 4. Evaluate the interactions between the system and its environment (diet).
The third step, involving the identification of interactions, usually involves some form of perturbation. Given that experimental perturbations are either not practical or are ethically impermissible in humans, one approach to this problem is to study natural human populations, where the system (microbiota, relevant tissues) is perturbed by common genetic and environmental factors. Such an approach, called “integrative genetics” or “systems genetics”, has a number of advantages (discussed in [121]). In particular, it allows an analysis of molecular interactions in the context that is most relevant to the clinical trait, namely, multiple perturbations on a variety of genetic backgrounds rather than an individual perturbation on a single genetic background (as in a transgenic mouse).
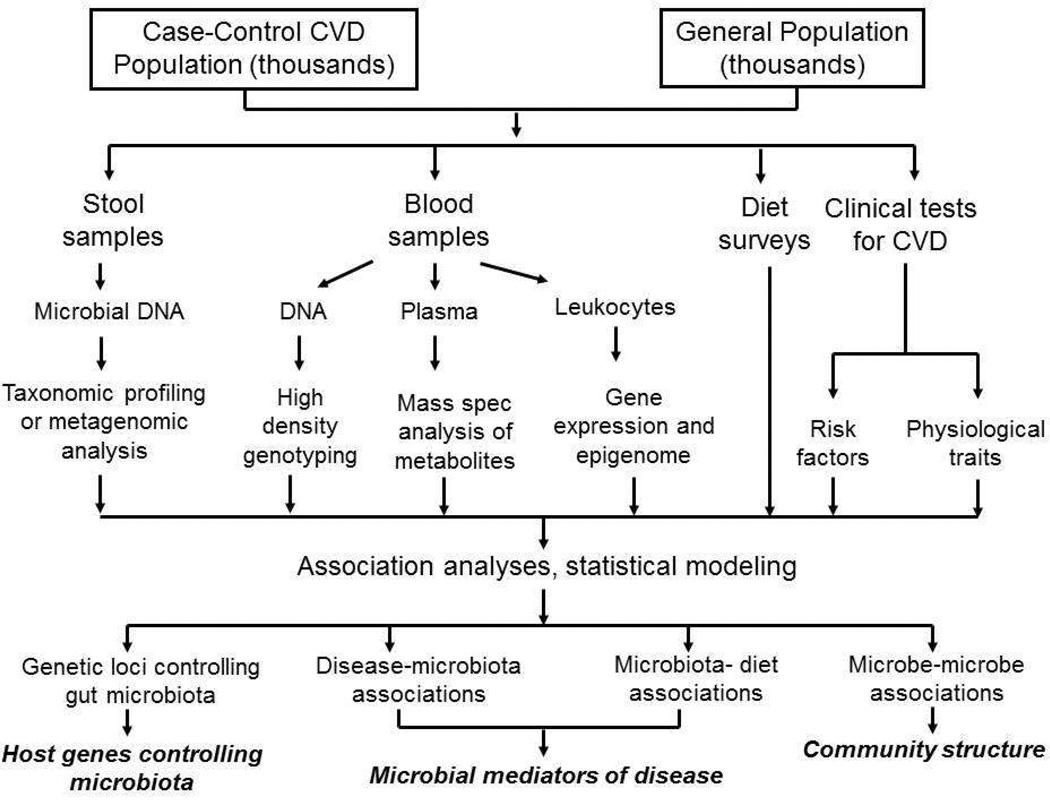
A possible systems biology study is outlined in Figure 9. Individuals in a human population would be studied for microbiome composition in stool samples using either 16S rRNA genes or whole genome sequencing. Catalogues of complete genomic sequences of species of microbes colonizing the gut and other body sites are available from the Human Microbiome Project in the United States [122] and the Metagenomics of the Human Intestinal Tract project in Europe [123]. The same individuals would also be examined for as many high throughput molecular traits as possible, particularly metabolite levels, as well as relevant physiologic and clinical traits. Ideally, detailed dietary surveys would be conducted as well. Such data would be integrated using appropriate statistical methods [124] to yield hypotheses about microbiota communities, microbiota effects on host physiology, dietary interactions, and molecular pathways perturbed by microbiota. Such hypotheses would then be tested in experimental organisms. If enough individuals (thousands, at least) could be recruited, genome-wide association analyses could conceivably also add information about the genetic control of microbiota composition. A similar strategy is being pursued by the Integrative Human Microbiome Project for the role of the microbiome in type 2 diabetes, inflammatory bowel disease, and premature birth (http://hmp2.org).
Figure 9. A systems genetics approach to the analysis of the role of gut microbiota in human disease.
This diagram outlines a potential population-based study that uses correlation, genetic mapping, and statistical modeling to understand the interactions among environmental factors, host genetics and gut microbiota, and how these contribute to CVD.
Therapeutic intervention
In addition to their implications for healthy nutrition [125–127], the findings linking microbiota to CVD suggest possible therapeutic approaches. First, since many of the effects of microbiota on CVD (and other disorders) are mediated by metabolites, it may be possible to specifically target their production, transport, or action. For example, possible interventions to block the TMA/TMAO pathway are discussed above. A second approach is to target the microbiota using probiotics, prebiotics, dietary constituents, or drugs [128, 129]. Unlike our genomes, our microbiomes can be readily perturbed. New approaches for the specific targeting of bacteria, such as “jamming” bacterial communication [130] or introduction of the RNA-guided nuclease CRISPR using a bacteriophage carrier [131] are being developed. It may also be possible to colonize our gut with genetically modified “smart” bacteria (as in the example of bile acid hydrolase) [129]. As noted above, our understanding of the interactions and functions of the gut microbiota is very incomplete and further studies in animal models and human subjects will be required to identify the precise microbial targets. It is worth noting that since gut microbiota contribute to many different disorders, it may be difficult to define a “healthy” microbiome, and there may be unforeseen consequences of long-term microbial disruption [132].
Highlights.
Gut microbiota vary widely in human populations and this variation influences lipid metabolism, insulin resistance, and systemic inflammation
Gut microbiota modulate the effects of dietary and, most likely genetic factors on atherosclerosis
Systems genetics approaches should be useful in dissecting the complexities of hostmicrobiota interactions
Acknowledgements
This work was supported by grants HL28481, HL30568, HL126753, D094311, and the Leducq Foundation. We thank Rosa Chen for preparing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holmes E, et al. Therapeutic modulation of microbiota-host metabolic interactions. Sci Transl Med. 2012;4(137):137rv6. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10(3):159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 5.Maynard CL, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmody RN, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17(1):72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolnick DI, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. doi: 10.1038/ncomms5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks BW, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes E, et al. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16(5):559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huse SM, et al. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics. 2014;15:41. doi: 10.1186/1471-2105-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TL, et al. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolars JC, et al. Yogurt--an autodigesting source of lactose. N Engl J Med. 1984;310(1):1–3. doi: 10.1056/NEJM198401053100101. [DOI] [PubMed] [Google Scholar]
- 21.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 22.Delzenne NM, Cani PD. Interaction between obesity and the gut microbiota: relevance in nutrition. Annu Rev Nutr. 2011;31:15–31. doi: 10.1146/annurev-nutr-072610-145146. [DOI] [PubMed] [Google Scholar]
- 23.Everard A, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8(10):2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright SD, et al. Infectious agents are not necessary for murine atherogenesis. J Exp Med. 2000;191(8):1437–1442. doi: 10.1084/jem.191.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepankova R, et al. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. J Atheroscler Thromb. 2010;17(8):796–804. doi: 10.5551/jat.3285. [DOI] [PubMed] [Google Scholar]
- 26.Koeth RA, et al. gamma-Butyrobetaine Is a Proatherogenic Intermediate in Gut Microbial Metabolism of L-Carnitine to TMAO. Cell Metab. 2014;20(5):799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanage WP. Microbiology: Microbiome science needs a healthy dose of scepticism. Nature. 2014;512(7514):247–248. doi: 10.1038/512247a. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert A, Lion G. Arterites infectueuses expérimentales. Comptes Rendus Hebdomadaires des Seances et Memoires de la Societe de Biologie. 1889;41:583–584. [Google Scholar]
- 30.Mattila KJ, et al. Association between dental health and acute myocardial infarction. BMJ. 1989;298(6676):779–781. doi: 10.1136/bmj.298.6676.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenfeld ME, Campbell LA. Pathogens and atherosclerosis: update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb Haemost. 2011;106(5):858–867. doi: 10.1160/TH11-06-0392. [DOI] [PubMed] [Google Scholar]
- 32.Saikku P, et al. Chronic Chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki Heart Study. Ann Intern Med. 1992;116(4):273–278. doi: 10.7326/0003-4819-116-4-273. [DOI] [PubMed] [Google Scholar]
- 33.Stassen FR, Vainas T, Bruggeman CA. Infection and atherosclerosis. An alternative view on an outdated hypothesis. Pharmacol Rep. 2008;60(1):85–92. [PubMed] [Google Scholar]
- 34.Godzik KL, et al. In vitro susceptibility of human vascular wall cells to infection with Chlamydia pneumoniae. J Clin Microbiol. 1995;33(9):2411–2414. doi: 10.1128/jcm.33.9.2411-2414.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blessing E, et al. Chlamydia pneumoniae infection accelerates hyperlipidemia induced atherosclerotic lesion development in C57BL/6J mice. Atherosclerosis. 2001;158(1):13–17. doi: 10.1016/s0021-9150(00)00758-9. [DOI] [PubMed] [Google Scholar]
- 36.Coombes BK, Mahony JB. Chlamydia pneumoniae infection of human endothelial cells induces proliferation of smooth muscle cells via an endothelial cell-derived soluble factor(s) Infect Immun. 1999;67(6):2909–2915. doi: 10.1128/iai.67.6.2909-2915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H, Pierce GN, Zhong G. The atherogenic effects of chlamydia are dependent on serum cholesterol and specific to Chlamydia pneumoniae. J Clin Invest. 1999;103(5):747–753. doi: 10.1172/JCI4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalayoglu MV, Byrne GI. Induction of macrophage foam cell formation by Chlamydia pneumoniae. J Infect Dis. 1998;177(3):725–729. doi: 10.1086/514241. [DOI] [PubMed] [Google Scholar]
- 39.Muhlestein JB, et al. Infection with Chlamydia pneumoniae accelerates the development of atherosclerosis and treatment with azithromycin prevents it in a rabbit model. Circulation. 1998;97(7):633–636. doi: 10.1161/01.cir.97.7.633. [DOI] [PubMed] [Google Scholar]
- 40.Rosenfeld ME. Inflammation and atherosclerosis: direct versus indirect mechanisms. Curr Opin Pharmacol. 2013;13(2):154–160. doi: 10.1016/j.coph.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston SC, et al. Chlamydia pneumoniae burden in carotid arteries is associated with upregulation of plaque interleukin-6 and elevated C-reactive protein in serum. Arterioscler Thromb Vasc Biol. 2005;25(12):2648–2653. doi: 10.1161/01.ATV.0000189157.88630.d1. [DOI] [PubMed] [Google Scholar]
- 42.Kol A, et al. Chlamydial heat shock protein 60 localizes in human atheroma and regulates macrophage tumor necrosis factor-alpha and matrix metalloproteinase expression. Circulation. 1998;98(4):300–307. doi: 10.1161/01.cir.98.4.300. [DOI] [PubMed] [Google Scholar]
- 43.Hogdahl M, Soderlund G, Kihlstrom E. Expression of chemokines and adhesion molecules in human coronary artery endothelial cells infected with Chlamydia (Chlamydophila) pneumoniae. APMIS. 2008;116(12):1082–1088. doi: 10.1111/j.1600-0463.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 44.Kol A, et al. Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J Clin Invest. 1999;103(4):571–577. doi: 10.1172/JCI5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464(7293):1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajamaki K, et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One. 2010;5(7):e11765. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robbins GR, Wen H, Ting JP. Inflammasomes and metabolic disorders: old genes in modern diseases. Mol Cell. 2014;54(2):297–308. doi: 10.1016/j.molcel.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridker PM, et al. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med. 2000;342(12):836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 49.Campbell LA, et al. The acute phase reactant response to respiratory infection with Chlamydia pneumoniae: implications for the pathogenesis of atherosclerosis. Microbes Infect. 2010;12(8–9):598–606. doi: 10.1016/j.micinf.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell LA, Rosenfeld ME. Persistent C. pneumoniae infection in atherosclerotic lesions: rethinking the clinical trials. Front Cell Infect Microbiol. 2014;4:34. doi: 10.3389/fcimb.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calandrini CA, et al. Microbial composition of atherosclerotic plaques. Oral Dis. 2012;20(3):e128–e134. doi: 10.1111/odi.12205. [DOI] [PubMed] [Google Scholar]
- 53.Wostmann BS, et al. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. 1983;33(1):46–50. [PubMed] [Google Scholar]
- 54.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 55.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Physiol Cell Physiol. 2014;307(11):C979–C985. doi: 10.1152/ajpcell.00228.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwanaga T, et al. Cellular expression of monocarboxylate transporters (MCT) in the digestive tract of the mouse, rat, and humans, with special reference to slc5a8. Biomed Res. 2006;27(5):243–254. doi: 10.2220/biomedres.27.243. [DOI] [PubMed] [Google Scholar]
- 59.Bjursell M, et al. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am J Physiol Endocrinol Metab. 2010;300(1):E211–E220. doi: 10.1152/ajpendo.00229.2010. [DOI] [PubMed] [Google Scholar]
- 60.Ge H, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149(9):4519–4526. doi: 10.1210/en.2008-0059. [DOI] [PubMed] [Google Scholar]
- 61.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110(11):4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin C, Henao-Mejia J, Flavell RA. Innate immune receptors: key regulators of metabolic disease progression. Cell Metab. 2013;17(6):873–882. doi: 10.1016/j.cmet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Ding Y, et al. Toll-like receptor 4 deficiency decreases atherosclerosis but does not protect against inflammation in obese low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2012;32(7):1596–1604. doi: 10.1161/ATVBAHA.112.249847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156(5):1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauernfeind F, et al. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol. 2011;187(2):613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Latz E. The inflammasomes: mechanisms of activation and function. Curr Opin Immunol. 2010;22(1):28–33. doi: 10.1016/j.coi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 71.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 72.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet. 2008;9(11):819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandek A, Anker SD, von Haehling S. The gut and intestinal bacteria in chronic heart failure. Curr Drug Metab. 2009;10(1):22–28. doi: 10.2174/138920009787048374. [DOI] [PubMed] [Google Scholar]
- 75.Ghosh SS, et al. Oral supplementation with non-absorbable antibiotics or curcumin attenuates western diet-induced atherosclerosis and glucose intolerance in LDLR−/− mice--role of intestinal permeability and macrophage activation. PLoS One. 2014;9(9):e108577. doi: 10.1371/journal.pone.0108577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harte AL, et al. High fat intake leads to acute postprandial exposure to circulating endotoxin in type 2 diabetic subjects. Diabetes Care. 2012;35(2):375–382. doi: 10.2337/dc11-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caricilli AM, et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9(12):e1001212. doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Clarke SF, et al. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes. 2012;3(3):186–202. doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Islam KB, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 80.Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol. 2015 doi: 10.1097/MOG.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velagapudi VR, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2009;51(5):1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swann JR, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wostmann BS. Intestinal bile acids and cholesterol absorption in the germfree rat. J Nutr. 1973;103(7):982–990. doi: 10.1093/jn/103.7.982. [DOI] [PubMed] [Google Scholar]
- 84.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Li F, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Joyce SA, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A. 2014;111(20):7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furet JP, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryan KK, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509(7499):183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang D, et al. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res. 2012;111(8):967–981. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124(10):4204–4211. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shih DM, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res. 2014;56(1):22–37. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao X, et al. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J Biosci Bioeng. 2014;118(4):476–481. doi: 10.1016/j.jbiosc.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 96.Tang WH, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ayesh R, et al. The fish odour syndrome: biochemical, familial, and clinical aspects. BMJ. 1993;307(6905):655–657. doi: 10.1136/bmj.307.6905.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.D'Angelo R, et al. FMO3 allelic variants in Sicilian and Sardinian populations: trimethylaminuria and absence of fish-like body odor. Gene. 2013;515(2):410–415. doi: 10.1016/j.gene.2012.12.047. [DOI] [PubMed] [Google Scholar]
- 99.Suska A, et al. G protein-coupled receptor mediated trimethylamine sensing. Biosens Bioelectron. 2009;25(4):715–720. doi: 10.1016/j.bios.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 100.Wallrabenstein I, et al. Human trace amine-associated receptor TAAR5 can be activated by trimethylamine. PLoS One. 2013;8(2):e54950. doi: 10.1371/journal.pone.0054950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hartiala J, et al. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler Thromb Vasc Biol. 2014;34(6):1307–1313. doi: 10.1161/ATVBAHA.114.303252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bennett BJ, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17(1):49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gregory JC, et al. Transmission of Atherosclerosis Susceptibility with Gut Microbial Transplantation. J Biol Chem. 2014 doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–359. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuka J, et al. Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting L-carnitine microbial degradation. Life Sci. 2014;117(2):84–92. doi: 10.1016/j.lfs.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 106.Brugere JF, et al. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes. 2014;5(1):5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Duncan SH, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73(4):1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Louis P, et al. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102(5):1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 110.Zhang C, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4(2):232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 111.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. e1–e2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Joller D, et al. Refined localization of the Escherichia coli F4ab/F4ac receptor locus on pig chromosome 13. Anim Genet. 2009;40(5):749–752. doi: 10.1111/j.1365-2052.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 113.McKnite AM, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7(6):e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meijerink E, et al. A DNA polymorphism influencing alpha(1,2)fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics. 2000;52(1–2):129–136. doi: 10.1007/s002510000263. [DOI] [PubMed] [Google Scholar]
- 115.Niu X, et al. Refined mapping of the Escherichia coli F4ab/F4ac receptor gene(s) on pig chromosome 13. Anim Genet. 2011;42(5):552–555. doi: 10.1111/j.1365-2052.2011.02176.x. [DOI] [PubMed] [Google Scholar]
- 116.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fritz JV, et al. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1(1):14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O'Connor A, et al. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm Genome. 2014;25(11–12):583–599. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim HJ, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12(12):2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 121.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15(1):34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2013;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li H. Systems biology approaches to epidemiological studies of complex diseases. Wiley Interdiscip Rev Syst Biol Med. 2013;5(6):677–686. doi: 10.1002/wsbm.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mitchell SC, Zhang AQ, Smith RL. Chemical and biological liberation of trimethylamine from foods. J Food Compos Anal. 2002;15:277–282. [Google Scholar]
- 126.Rohrmann S, et al. Meat consumption and mortality--results from the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2013;11:63. doi: 10.1186/1741-7015-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ussher JR, Lopaschuk GD, Arduini A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis. 2013;231(2):456–461. doi: 10.1016/j.atherosclerosis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 128.Knight R. Why microbiome treatments could pay off soon. Nature. 2015;518:55. doi: 10.1038/518S5a. [DOI] [PubMed] [Google Scholar]
- 129.Sonnenburg JL. Microbiome engineering: Synthetic biology may lead to the creation of smart microbes that can detect and treat disease. Nature. 2015;518:510. doi: 10.1038/518S10a. [DOI] [PubMed] [Google Scholar]
- 130.Njoroge J, Sperandio V. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol Med. 2009;1(4):201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32(11):1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol. 2015;11(3):182–190. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hofmann AF. Bile acid sequestrants improve glycemic control in type 2 diabetes: a proposed mechanism implicating glucagon-like peptide 1 release. Hepatology. 2011;53(5):1784. doi: 10.1002/hep.24100. [DOI] [PubMed] [Google Scholar]
- 134.Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev. 2007;87(4):1409–1439. doi: 10.1152/physrev.00034.2006. [DOI] [PubMed] [Google Scholar]
- 135.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Harach T, et al. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci Rep. 2012;2:430. doi: 10.1038/srep00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhu Y, et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A. 2014;111(11):4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]