Abstract
Introduction
L-selectin (CD62L) is a vascular adhesion molecule constitutively expressed on leucocytes with a primary function of directing leucocyte migration and homing of lymphocytes to lymph nodes (LNs). In a gene expression microarray study comparing laser captured micro-dissected (LCM) high grade muscle invasive bladder cancer (MIBC) without prior treatment and low grade bladder cancer (LGBC) human samples, we found CD62L to be the highest differentially expressed gene. We sought to examine the differential expression of CD62L in MIBCs and its clinical relevance.
Methods
Unfixed fresh and formalin fixed paraffin-embedded human bladder cancer specimens and serum samples were obtained from the UCHC tumor bank. Tumor cells were isolated from frozen tumor tissue sections by LCM followed by RNA isolation. qPCR was used to validate the level of CD62L transcripts. Immunohistochemistry and ELISA were performed to evaluate the CD62L protein localization and expression level. Flow cytometry was used to identify the relative number of cells expressing CD62L in fresh tumor tissue. In silico studies were performed using the Oncomine Database.
Results
Immunostaining showed a uniformly higher expression of CD62L in MIBC specimens vs. LGBCs specimens. Further, CD62L localization was seen in foci of metastatic tumor cells in LN specimens from patients with high grade MIBC and known nodal involvement. Upregulated expression of CD62L was also observed by flow cytometric analysis of freshly isolated tumor cells from biopsies of high grade cancers vs. LGBC specimens. Circulating CD62L levels were also found to be higher in serum samples from patients with high grade metastatic vs. high grade non-metastatic MIBC. In addition, in silico analysis of Oncomine Microarray Database showed a significant correlation between CD62L expression and tumor aggressiveness and clinical outcomes.
Conclusion
These data confirm the expression of CD62L on urothelial carcinoma cells and suggest that CD62L may serve as biomarker to predict the presence of or risk for developing metastatic disease in patients with bladder cancer.
Keywords: bladder cancer, lymph node, metastasis, Oncomine
1. Introduction
Bladder cancer is the 3rd most common solid tumor in men and the 8th in women. In the United States there will be an estimated 74,690 new cases and 15,580 deaths in 2014 [1]. Five year survival rates for advanced regional and distant disease are 36% and 6%, respectively [2, 3]. Only modest improvements in survival are seen even with aggressive surgical or medical treatments. Presently, there are no defined tumor markers available for stratifying patients with respect to disease progression, prognosis or treatment, and no predictors of metastatic potential. Most patients with advanced cancer ultimately succumb to their disease with the most significant prognostic factor being the presence of LN metastasis [4]. While much work has been done on identifying the presence of metastatic tumor cells in LNs [reviewed in Watts el al., [4]], little research currently explores the ability to predict the likelihood of harboring or developing metastasis based on primary tumor characteristics.
The majority of bladder cancers present as superficial, non-invasive disease. While both high and low grade tumors have high risk of recurrence, only high grade tumors possess the ability to invade the muscle wall of the bladder and spread to distant organ sites. Given that patients do not die from loco-regional disease but rather from the sequelae of metastasis, a focus of recent research has been to understand the basic biological processes that promote cancer progression. Due to lack of curative therapy and poor clinical outcome of patients who present with, or develop, metastatic disease, there is an urgent need to identify biomarkers that are capable of predicting the events that lead to metastasis of high grade tumors to LNs.
Selectins (cluster of differentiation 62 or CD62) are a family of mammalian vascular adhesion molecules involved in the tethering and deceleration of cells in lymphatic/blood flow on capillary endothelium [5]. They are type I transmembrane proteins, which are classified into three categories: E-selectin and L-selectin, which have been shown to be present on activated endothelial cells and leucocytes respectively, and P-selectin, which has been detected on platelets and endothelial cells. Natural ligands for selectins are carbohydrate moieties such as O-linked glycan derivative forms localized on surface glycoproteins [5, 6].
CD62L (also called L-selectin) is constitutively expressed on a wide variety of inflammatory cells including monocytes, neutrophils, eosinophils, B cells, and subsets of T cells. Ligands for this receptor are found on high endothelial venules (HEVs) in LNs. The interaction of CD62L with a set of sialomucins ligands, also called peripheral node addressins, on the HEVs is a major mechanism for lymphocyte homing to LNs [7]. CD62L knockout mice have been shown to display defective homing and migration of lymphocytes to LNs [8, 9]. In addition, previous studies have shown that engineered overexpression of CD62L on non-lymphoma tumor cells, which normally do not express CD62L, increases their metastatic capacity to LNs [10, 11]. We hypothesize that CD62L expression on urothelial carcinoma cells plays a critical role in determining the potential for LN metastasis. The major objective of this study was to examine the differential expression of CD62L in high grade tumors and its potential clinical relevance.
2. Material and Methods
2.1. Materials
Monoclonal CD62L antibody was purchased from Abcam (Cambridge, MA). Fluorescein isothiocyanate (FITC)-conjugated CD62L and phycoerythrin (PE)-conjugated CD45 were obtained from Santa-Cruz, Inc. (Dallas, TX). CD62L ELISA kit was purchased from R & D Systems, Inc. (Minneapolis, MN). Cytokeratin AE1/3 antibody was from Life technologies (Grand Island, NY). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise stated.
2.2. Tumor specimens
Unfixed fresh, Optimal Cutting Temperature compound (OCT)-embedded, or formalin fixed paraffin-embedded (FFPE) human bladder cancer samples were obtained from the University of Connecticut Health Center, Institutional Review Board approved, Biorepository Facility. Clinical and pathological characteristics are maintained in the tumor bank database and are provided with the samples in a de-identified manner. In addition, all specimens were reviewed by a single pathologist (PH) in a blinded manner to confirm stage and grade based on the 2003 TNM and WHO classifications.
2.3. Laser capture micro-dissection (LCM)
Banked de-identified OCT-embedded fresh frozen tumor samples were used to isolate tumor cells from 5-10 μm thick tissue sections using UV laser cutting on Veritas LCM system (Arcturus, Mountain View, CA) following the manufacturer’s instructions. Briefly, one slide was stained for H&E and independently evaluated by a pathologist (PH) and urologist (JT) for use as a reference guide for selecting the region of interest. Additional slides with 3 sections per slide were used for LCM. On average, 0.25 mm2 (0.5 mm × 0.5mm) per slide was captured and pooled for each sample in a capped collection vial. RNA was extracted using Picopure RNA extraction kit following the manufacturer’s guidelines (Arcturus, Life Technologies).
2.4. Microarray gene expression profiling
Total RNA extracted from captured tumor cells was sent to GenUS Biosystems (Northbrook, IL) for gene expression microarray study. The concentration of total RNA was measured by spectrophotometry at OD260/280 and the quality of RNA was assessed by Agilent Bioanalyzer RNA6000 Nano Lab Chip (Agilent Technologies, Santa Clara, CA). Labeled cRNA was prepared by linear amplification of the Poly(A)+ RNA population within the total RNA sample. Briefly, <1 μg of total RNA was reverse transcribed after priming with a DNA oligonucleotide containing the T7 RNA polymerase promoter 5’ to a d(T)24 sequence. After second-strand cDNA synthesis and purification of double-stranded cDNA, in vitro transcription was performed using T7 RNA polymerase. The quantity and quality of the labeled cRNA was again assessed on an Agilent Bioanalyzer. One μg of purified cRNA was fragmented to uniform size and applied to an Agilent Human GE 4×44K v2 Microarray (Design ID 026652, Agilent Technologies) in hybridization buffer. Arrays were hybridized at 65° C for 17 h in a shaking incubator and washed at 37° C for 1 min. Arrays were scanned with an Agilent G2565 Microarray Scanner (Agilent Technologies) at 5 μm resolution. Agilent Feature Extraction software was used to process the scanned images from arrays (gridding and feature intensity extraction) and the data generated for each probe on the array was analyzed using GeneSpring GX software (Agilent Technologies) by GenUS Biosystems (Northbrook, IL). To compare individual expression values across arrays, raw intensity data from each gene was normalized to the 75th percentile intensity of each array. Only genes with values greater than background intensity for all samples within each group were used for further analysis. Differentially expressed genes were identified by >2-fold change and Welch T-test p-values < 0.05 between the two condition groups.
2.5. Culture of urothelial carcinoma cell lines
High-grade human bladder cancer cell lines HTB-5, HT-1376, HTB-9 and HTB-4 were obtained from ATCC (Mannasas, VA). The UROtsa (benign) urothelial cell line was a gift from Dr. Brian Philips, University of Pittsburgh and was originally developed by Dr. Masters, University College, London, UK [12, 13]. HTB-5 and HT-1376 cells were cultured in Eagle’s MEM (103700-021, Invitrogen, Grand Island, NY), HTB-9 cells were cultured in RPMI (Invitrogen) and HTB-4 cells were cultured in McCoy’s (Invitrogen) media. UROtsa cells were cultured in DMEM F/12 medium. All cultures were supplemented with 10% heat-inactivated fetal calf serum, 1 mM sodium pyruvate, 2 mM L-glutamine, 100 U/ml penicillin and 50 μg/ml streptomycin and grown at 37°C in a 5% CO2 in air atmosphere.
2.6. Quantitative PCR (qPCR)
Total RNA was extracted using either Trizol (Invitrogen) or a Picopure Kit. RNA was DNase treated (Ambion, Grand Island, NY) and converted to cDNA using a High Capacity cDNA Archive Kit (Applied Biosystems, Grand Island, NY). Quantitative PCR was performed in 96-well plates using Assays-on-Demand Gene Expression system on a 7300 Sequence Detection System instrument utilizing universal thermal cycling parameters (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the endogenous control. Data analyses were performed using relative quantification (RQ, ΔΔCt) or relative standard curve method.
2.7. Immunohistochemistry (IHC)
FFPE samples were used for IHC. Sections were deparaffinized and rehydrated in graduated levels of alcohol. A monoclonal antibody was used for IHC that is specific for the C-terminal domain and hence can detect the CD62L protein presence even after shedding of the N-terminal extracellular domain which is known to occur under physiological conditions [14, 15]. After incubation for 5 min in 3% hydrogen peroxide/methanol, sections were incubated for 2 hr at room temperature with a mouse monoclonal anti-human CD62L antibody (ab49508; Abcam) at a 1:10 dilution. Bound anti-CD62L antibody was detected with a secondary anti-mouse EnVision+ system – HRP labeled polymer (Dako Inc., Carpinteria, CA) and visualized using diaminobenzidine (DAB). For negative controls, buffer was substituted for the primary antibody. Cytokeratin staining was performed using purified pan-cytokeratin (AE1/AE3) monoclonal antibody (Covance, Berkeley, CA) at a 1:1000 dilution and a pretreatment with Bond epitope retrieval solution (Leica Microsystems, CA) for 20 min. The staining protocol was performed on Leica Bond-Max automated immunostainer. Sections were evaluated for degree of cytoplasmic staining and scored from 0–3 corresponding to negative, weak, intermediate and strong staining respectively. The number of cells staining at each intensity was evaluated and semiquantified by determining the H-score using the formula [1× (% of cells showing weak staining) + 2×(% of cells showing moderate staining) + 3×(% of cells showing strong staining)] [16].
2.8. Isolation of viable tumor cells from fresh surgically removed unfixed tumor samples
Freshly obtained tumor samples were immediately placed in DMEM/F12 medium with ROCK inhibitor (Y-27632). Tumor tissue was mechanically divided into 1 mm3 pieces followed by digestion with collagenase (0.5 gm/L) for 30 min at 37 °C. The digested tissue pieces were passed through a 100 μm sieve into a 50-ml centrifuge tube to eliminate undigested tissue.
2.9. Flow cytometry
The freshly isolated tumor cells were fluorescently labeled using a standard protocol. Briefly, ~2×105 cells were incubated with mouse anti-human CD62L (DREG-56) primary antibody conjugated with FITC and mouse anti-human CD45 primary antibody conjugated with PE in fluorescence-activated cell sorting (FACS) staining medium (PBS + 1% FCS) for 30 min at 4 °C. Cells were washed 3 times with FACS staining medium and flow cytometry was performed using a FACS LSRII-A flow cytometer (BD Biosciences, San Jose, California) and analyzed using BD FACS Diva analysis software (BD Biosciences). Gates for single cells and debris exclusion were made based on light scatter properties. For each stained sample, a minimum of 20,000 events were collected. Compensation of spectral overlap was calculated using a combination of unstained cells, PI-labeled cells, and BD Calibrite beads (BD Biosciences). Gates for PI, CD45, or CD62L positivity were made using fluorescence minus one control samples. The calculated percent of CD45−CD62L+ cells in each sample were normalized to 106 live cell populations.
2.10. Enzyme-linked immunosorbent assay (ELISA)
Levels of circulating CD62L in serum samples were measured by a commercially available ELISA kit (R & D Systems, Inc) following manufacturer’s instructions. Light emission was measured at 450 nm using an ELISA microplate reader (EMD Millipore). Concentrations were determined from a standard curve range.
2.11. Bioinformatic analysis of CD62L expression using Oncomine Cancer Gene Microarray Database
Datasets in the Oncomine database (Compendia Biosciences; Ann Arbor, MI, USA; www.oncomine.org) contain information from gene microarrays of cancers and corresponding benign adjacent tissues, thereby generating a ratio of gene expression in tumor vs. benign [17]. The raw data of signal intensity values were processed to a minimum value of 10, followed by normalization of the data by subtracting the log ratios from the median log ratio of a specific microarray database. Significance values were considered at p<0.05.
2.12. Statistical analysis
Statistical differences were determined using SPSS software (IBM Corp, Somers, NY). Data are means ± SEM. T-test was used for comparison of two variables, one-way ANOVA for comparison of more than two independent variables and one dependent variable and two-way ANOVA for comparison of two independent and two or more dependent variables. Post-hoc testing was performed with Bonferroni correction.
3. Results
3.1. Differential expression of CD62L transcripts in high-grade vs. low-grade human bladder cancer specimens
In a pilot study, we performed microarray analysis of samples obtained by LCM from 3 untreated MIBC and 3 LGBC specimens. The LCM protocol allowed us to focus on a near pure population of tumor cells. Two-hundred fifty genes were differentially expressed (≥ 2-fold) between the groups with 86 overexpressed in MIBC compared to LGBC. Table 1 shows a list of the genes that were overexpressed > 5 fold (p<0.05) in MIBC specimens. The most highly elevated transcript (34-fold) was L-selectin, with individual normalized expression values of 0.58, 1.16 and 9.69 for MIBC specimens compared to 0.03, 0.07 and 0.08 for LGBC specimens. We confirmed the upregulated expression of CD62L by performing qPCR on the same RNA used for the microarray experiment. There was a similar pattern of elevated expression for CD62L with individual RQ values of 0.69, 1.0, and 68.24 for MIBCs and 0.10, 0.16 and 0.17 for LGBCs (23.31 ± 22.47 vs. 0.14 ± 0.04).
Table 1.
Differentially expressed genes in high grade tumors vs. low grade (ratio > 5 fold and p>0.05).
| Gene Symbol |
Primary Accession |
UniGene ID |
EntrezGene ID |
Ratio | P-value | Description |
|---|---|---|---|---|---|---|
| SELL | NM_000655 | Hs.82848 | 6402 | 33.71 | 4.13E-02 | selectin L |
| PDK4 | NM_002612 | Hs.8364 | 5166 | 32.97 | 4.88E-02 | pyruvate dehydrogenase kinase, isozyme 4 |
| APBB1IP | NM_019043 | Hs.310421 | 54518 | 32.67 | 1.21E-02 | amyloid beta (A4) precursor protein- binding, family B, member 1 interacting protein |
| ENST00000390634 | Hs.547094 | 23.59 | 4.10E-02 | immunoglobulin heavy variable 2-70 [Source:HGNC Symbol;Acc:5577] [ENST00000390634] |
||
| SIDT1 | NM_017699 | Hs.591291 | 54847 | 21.78 | 2.00E-02 | SID1 transmembrane family, member 1 |
| APBB1IP | NM_019043 | Hs.310421 | 54518 | 20.13 | 5.86E-03 | amyloid beta (A4) precursor protein- binding, family B, member 1 interacting protein |
| DDX43 | NM_018665 | Hs.125507 | 55510 | 14.90 | 4.76E-02 | DEAD (Asp-Glu-Ala- Asp) box polypeptide 43 |
| MAN1C1 | NM_020379 | Hs.197043 | 57134 | 14.63 | 3.33E-02 | mannosidase, alpha, class 1C, member 1 |
| CNIH3 | NM_152495 | Hs.28659 | 149111 | 13.52 | 2.87E-02 | cornichon homolog 3 (Drosophila) |
| ACAA2 | NM_006111 | Hs.200136 | 10449 | 11.30 | 4.38E-02 | acetyl-Coenzyme A acyltransferase 2 |
| TMIGD2 | NM_144615 | Hs.263928 | 126259 | 8.87 | 5.32E-03 | transmembrane and immunoglobulin domain containing 2 |
| AK126778 | Hs.529357 | 8.73 | 1.30E-02 | Homo sapiens cDNA FLJ44826 fis, clone BRACE3046762. |
||
| SHD | NM_020209 | Hs.7423 | 56961 | 8.32 | 4.54E-02 | Src homology 2 domain containing transforming protein D |
| PRDM6 | NM_001136239 | Hs.135118 | 93166 | 8.24 | 2.62E-02 | PR domain containing 6 |
| PLAGL1 | NM_006718 | Hs.444975 | 5325 | 7.86 | 4.46E-02 | pleiomorphic adenoma gene-like 1 |
| TMEM22 | NM_025246 | Hs.477692 | 80723 | 7.43 | 4.21E-02 | transmembrane protein 22 |
| RORC | NM_005060 | Hs.256022 | 6097 | 6.78 | 2.08E-02 | RAR-related orphan receptor C |
| NUPR1 | NM_001042483 | Hs.513463 | 26471 | 6.70 | 4.61E-02 | nuclear protein, transcriptional regulator, 1 |
| BMP5 | NM_021073 | Hs.296648 | 653 | 6.53 | 3.04E-02 | bone morphogenetic protein 5 |
| FGF7 | NM_002009 | Hs.567268 | 2252 | 6.50 | 3.91E-02 | fibroblast growth factor 7 (keratinocyte growth factor) |
| BHLHE22 | NM_152414 | Hs.591870 | 27319 | 6.39 | 4.70E-02 | basic helix-loop-helix family, member e22 |
| LOC100132345 | XM_001716679 | Hs.543235 | 100132345 | 6.23 | 5.01E-03 | hypothetical LOC100132345 |
| PTPRM | NM_002845 | Hs.49774 | 5797 | 5.89 | 4.68E-02 | protein tyrosine phosphatase, receptor type, M |
| C1orf115 | NM_024709 | Hs.519839 | 79762 | 5.68 | 1.25E-02 | chromosome 1 open reading frame 115 |
| KCNK17 | NM_031460 | Hs.162282 | 89822 | 5.60 | 3.47E-02 | potassium channel, subfamily K, member 17 |
| FBLN1 | NM_006486 | Hs.24601 | 2192 | 5.49 | 4.15E-02 | fibulin 1 |
| WNT11 | NM_004626 | Hs.108219 | 7481 | 5.26 | 1.31E-02 | wingless-type MMTV integration site family, member 11 |
3.2. Increased expression of CD62L protein in LNs and bladder tumor sections
Based on the hypothesis that CD62L may play a role in homing of tumor cells to the LNs, we obtained 6 sets of matched tissue blocks (primary tumor and LNs with metastases) from patients with high grade metastatic MIBC. Immunostaining was performed for CD62L and cytokeratin AE1/3 (epithelial/tumor cell marker). Grossly, CD62L expression was weakly observed in all the discrete foci of tumor cells in LNs compared to non-uniform staining in some lymphocytes. Tumor cells were confirmed by pan-cytokeratin (AE1/AE3) staining, a marker for urothelial cells (Figure 1A).
Figure 1.
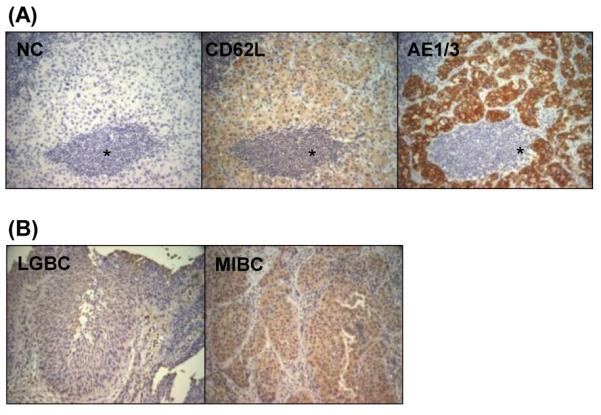
Immunohistochemical staining of (A) CD62L and AE 1/3 in metastatic lymph nodes, NC-negative control, *Lymphoid aggregate surrounded by tumor cells surrounding; (B) CD62L staining in low grade and muscle-invasive bladder tumors.
We then compared the expression level of CD62L protein in 12 human bladder tumor samples representing 6 MIBC and 6 LGBC. We found higher expression levels of CD62L in MIBCs samples (Figure 1B). CD62L was predominantly localized in the cytoplasm in LGBC while MIBC displayed both nuclear and cytoplasmic accumulation. In LGBC, a significant proportion (20-50%) of the cells were not immunoreactive and showed variability in few fields with high reactivity. The comparative H-score values for LGBC and MIBC were 96.6 ± 11.2 and 133.3 ± 14.9, (n=6) (P=0.07) respectively.
3.3. Expression of CD62L transcripts in high grade urothelial carcinoma cell lines
To determine if CD62L expression is elevated in high grade bladder cancer lines, we screened four cell lines (HTB4, HTB5, HTB9, CRL-1472) representing different tumor grades (2-4) [18], as well as a benign cell line UROtsa. The HTB-9 cell line had a significantly (P<0.001) higher level of CD62L mRNA expression (13-fold) compared to UROtsa cells (Figure 2).
Figure 2.
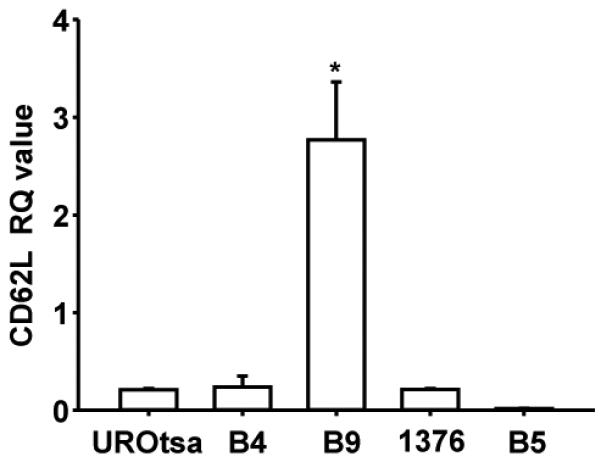
CD62L transcript expression levels in four bladder cancer cell lines (HTB4, HTB9, 1376, HTB5) and a benign cell line (UROtsa). (* P<0.001).
3.4. Higher expression of CD62L in freshly isolated high grade vs. low grade human bladder tumor cells
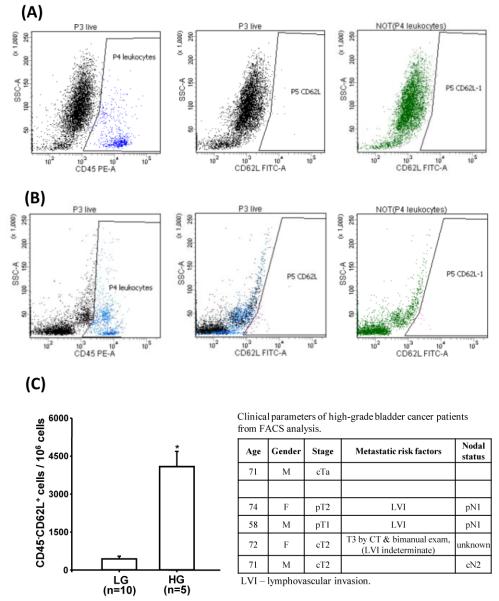
Freshly isolated human bladder tumor cells were analyzed by flow cytometry. They were stained with CD62L and CD45 antibodies to gate for leucocytes which are the only other cell population reported in the literature to express CD62L (Figure 3A and 3B). After gating, there was a 9.2 fold (P<0.001) increase in CD62L expression in high-grade compared to low-grade tumor samples (Figure 3C). The highest expression levels were seen in tissue from patients ultimately found to have metastatic disease at cystectomy or high suspicion of metastatic disease based on tumor characteristics or clinical staging studies.
Figure 3.
Representative flow cytometry dot plots of CD45 and CD62L labeled cells in freshly isolated (A) low grade (LG) bladder tumor, and (B) high grade (HG) bladder tumor specimen. (C) Flow cytometric data analysis of normalized live CD45− CD62+ cells per million cells from 15 individual samples (*P<0.001) and associated clinical parameters of HG bladder tumor samples.
3.5. Increased circulating CD62L level in serum
CD62L is known to undergo shedding by cleaving at the membrane proximal region [19, 20]. Serum levels of released CD62L have been used as a biomarker of leukemia relapse and are associated with poor prognosis [15, 21]. Based on these studies, we evaluated the constitutive levels of CD62L protein in twenty serum samples of age/sex matched MIBC patients with and without LN metastasis by commercially available ELISA (R & D Systems, Inc. Minneapolis, MN). We found a higher level (640 ± 75 vs. 558 ± 34 ng/ml; n=10) (P=0.34) in metastatic vs. non-metastatic MIBC.
3.6. In silico analysis of CD62L expression using Oncomine database
The Oncomine database provides a systematic approach to meta-analyses of independent microarray datasets based on validated normalizing statistical methods. Before initiating our search, we defined threshold parameters for gene expression analysis at; p<0.05, fold change <1.5 and median gene rank in the top 10%. CD62L gene expression was detected in 20 datasets comprising a total of 812 patient samples. The significant differential expression was detected in five datasets with highest expression in a study by Dyrskjot et al., (2003) [22] showing a 7.9 fold overexpression in infiltrating as compared to superficial tumors [22] (Figure 4). There were two additional studies [23, 24] that showed a high expression of CD62L in invasive bladder cancer (Table 2A). A meta-analysis of combined normalized values from these three independent datasets gave a significant median gene rank value of 127 for CD62L overexpression in invasive bladder cancers, which indicate that CD62L expression lies in top 3% of the total genes analyzed across the selected datasets. In contrast, only one study showed an upregulation of CD62L in superficial bladder cancer [25] (Figure 4).
Figure 4.
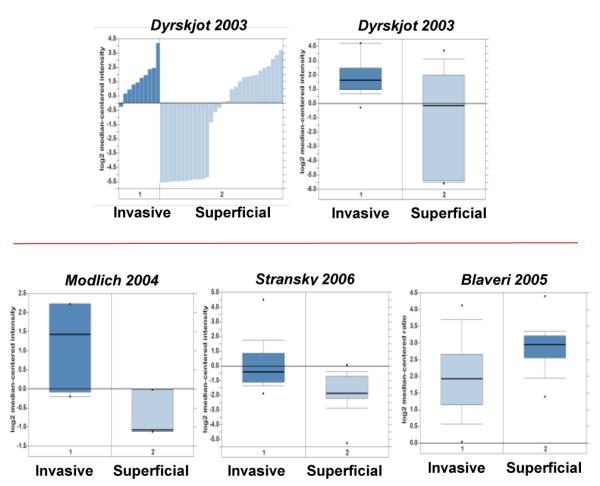
Bar chart and box plot of CD62L overexpression in invasive bladder cancer samples from Dyrskjot (2003) dataset obtained from Oncomine. Box plot of Modlich (2004) and Stransky (2006) datasets showing overexpression of CD62L in invasive bladder cancer samples and Blaveri (2005) showing overexpression in superficial bladder cancer samples.
Table 2A.
Over expression of CD62L in bladder cancers
| Dataset | Fold change |
Gene Rank |
Number of samples (Invasive, Superficial) |
P-value | Meta-analysis |
|---|---|---|---|---|---|
| Upregulated expression in invasive bladder cancers | |||||
| Dyrskjot - 2003 | 7.88 | 110 | 40 (10, 30) | 1.88 E-4 | Median Rank = 127 P=4.21 E-6 |
| Modlich – 2004 | 3.62 | 462 | 9 (6, 3) | 7.00 E-3 | |
| Stransky – 2006 | 3.24 | 127 | 57 (32, 25) | 4.21 E-6 | |
| Upregulated expression in superficial bladder cancers | |||||
| Blaveri – 2005 | 1.80 | 411 | 65 (41, 24) | 1.11 E-4 | |
| Dyrskjot - 2004 | 1.60 | 857 | 27 (8, 19)* | 1.90 E-2 | |
(Grade 2, Grade 3)
The Oncomine database can also be used to stratify data into subgroups depending upon clinico-pathological characteristics such as tumor grade, metastatic LN status and survival endpoints. Data was available for survival, recurrence and metastatic event status in 7, 5 and 2 microarray datasets, respectively. CD62L overexpression correlated with survival in 5 out of 7, recurrence in 3 out of 5 and metastatic satus in 1 out of 2 of these microarray datasets. Specifically, Dyrskjot et al. (2004) [26] showed a significant increase of CD62L expression with increasing tumor grade [26] (Figure 5). In another dataset, CD62L overexpression positively correlated with recurrence rate in superficial cancers (Table 2B) [24]. Additionally, high CD62L expression levels were found to be significantly associated with decreased survival [25]. None of the publications associated with these datasets specifically mention or suggest a role for CD62L in bladder cancer. The current study is the first to report on the correlation of CD62L expression with clinical outcomes in bladder cancer.
Figure 5.
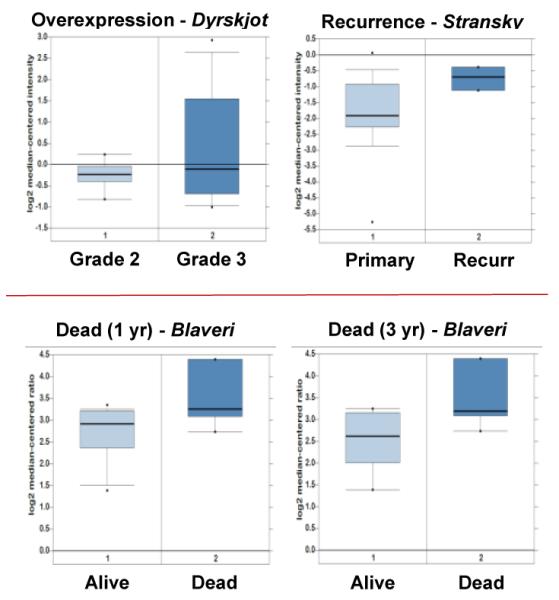
Box plots of CD62L overexpression and their relationship to clinical outcome parameters.
Table 2B.
Correlation of overexpression of CD62L with clinical outcome
| Dataset | Fold change |
Gene Rank |
Number of samples (Dead, Alive) |
P-value |
|---|---|---|---|---|
| Recurrence in superficial bladder cancers | ||||
| Stransky – 2006 | 2.22 | 141 | 25 (22, 3)* | 3.00 E-3 |
| Dead at 1 yr in superficial bladder cancers | ||||
| Blaveri -2005 | 1.67 | 291 | 19 (14, 5) | |
| Dead at 3 yr in superficial bladder cancers | ||||
| Blaveri -2005 | 1.89 | 94 | ||
(primary recurrence, recurrence)
4. Discussion
Metastasis is a multistep process involving separation of tumor cells from the primary localized proliferative zone, intravasation into blood vessels or lymphatics and extravasation into tissues with resultant hematogenous or lymphatic spread respectively. LNs are commonly the first site of metastasis for many carcinomas. The presence of LN involvement has been shown to be the most significant, independent prognostic factor for mortality and dictates the choice of therapy in bladder cancer [27, 28]. While the goal of treatment is to eradicate the primary tumor and risk for recurrence prior to spread, several studies have shown that 20–40% of patients with organ confined, LN-negative disease ultimately have recurrence of their bladder cancer, strongly suggesting pathological under staging [28, 29].
Our study is the first to evaluate the expression of CD62L protein in patients with high-grade bladder cancer. CD62L showed the highest differential gene expression in untreated MIBC vs. LGBC banked samples, and there was concordant IHC staining on primary and LN-matched metastatic tumor. There was an approximate 9.2 fold increase in the number of CD62L positive cells in freshly isolated high-grade tumor samples compared to low-grade tumors with highest levels noted in patients with documented or high suspicion for metastatic disease. Finally, serum levels of the CD62L were higher in banked samples from patients with LN positive MIBC as compared to samples from MIBC patients without metastatic disease. These findings suggest a possible utility of CD62L as a marker of risk for metastasis.
Given that CD62L is a transmembrane receptor we expected to find IHC staining localized to the cell membrane. However, we chose to use a monoclonal antibody made against the C-terminal domain of the human CD62L protein, as the extracellular domain is known to undergo shedding. It is possible that the C-terminal domain accumulates in the cytoplasm after extracellular shedding as has been reported in adhesion molecules such as EpCAM [30, 31]. It is also possible that after LN localization and CD62L shedding tumor cells undergo a series of phenotypic and molecular changes (eg. mesenchymal to epithelium transition) which may result in decreased level of CD62L .
CD62L is characteristically found on cells of the immune system and has not generally been reported in solid organ tumors. In non-solid tumors such as lymphoma, CD62L has been shown to play a critical role in the development of metastasis. In a study evaluating the role of CD62L on lymphomagenesis and dissemination, Belanger et al. (2005) [10] reported that L-selectin deficiency resulted in resistance to the development of metastatic spread [10, 11]. In solid organ cancers, the presence of CD62L ligands on carcinoma cells and their interaction with CD62L on leucocytes has been suggested as an indirect mechanism to facilitate cancer cell migration and LN homing [32]. This route may provide a shield of leucocytes (with CD62L) around disseminated cancer cells (with CD62L ligand) to protect them from circulating immune cell recognition and attack. Our data show that bladder cancer cells directly express CD62L possibly leading to interaction with its ligands in LNs thus facilitating the colonization process. Consistent with this hypothesis, engineered overexpression of CD62L in a mouse model of islet cell tumors, which usually spread almost exclusively to the liver via portal vein dissemination and are rarely present in LNs, resulted in novel LN-specific spread [11]. A recent published study also reported upregulated CD62L expression in primary and metastatic oral pharyngeal squamous cell carcinoma samples [33].
The presence of CD62L mRNA in urothelial carcinoma was confirmed by performing a meta-analysis of mRNA expression profile datasets available in the ONCOMINE microarray database (www.oncomine.org) [17]. This showed a significantly higher level (3- to 8-fold) of CD62L in high grade, invasive bladder cancer samples as compared to low grade samples [22-24]. The higher degree of overexpression found in our study (30 fold), might be explained by techniques utilized. In the studies evaluated in ONCOMINE, tumor tissues were selected for the presence of at least 70-80% tumor cells [24, 25, 34]. This can result in significant contribution from non-tumor cells (i.e. endothelial, immune, fibroblasts). In our study we used LCM that should reduce contamination from other cell types. Further, CD45, a marker of leucocytes was not found to be differentially regulated in our gene microarray dataset.
ONCOMINE profiles, like our data, show that not all samples had uniform upregulation (Bar chart of Dyrskjot-2003-Figure 4). In our study, IHC revealed clusters of cells with increased CD62L protein expression within tumors suggesting areas of phenotypic/clonal expansion. This also could contribute to the variability observed in microarray studies. In addition, we found only 1 of 4 high grade urothelial cancer cell lines to have elevated CD62L mRNA expression relative to a benign cell line, agreeing with heterogeneity in expression level of CD62L observed in clinical samples. This cell line will be used in future studies as an in vitro model to investigate the relationship between ligand and receptor by employing cell adhesion and transmigration assays under static and flow conditions.
CD62L has been used previously as a marker of disease. Elevated serum levels have been used as a biomarker of leukemia relapse and were shown to be associated with poor prognosis [15, 21]. In a small cohort of metastatic relative to non-metastatic MIBC samples, we observed enhanced CD62L expression suggesting the possibility for use as a marker of metastatic disease. However, it is to be noted that our study could not identify the source of CD62L in serum, which generally has a significant contribution from activated leucocytes under constitutive conditions [14] and may be induced further under tumorogenic environment [15]. One of the limitations of the current study is the small sample size. However, our observation of upregulated CD62L expression using different methodologies provides a rationale for doing larger studies which will evaluate its relationship to disease outcomes.
5. Conclusions
In summary, our preliminary findings suggest a novel role of CD62L in metastatic spread of a solid tumor cells to LNs. This CD62L-mediated adhesion of disseminated cells to ligands present in LNs or vasculature may explain the direction of their migration and preferential localization to a specific site. Our study suggests that CD62L is a potential candidate for identifying patients who are at higher risk for the presence or development of LN metastasis thereby directing therapy appropriate for advanced disease.
Acknowledgements
The authors thank Dr. Pramod Srivastava (UCHC) for kindly providing critical comments on the manuscript and access to Oncomine database. This work was supported by: ACS-MRSG 08-270-01-CCE (JAT), RO1-AR-060286 (CCP), Connecticut Institute for Clinical and Translational Science (CICATS) and The Leo and Anne Albert Charitable Trust (JAT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: none
References
- [1].American Cancer Society . Cancer Facts and Figures. American Cancer Society; Atlanta: 2014. [Google Scholar]
- [2].Herr HW, Donat SM. Outcome of patients with grossly node positive bladder cancer after pelvic lymph node dissection and radical cystectomy. J Urol. 2001;165:62–4. doi: 10.1097/00005392-200101000-00015. discussion 4. [DOI] [PubMed] [Google Scholar]
- [3].Ries LAG, Krapcho M, Stinchcomb DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, Lewis DR, Clegg L, Eisner MP, Reichman M, Edwards BK, editors. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 1975-2005. MD. http://seer.cancer.gov/csr/1975_2005/, based on November 2007 SEER data submission, posted to the SEER web site; 2008. [Google Scholar]
- [4].Watts KL, Ristau BT, Yamase HT, Taylor JA., 3rd Prognostic implications of lymph node involvement in bladder cancer: are we understaging using current methods? BJU Int. 2011;108:484–92. doi: 10.1111/j.1464-410X.2011.10330.x. [DOI] [PubMed] [Google Scholar]
- [5].McEver RP. Selectin-carbohydrate interactions during inflammation and metastasis. Glycoconj J. 1997;14:585–91. doi: 10.1023/a:1018584425879. [DOI] [PubMed] [Google Scholar]
- [6].Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–56. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- [7].Michie SA, Streeter PR, Bolt PA, Butcher EC, Picker LJ. The human peripheral lymph node vascular addressin. An inducible endothelial antigen involved in lymphocyte homing. Am J Pathol. 1993;143:1688–98. [PMC free article] [PubMed] [Google Scholar]
- [8].Arbones ML, Ord DC, Ley K, Ratech H, Maynard-Curry C, Otten G, et al. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity. 1994;1:247–60. doi: 10.1016/1074-7613(94)90076-0. [DOI] [PubMed] [Google Scholar]
- [9].Tedder TF, Steeber DA, Pizcueta P. L-selectin-deficient mice have impaired leukocyte recruitment into inflammatory sites. J Exp Med. 1995;181:2259–64. doi: 10.1084/jem.181.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Belanger SD, St-Pierre Y. Role of selectins in the triggering, growth, and dissemination of T-lymphoma cells: implication of L-selectin in the growth of thymic lymphoma. Blood. 2005;105:4800–6. doi: 10.1182/blood-2004-04-1406. [DOI] [PubMed] [Google Scholar]
- [11].Qian F, Hanahan D, Weissman IL. L-selectin can facilitate metastasis to lymph nodes in a transgenic mouse model of carcinogenesis. Proc Natl Acad Sci U S A. 2001;98:3976–81. doi: 10.1073/pnas.061633698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Petzoldt JL, Leigh IM, Duffy PG, Sexton C, Masters JR. Immortalisation of human urothelial cells. Urol Res. 1995;23:377–80. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- [13].Rossi MR, Masters JR, Park S, Todd JH, Garrett SH, Sens MA, et al. The immortalized UROtsa cell line as a potential cell culture model of human urothelium. Environ Health Perspect. 2001;109:801–8. doi: 10.1289/ehp.01109801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schleiffenbaum B, Spertini O, Tedder TF. Soluble L-selectin is present in human plasma at high levels and retains functional activity. J Cell Biol. 1992;119:229–38. doi: 10.1083/jcb.119.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spertini O, Callegari P, Cordey AS, Hauert J, Joggi J, von Fliedner V, et al. High levels of the shed form of L-selectin are present in patients with acute leukemia and inhibit blast cell adhesion to activated endothelium. Blood. 1994;84:1249–56. [PubMed] [Google Scholar]
- [16].Taylor JA, 3rd, Kuchel GA, Hegde P, Voznesensky OS, Claffey K, Tsimikas J, et al. Null mutation for macrophage migration inhibitory factor (MIF) is associated with less aggressive bladder cancer in mice. BMC Cancer. 2007;7:135. doi: 10.1186/1471-2407-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].DeGraff DJ, Robinson VL, Shah JB, Brandt WD, Sonpavde G, Kang Y, et al. Current preclinical models for the advancement of translational bladder cancer research. Mol Cancer Ther. 2013;12:121–30. doi: 10.1158/1535-7163.MCT-12-0508. [DOI] [PubMed] [Google Scholar]
- [19].Smalley DM, Ley K. L-selectin: mechanisms and physiological significance of ectodomain cleavage. J Cell Mol Med. 2005;9:255–66. doi: 10.1111/j.1582-4934.2005.tb00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang S, Liu F, Wang QJ, Rosenberg SA, Morgan RA. The shedding of CD62L (L-selectin) regulates the acquisition of lytic activity in human tumor reactive T lymphocytes. PLoS One. 2011;6:e22560. doi: 10.1371/journal.pone.0022560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Extermann M, Bacchi M, Monai N, Fopp M, Fey M, Tichelli A, et al. Relationship between cleaved L-selectin levels and the outcome of acute myeloid leukemia. Blood. 1998;92:3115–22. [PubMed] [Google Scholar]
- [22].Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–6. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- [23].Modlich O, Prisack HB, Pitschke G, Ramp U, Ackermann R, Bojar H, et al. Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: use of cDNA array analysis of gene expression profiles. Clin Cancer Res. 2004;10:3410–21. doi: 10.1158/1078-0432.CCR-03-0134. [DOI] [PubMed] [Google Scholar]
- [24].Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–96. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- [25].Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–55. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- [26].Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–8. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- [27].Shariat SF, Karakiewicz PI, Palapattu GS, Lotan Y, Rogers CG, Amiel GE, et al. Outcomes of radical cystectomy for transitional cell carcinoma of the bladder: a contemporary series from the Bladder Cancer Research Consortium. J Urol. 2006;176:2414–22. doi: 10.1016/j.juro.2006.08.004. discussion 22. [DOI] [PubMed] [Google Scholar]
- [28].Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol. 2001;19:666–75. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- [29].Bassi P, Ferrante GD, Piazza N, Spinadin R, Carando R, Pappagallo G, et al. Prognostic factors of outcome after radical cystectomy for bladder cancer: a retrospective study of a homogeneous patient cohort. J Urol. 1999;161:1494–7. [PubMed] [Google Scholar]
- [30].Ralhan R, He HC, So AK, Tripathi SC, Kumar M, Hasan MR, et al. Nuclear and cytoplasmic accumulation of Ep-ICD is frequently detected in human epithelial cancers. PLoS One. 2010;5:e14130. doi: 10.1371/journal.pone.0014130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jachin S, Bae JS, Sung JJ, Park HS, Jang KY, Chung MJ, et al. The role of nuclear EpICD in extrahepatic cholangiocarcinoma: association with beta-catenin. Int J Oncol. 2014;45:691–8. doi: 10.3892/ijo.2014.2472. [DOI] [PubMed] [Google Scholar]
- [32].Laubli H, Borsig L. Selectins promote tumor metastasis. Semin Cancer Biol. 2010;20:169–77. doi: 10.1016/j.semcancer.2010.04.005. [DOI] [PubMed] [Google Scholar]
- [33].Shen H, Wang X, Shao Z, Liu K, Xia XY, Zhang HZ, et al. Alterations of high endothelial venules in primary and metastatic tumors are correlated with lymph node metastasis of oral and pharyngeal carcinoma. Cancer Biol Ther. 2013;15:342–9. doi: 10.4161/cbt.27328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sanchez-Carbayo M. Use of high-throughput DNA microarrays to identify biomarkers for bladder cancer. Clin Chem. 2003;49:23–31. doi: 10.1373/49.1.23. [DOI] [PubMed] [Google Scholar]